Introduction
Non-small cell lung cancer (NSCLC) is one of the
most common cancers and the leading cause of cancer-related deaths
in China (1). Although several
targeted therapies (EGFR and ALK) have been developed, NSCLCs still
have a tendency for recurrence and metastasis (2,3).
Furthermore, our understanding of lung cancer is very limited,
which has resulted in poor patient outcomes. Discovery of new
targeted biomarkers for prognosis is important in cancer research
(4–6). Recently, accumulating studies have
revealed that angiogenesis-related genes, including DLL4 are
dysregulated in lung cancer and they act as oncogenes or tumor
suppressors.
DLL4 is a member of the Notch signaling family and
plays an important role in angiogenesis (7–9).
Various studies have found that DLL4 regulates vessel sprouting via
angiogenic stimuli (10–13). Promotion of new vessel sprouting is
a very fundamental factor in tumor growth and metastasis (12). Therefore, DLL4 may function as an
oncogene in bladder cancer and breast cancer (12,13).
However, other reports have indicated that DLL4 acts as a tumor
suppressor in other cancer cell types due to deregulated vascular
development (14–16). Based on these reports, our knowledge
of the roles of DLL4 in NSCLC is conflicting and limited.
In the present study, expression levels of DLL4 in
NSCLC patients and lung cancer cell lines were determined, and its
clinical significance of prognosis was analyzed. The effects of
DLL4 on cell proliferation and invasion of lung cancer cell lines
were also determined. Understanding the DLL4 functions will
hopefully provide a new prognostic biomarker for lung cancer.
Materials and methods
Patients and Ethics statement
One hundred and two formalin-fixed,
paraffin-embedded lung tissues and non-cancerous lung tissues were
collected before any patient treatment from NSCLC patients, who
were enrolled in the study between January 2007 and January 2012 at
the Central Hospital (Table I).
This study was approved by the Ethics and Scientific Committees of
the Central Hospital (Wuhan, China) and complied with the
Declaration of Helsinki. Written informed consent was obtained from
all patients.
 | Table I.Characteristics of the NSCLC patients
(N=102). |
Table I.
Characteristics of the NSCLC patients
(N=102).
|
Characteristics | Data |
|---|
| Mean age (range) in
years | 52 (24–76) |
| Sex, n (%) |
|
|
Male | 63 (61.8) |
|
Female | 39 (38.2) |
| Survival status, n
(%) |
|
|
Dead | 53 (52.0) |
|
Surviving | 49 (48.0) |
| Depth of invasion
(T), n (%) |
|
| T1 | 21 (20.6) |
| T2 | 71 (69.6) |
| T3 | 8 (7.8) |
| T4 | 2 (2.0) |
| Lymph node
metastasis (N), n (%) |
|
| N0 | 66 (64.7) |
| N1 | 23 (22.6) |
| N2 | 4 (3.9) |
| NX | 9 (8.8) |
| Distant metastasis
(M), n (%) |
|
| M0 | 99 (97.1) |
| M1 | 3 (2.9) |
| TNM stage, n
(%) |
|
|
Ia/Ib | 19/12 (30.4) |
|
IIa/IIb | 38/18 (54.9) |
|
IIIa/IIIb | 11/1 (11.8) |
| IV | 3 (2.9) |
| Total | 102 (100) |
A total of 63 men and 39 women with a mean age of 52
(range, 24–76 years) years were included. All patients were
followed up from the date of surgery to December, 2014.
Pathological features, such as age and sex are shown in Table I. All of the NSCLC lung tissue
samples were classified according to the 7th edition of the TNM
classification by the International Association for the Study of
Lung Cancer (IASLC) (17,18). Overall survival (OS) was calculated,
which was the period from the date of initial diagnosis to death or
the last follow-up. At the end of the study, 53 patients (52.0%)
were still alive and 49 patients (48.0%) died of NSCLCs.
Data involving gene mutations were not obtained. No
patients received new adjuvant therapy before or after surgery. The
data of patients who received chemotherapy and radiotherapy were
not fully collected; 61 patients received chemotherapy prior to or
after surgery and 14 patients received radiotherapy before surgery.
The clinical data of other patients were not collected.
Interactions of these clinical data were not evaluated.
Tissue microarray construction and
QDs-IHC
Initially, hematoxylin and eosin-staining was
performed and screened for tumor tissues and matched non-cancerous
tissues. Two tissue microarray (TMA) slides, which consisted of 102
NSCLC tissues and adjacent non-cancerous lung tissues, were
constructed with a diameter of 1.5 mm and technological support was
provided from Beijing Do Biotech Co., Ltd. (19,20).
The expression of Atg4C and DLL4 was assessed by
QDs-IHC staining according to the manufacturers instructions and
Wuhan Jiayang Quantum Dots Co., Ltd. (Wuhan, China) provided the
technological support. In brief, the TMAs were prepared in xylene
and in graded alcohol. Antigen retrieval of Atg4C was performed in
EDTA buffer (1 mM, pH 8.0) at microwave oven for 20 min, while DLL4
was in EDTA buffer (1 mM, pH 8.0) using autoclave for 4 min.
Tris-buffered saline (TBS) was used for dilution (antibodies and
QDs), containing 2% bovine serum albumin (BSA; Sigma-Aldrich; Merck
KGaA, Darmstadt, Germany). At first, TMAs were incubated in 2% BSA
buffer, and then, TMAs were incubated with primary antibodies,
which included rabbit anti-Atg4C (diluted 1:200; cat. no. ab191705;
Abcam, Cambridge, MA, USA) and rabbit anti-DLL4 (diluted 1:200;
cat. no. ab7280; Abcam). Then, TBS-T (0.5% Tween in TBS) was used
for washing the TMAs. Goat anti-rabbit IgG was used as a secondary
antibody (1:400; cat. no. 7074; Cell Signaling Technology, Inc.,
Danvers, MA, USA). Finally, the TMAs were incubated in QDs (605 nm)
conjugated to streptavidin (1:300; Wuhan Jiayang Quantum Dots Co.,
Ltd.), and TMAs were sealed in 90% glycerin (Sigma-Aldrich; Merck
KGaA). TBS instead of two primary antibodies was used for negative
control, which showed auto-fluorescence signal.
Scoring of QDs-IHC staining
The signals of QDs-IHC staining were detected using
Olympus BX53 fluorescence microscopy (Olympus Corp., Tokyo, Japan)
at 605 nm and the results were evaluated by two independent
researchers. They were also blinded to the clinical parameters of
the patients. The scoring was calculated using the positive area
and the staining intensity. The area of positivity (AD) was
calculated as 0 (no positive area or positive area <5%), 1
(5–25%), 2 (26–50%), 3 (51–75%) and 4 (>75%), while the
intensity of staining (IS) was scored as 1 (weak), 2 (moderate) and
3 (strong) (2). Intensity
distribution (ID) = AP × IS, with the ID score being the final
expression level of protein, which ranged from 0 to 12. The cutoff
point of high or low expression of DLL4 protein was determined on
the receiver operating characteristic (ROC) curve analysis with
respect to OS.
Cell culture and transfection
A549, H1299 and A427 cell lines were obtained from
the Cell Bank of Shanghai Institutes for Biological Sciences
(Shanghai, China). They were cultured in RPMI-1640 media with 10%
fetal bovine serum (FBS) (Life Technologies, Beijing, China) in 5%
CO2. A549 and A427 cell lines were seeded in 6-well
plates at 106 cells/wells. The pCMV-myc vector, and
pCMV-myc-DLL4 (Life Technologies, Shanghai, China) were used for
overexpression. Total proteins were isolated for Western blot
analysis at 48 h after transfection using RIPA lysis buffer
(Beyotime, Shanghai, China). The pSILENCE vector, pSILENCE-A and
pSILENCE-B (Life Technologies, Shanghai, China) were used for
knockdown with Invitrogen™ Lipofectamine 2000 (Thermo Fisher
Scientific, Inc., Waltham, MA, USA).
Quantitative real-time PCR (qPCR)
cDNA was obtained using the RevertAid First Strand
cDNA Synthesis kit (Fermentas, Burlington, ON, Canada). The
relative expression of DLL4 (reference gene transcript ID:
NM_019074.3) mRNA was measured using qRT-PCR in a CFX 96 Real-Time
PCR system (Bio-Rad Laboratories, Shanghai, China) using a
SYBR-Green kit (Takara Bio Co., Ltd., Japan), and the relative
changes were quantified. Forward primer of DLL4 was,
5-CTAGCTGTGGGTCAGAACTGGTTATT-3 and the reverse primer was,
5-ATGACAGCCCGAAAGACAGAT-3. GAPDH was used as a control. The primers
were as follows: Forward primer, 5-GGAGTCAACGGATTTGGTCGTA-3 and
reverse primer, 5-GGCAACAATATCCACTTTACCAGAGT-3. Relative gene
expression levels were determined by the 2−ΔΔCq [2
− (testCq {DLL4} - testCq {GAPDH}) - (controlCq {DLL4} -
control Cq{GAPDH})] method (2). qPCR conditions: SYBR Green (2×) 10 µl,
forward primer and reverse primer (10 pmol) 1 µl, cDNA 2 µl,
ddH2O 6 µl; Initial denaturation 94°C for 5 min;
denaturation 94°C for 30 sec, annealing 64°C for 30 sec, extension
72°C for 45 sec, cycle 35; extension 72°C for 5 min.
Cell proliferation
Cell proliferation was assessed using an MTT assay.
Cells were plated in 24-well plates at 3×105 cells/well.
Then cells were incubated with 100 µl MTT dye (0.5 mg/ml;
Sigma-Aldrich; Merck KGaA) for 4 h and 150 µl DMSO (Sigma-Aldrich;
Merck KGaA) was added after the supernatant was removed. The
absorbance was detected at 570 and 655 nm was used as the reference
wavelength. The absorbance was determined at 12, 24, 36, 48, 60 and
72 h after transfection and the MTT assay was performed in
triplicate.
Cell migration and invasion
Cell migration and invasion abilities were detected
using wound healing a and Transwell chamber assays (Corning,
Beijing, China) with or without Matrigel (Invitrogen; Thermo Fisher
Scientific, Inc., Beijing, China). For the determination of cell
migration, a wound was produced using a plastic pipette tip when
90–100% cell confluence was reached. Then the migrated cells were
washed and cultured in low serum (2.5%) media for 48 h. Wound
closure (%) was defined as the area of migrated cells at 48 h
divided by the area at 0 h.
Transwell chambers (Corning, Beijing, China) with
Matrigel were used for detection of cell invasion. Transwell
chambers were placed into 6-well plates, and coated with Matrigel.
A total of 4×104 cells were seeded in the upper chambers
into serum-free media at 24 h after transfection. Meanwhile media
of 10% FBS/DMEM (Gibco; Thermo Fisher Scientific) was added to the
lower chambers. After 48 h, the cells which had invaded through the
membrane were fixed in 20% methanol and stained with 0.1% crystal
violet. The non-migrated A549 and A427 cells were removed by cotton
swabs. Other cells on the upper surface of the membrane were
removed by cotton swabs. Images were captured using microscope
(Olympus Corp., Tokyo, Japan) for calculating the number of
migrated cells at ×200 magnification.
Western blot analysis
Protein samples were isolated from A549 and A427
cells using RIPA lysis buffer and protein concentrations were
detected using the BCA kit (Beyotime Institute of Biotehnology,
Shanghai, China). The cellular extracts were separated on 10%
SDS-PAGE gel and transferred onto PVDF membranes (Bio-Rad
Laboratories). Membranes were blocked using 1% non-fat milk and
incubated with the primary DLL4 antibody (cat. no. ab7280; Abcam)
or GAPDH antibody (cat. no. ab9485; Abcam) overnight. Next, the
secondary antibody (cat. no. 7074; Cell Signaling Technology) was
added and incubation was carried out. Finally, protein bands were
visualized using the enhanced chemiluminescence (ECL) assay.
Statistical analysis
Statistical analyses were performed using SPSS19.0
software (IBM Corp., Armonk, NY, USA). Data are expressed as means
± SD and the differences between groups were assessed with the
Student's t-test. Comparisons of multiple groups were performed
using ANOVA and the S-N-K test as a post hoc test was used. The
association between protein levels and clinical parameters were
estimated using the Chi-square test. Kaplan-Meier test and log-rank
test were performed for survival analysis. K-M plotter database was
used for NSCLC survival analysis. Statistically significant
differences were considered when two-tailed P-values <0.05.
Results
Expression of DLL4 in clinical
specimens and lung cancer cell lines
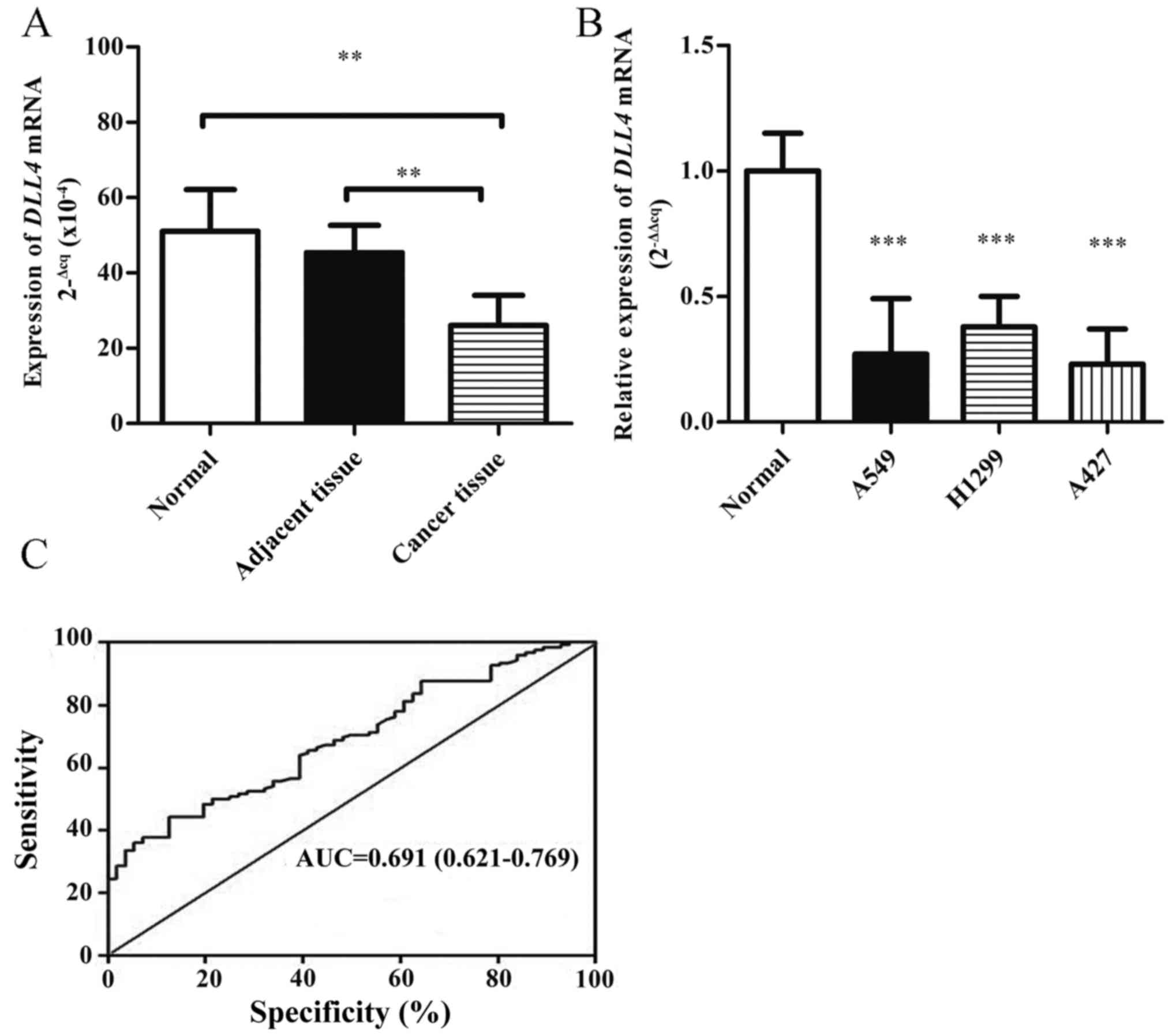
DLL4 mRNA levels were determined using qPCR in lung
tissues of 22 NSCLC patients and 20 healthy controls. As shown in
Fig. 1A, the DLL4 mRNA levels were
downregulated in the lung tissues of NSCLC patients compared with
these levels in the non-cancerous tissues and healthy controls.
DLL4 mRNA levels were also detected in lung cancer cell lines. The
expression of DLL4 was decreased (0.25-fold) in three lung cancer
cell lines compared with that noted in primary human alveolar
epithelial cells (Fig. 1B). The
cutoff value of DLL4 expression levels was determined using ROC
curve analysis (Fig. 1C); 4.2 was
defined as the cutoff point of DLL4 in NSCLC patients (an ID score
≥4.2 defined high expression and ID <4.2 indicated low
expression). The cutoff value of DLL4 expression had optimal
sensitivity and specificity. The area under the curve was 0.691 and
the 95% confidence interval (CI) was 0.621–0.769.
Expression of DLL4 protein in TMA and
overall survival analysis
To validated the results that levels of DLL4 mRNA
were downregulated in lung cancer tissues and lung cancer cell
lines, we performed ODs-IHC staining in a larger cohort of NSCLC
patients (n=102). DLL4 was expressed in both cancer tissues and
adjacent non-cancerous lung tissues including vascular endothelial
cells (Fig. 2). DLL4 was located in
the cell membrane and cytoplasm (Fig.
2A). In tumor tissues, expression of DLL4 was significantly
decreased (Fig. 2B-D). Forty-one
(40.2%) patients showed high DLL4 expression and 61 (59.8%)
patients showed low DLL4 expression in tumor tissues. Subsequently,
the prognostic value of DLL4 expression was investigated in the
NSCLC patients. The results demonstrated that high DLL4 protein
expression predicted a prolonged survival rate (Fig. 2E, P=0.026) using Kaplan-Meier
analysis and log-rank test. K-M plotter database of lung cancer
patients (n=1422) was used and the result of survival analysis
supported our conclusion (Fig. 2F,
P<0.001). This was in contrast to the results in breast cancer
and this finding warrants further research. Atg4C expression was
also determined. However, no significant difference was observed in
this study (data not shown).
Clinical significance of DLL4
expression
As shown in Table
II, the association between the level of DLL4 protein and
clinicopathological variables was analyzed. The expression level of
DLL4 was not significantly associated with sex, age, T, M, or TNM
stage of the NSCLC patients. Notably, a significant association
between lymph node metastasis (N) status and DLL4 expression was
observed.
 | Table II.Association between DLL4 expression
and clinicopathological parameters of the NSCLC patients. |
Table II.
Association between DLL4 expression
and clinicopathological parameters of the NSCLC patients.
|
|
| DLL4
expression |
|
|---|
|
Characteristics | n | Low n (%) | High n (%) | P-value |
|---|
| Age (years) |
|
|
| >0.05 |
|
<60 | 72 | 42 (41.2) | 30 (29.4) |
|
|
≥60 | 30 | 19 (18.6) | 11 (10.8) |
|
| Sex |
|
|
| >0.05 |
|
Male | 63 | 35 (34.3) | 28 (27.5) |
|
|
Female | 39 | 26 (25.5) | 13 (12.7) |
|
| Depth of invasion
(T) |
|
|
| >0.05 |
|
T1-T2 | 92 | 53 (52.0) | 39 (38.2) |
|
|
T3-T4 | 10 | 8 (7.8) | 2 (2.0) |
|
| Lymph node
metastasis (N) |
|
|
| <0.05 |
| N0 | 66 | 31 (30.4) | 35 (34.3) |
|
| N1, N2,
NX | 36 | 30 (29.4) | 6 (5.9) |
|
| Distant metastasis
(M) |
|
|
| >0.05 |
| M0 | 99 | 59 (57.8) | 40 (39.3) |
|
| M1 | 3 | 2 (2.0) | 1 (0.9) |
|
| TNM stage |
|
|
| >0.05 |
|
I–II | 87 | 50 (49.0) | 37 (36.3) |
|
|
III–IV | 15 | 11 (10.8) | 4 (3.9) |
|
| Total |
| 61 (59.8) | 41 (40.2) |
|
Overexpression of DLL4 reduces cell
proliferation in A549 and A427 cell lines
Cell viability was detected using the MTT assay.
Transfection with pCMV-myc-DLL4 significantly increased DLL4 mRNA
and protein levels in the A549 and A427 cell lines (Fig. 3A-C). Transfection efficiency was
~30–40% (data not shown). Compared with the control vector, cell
viability and proliferation were significantly decreased in the
A549 cells transfected with the pCMV-DLL4 vector (Fig. 3D). Identical results were also
observed in the A427 cells (Fig.
3E). These results demonstrated that DLL4 overexpression
inhibited cell viability and proliferation in lung cancer cell
lines. They also indicated that DLL4 acted as a tumor suppressor in
NSCLC cell lines.
Overexpression of DLL4 inhibits cell
migration and invasion
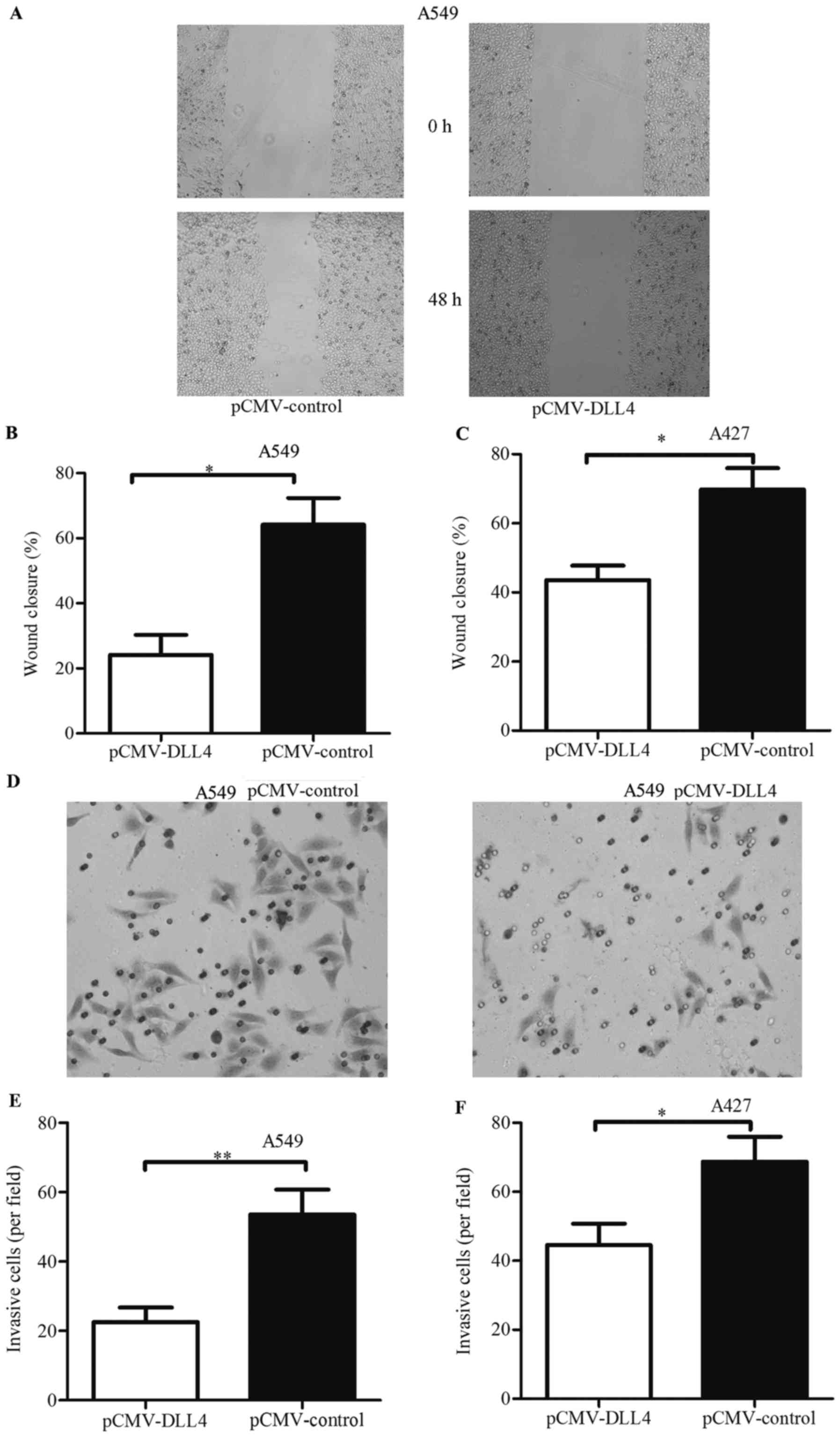
Wound healing assay and Transwell invasion assay
were used to detect the effects of DLL4 overexpression on the
migration and invasion of NSCLC cell lines. As shown in Fig. 4A, the closure rate of cells
transfected with pCMV-DLL4 was less than the rate of cells
transfected with pCMV-control in the A549 cell line. The rate of
cells transfected with the control vector was 0.64-fold, and the
rate with pCMV-DLL4 was 0.37-fold (Fig.
4B). Similarly, identical results were also observed in the
A427 cell line (Fig. 4C). These
results demonstrated that cell migration was inhibited in cancer
cells with DLL4 overexpression.
As shown in Fig. 4D,
the number of invaded cells were decreased in the A549 cells
transfecting with pCMV-DLL4. Compared with the control vector, the
ability of invasion was decreased to 0.22-fold (Fig. 4E). These results were also observed
in the A427 cells (Fig. 4F). These
results indicated that DLL4 functioned as a tumor suppressor that
inhibited cell viability, migration and invasion.
Discussion
Many studies have reported that DLL4 expression in
lung cancer may be associated with tumor metastasis and prognosis
(3,20–25).
In the present study, DLL4 expression was assessed using tissue
microarray and the prognostic value was examined in NSCLC patients.
We found that DLL4 expression was significantly decreased in NSCLC
patients compared with that noted in normal subjects, and low DLL4
expression predicted a poor survival rate and was significantly
correlated with lymph node metastasis. DLL4 expression was
downregulated in A549, H1299 and A427 cells. Furthermore,
overexpression of DLL4 reduced cell proliferation and invasion in
both A549 and A427 cells. Our results suggest that DLL4 is an
independent prognostic biomarker for lung cancer.
There are many preclinical models which have focused
on dll4 allele deletion and systemic application of DLL4/Notch
inhibitors, which have been found to result in significant
suppression of tumor growth (14,26).
Our findings were completely contrasting to the results in other
tumor types where upregulation of DLL4 correlates with tumor
promotion (16–29). In an attempt to determine the role
of DLL4 in lung cancer cells, DLL4 was overexpressed in two NSCLC
cell lines. The results revealed that DLL4 overexpression had a
negative effect on the growth, migration and invasion of lung
cancer cells. Although bioinformatics analysis using K-M plotter
supported our conclusion, these conflicting results need further
research and should be explained carefully.
On the one hand, as a member of Notch signaling,
DLL4 plays an important role in vessel sprouting (30). Expression of DLL4 was found to
stimulate Notch signaling and regulate the ratio of tip cells to
stalk cells (30,31). When DLL4 was inhibited, tip-cell
specification was not able to be controlled and excessive sprouting
occurred, leading to tumor migration. DLL4 was considered as a
tumor suppressor due to reducing endothelial sensitivity to VEGF
and increased DLL4 could reduce tumor growth and VEGF-induced
overall tumor blood supply (16,32).
DLL4 overexpression was found to prevent metastasis formation and
allow for increased delivery to the tumor of concomitant
chemotherapy and improve its efficacy (32).
On the other hand, DLL4 expression was found
to be downregulated in NSCLC patients due to posttranscriptional
mechanisms, due to upregulation of the miR-30 family. microRNAs
(miRNAs) are small non-coding RNAs, which regulate target gene
expression by mRNA degradation and translational inhibition
(1). Numerous miRNAs are found to
play roles in carcinogenesis of NSCLC, such as the miR-30 family
(21–23). Furthermore, the miR-30 family and
miR-27b are implicated in DLL4 regulation (24,25).
It is unclear whether or not these miRNAs believed to suppress DLL4
specifically lead to tumor growth and invasion in NSCLC
patients.
There are some limitations to this study. All the
patients were diagnosed and treated between 2007 and 2012 according
to the 7th edition of the TNM classification by IASLC. However, it
was difficult to reappraise according to the 8th edition of the TNM
classification by UICC/AJCC. In vivo xenograft study should
be conducted for further research. The data of the patients'
pulmonary function test, histological classification, and gene
mutations could not obtain and interactions of these clinical data
were not evaluated. An experiment using knockdown was not
performed, as the expression of DLL4 was difficult to silence.
pSILENCE-A and pSILENCE-B failed to knock down DLL4. Thus, we did
not discuss it in the results. We will perform this again in
further research. The use of the MTT assay in the growth studies
should be explained carefully. Overexpression for DLL4 could partly
support the conclusions. However, it may result in loss of
viability (ie. cell death) which could explain the apparent effects
on growth, migration and invasion. Therefore, further research is
needed.
In conclusion, we identified low expression of DLL4
in NSCLC patients. Downregulation of DLL4 was found to be
associated with poor OS and overexpression of DLL4 inhibited
proliferation, migration and invasion of cancer cells.
Acknowledgements
Authors thank all patients enrolled in the study and
staff at the Department of Pathology, Zhongnan Hospital of Wuhan
University.
Funding
No funding was received.
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
HL, ZL and SL conceived and designed the study. HL,
JP, MZ, PM and JZ performed the experiments. HL, ZL and MZ wrote
the paper. SL, JP, MZ, ZL and PM reviewed and edited the
manuscript. All authors read and approved the manuscript and agree
to be accountable for all aspects of the research in ensuring that
the accuracy or integrity of any part of the work are appropriately
investigated and resolved.
Ethics approval and consent to
participate
The present study was approved by the Ethics and
Scientific Committees of the Central Hospital (Wuhan, China) and
complied with the Declaration of Helsinki. Written informed consent
was obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
All authors declare that they have no competing
interests.
References
|
1
|
Peng J, Liu HZ, Zhong J, Deng ZF, Tie CR,
Rao Q, Xu W, You T, Li J, Cai CB, et al: MicroRNA-187 is an
independent prognostic factor in lung cancer and promotes lung
cancer cell invasion via targeting of PTRF. Oncol Rep.
36:2609–2618. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gkolfinopoulos S and Mountzios G: Beyond
EGFR and ALK: targeting rare mutations in advanced non-small cell
lung cancer. Ann Transl Med. 6:1422018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gallant JN and Lovly CM: Established,
emerging and elusive molecular targets in the treatment of lung
cancer. J Pathol. 244:565–577. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chen C, Zhao Z, Liu Y and Mu D:
MicroRNA-99a is downregulated and promotes proliferation, migration
and invasion in non-small cell lung cancer A549 and H1299 cells.
Oncol Lett. 9:1128–1134. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Liu HZ, Du CX, Luo J, Qiu XP, Li ZH, Lou
QY, Yin Z and Zheng F: A novel mutation in nuclear prelamin a
recognition factor-like causes diffuse pulmonary arteriovenous
malformations. Oncotarget. 8:2708–2718. 2017.PubMed/NCBI
|
|
6
|
Mishra N, Timilsina U, Ghimire D, Dubey RC
and Gaur R: Downregulation of cytochrome c oxidase subunit 7A1
expression is important in enhancing cell proliferation in
adenocarcinoma cells. Biochem Biophys Res Commun. 482:713–719.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gridley T: Notch signaling in the
vasculature. Curr Top Dev Biol. 92:277–309. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Phng LK and Gerhardt H: Angiogenesis: A
team effort coordinated by notch. Dev Cell. 16:196–208. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gale NW, Dominguez MG, Noguera I, Pan L,
Hughes V, Valenzuela DM, Murphy AJ, Adams NC, Lin HC, Holash J, et
al: Haploinsufficiency of delta-like 4 ligand results in embryonic
lethality due to major defects in arterial and vascular
development. Proc Natl Acad Sci USA. 101:15949–15954. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Siekmann AF and Lawson ND: Notch
signalling limits angiogenic cell behaviour in developing zebrafish
arteries. Nature. 445:781–784. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Patel NS, Li JL, Generali D, Poulsom R,
Cranston DW and Harris AL: Up-regulation of Delta-like 4 ligand in
human tumor vasculature and the role of basal expression in
endothelial cell function. Cancer Res. 65:8690–8697. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Patel NS, Dobbie MS, Rochester M, Steers
G, Poulsom R, Le Monnier K, Cranston DW, Li JL and Harris AL:
Upregulation of endothelial Delta-like 4 expressioncorrelates with
vessel maturation in bladder cancer. Clin Cancer Res. 12:4836–4844.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jubb AM, Soilleux EJ, Turley H, Steers G,
Parker A, Low I, Blades J, Li JL, Allen P, Leek R, et al:
Expression of vascular Notch ligand Delta-like 4 and inflammatory
markers in breast cancer. Am J Pathol. 176:2019–2028. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Noguera-Troise I, Daly C, Papadopoulos NJ,
Coetzee S, Boland P, Gale NW, Lin HC, Yancopoulos GD and Thurston
G: Blockade of Dll4 inhibits tumour growth by promoting
non-productive angiogenesis. Nature. 444:1032–1037. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ridgway J, Zhang G, Wu Y, Stawicki S,
Liang WC, Chanthery Y, Kowalski J, Watts RJ, Callahan C, Kasman I,
et al: Inhibition of Dll4 signalling inhibits tumour growth by
deregulating angiogenesis. Nature. 444:1083–1087. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li JL, Sainson RCA, Shi W, Leek R,
Harrington LS, Preusser M, Biswas S, Turley H, Heikamp E,
Hainfellner JA and Harris AL: Delta-like 4 Notch ligand regulates
tumor angiogenesis, improves tumor vascular function, and promotes
tumor growth in vivo. Cancer Res. 67:11244–11253. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
International Union against Cancer: TNM
Classification of Malignant Tumours. Sobin LH, Gospodarowicz MK and
Wittekind C: 7th edition. Wiley-Blackwell; Hoboken, NJ: 2009
|
|
18
|
Travis WD, Brambilla E, Noguchi M,
Nicholson AG, Geisinger K, Yatabe Y, Powell CA, Beer D, Riely G,
Garg K, et al: International association for the study of lung
cancer/american thoracic society/european respiratory society:
International multidisciplinary classification of lung
adenocarcinoma: Executive summary. Proc Am Thorac Soc. 8:381–385.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kononen J, Bubendorf L, Kallioniemi A,
Bärlund M, Schraml P, Leighton S, Torhorst J, Mihatsch MJ, Sauter G
and Kallioniemi OP: Tissue microarrays for high-throughput
molecular profiling of tumor specimens. Nat Med. 4:844–847. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhong Z, Xia Y, Wang P, Liu B and Chen Y:
Low expression of microRNA-30c promotes invasion by inducing
epithelial mesenchymal transition in non-small cell lung cancer.
Mol Med Rep. 10:2575–2579. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yu G, Herazo-Maya JD, Nukui T, Romkes M,
Parwani A, Juan-Guardela BM, Robertson J, Gauldie J, Siegfried JM,
Kaminski N, et al: Matrix metalloproteinase-19 promotes metastatic
behavior in vitro and is associated with increased mortality in
non-small cell lung cancer. Am J Respir Crit Care Med. 190:780–790.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhong K, Chen K, Han L and Li B:
MicroRNA-30b/c inhibits non-small cell lung cancer cell
proliferation by targeting Rab18. BMC Cancer. 14:7032014.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Biyashev D, Veliceasa D, Topczewski J,
Topczewska JM, Mizgirev I, Vinokour E, Reddi AL, Licht JD, Revskoy
SY and Volpert OV: miR-27b controls venous specification and tip
cell fate. Blood. 119:2679–2687. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bridge G, Monteiro R, Henderson S, Emuss
V, Lagos D, Georgopoulou D, Patient R and Boshoff C: The
microRNA-30 family targets DLL4 to modulate endothelial cell
behavior during angiogenesis. Blood. 120:5063–5072. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Scehnet JS, Jiang W, Kumar SR, Krasnoperov
V, Trindade A, Benedito R, Djokovic D, Borges C, Ley EJ, Duarte A,
et al: Inhibition of Dll4-mediated signaling induces proliferation
of immature vessels and results in poor tissue perfusion. Blood.
109:4753–4760. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Haller BK, Bråve A, Wallgard E, Roswall P,
Sunkari VG, Mattson U, Hallengärd D, Catrina SB, Hellström M and
Pietras K: Therapeutic efficacy of a DNA vaccine targeting the
endothelial tip cell antigen delta-like ligand 4 in mammary
carcinoma. Oncogene. 29:4276–4286. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kalén M, Heikura T, Karvinen H, Nitzsche
A, Weber H, Esser N, Ylä-Herttuala S and Hellström M:
Gamma-secretase inhibitor treatment promotes VEGF-A-driven blood
vessel growth and vascular leakage but disrupts neovascular
perfusion. PLoS One. 6:e187092011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Segarra M, Williams CK, de la Luz SM,
Bernardo M, McCormick PJ, Maric D, Regino C, Choyke P and Tosato G:
Dll4 activation of Notch signaling reduces tumor vascularity and
inhibits tumor growth. Blood. 112:1904–1911. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hellström M, Phng LK, Hofmann JJ, Wallgard
E, Coultas L, Lindblom P, Alva J, Nilsson AK, Karlsson L, Gaiano N,
et al: Dll4 signalling through Notch1 regulates formation of tip
cells during angiogenesis. Nature. 445:776–780. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Leslie JD, Ariza-McNaughton L, Bermange
AL, McAdow R, Johnson SL and Lewis J: Endothelial signalling by the
Notch ligand Delta-like 4 restricts angiogenesis. Development.
134:839–844. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Trindade A, Djokovic D, Gigante J,
Mendonça L and Duarte A: Endothelial Dll4 overexpression reduces
vascular response and inhibits tumor growth and metastasization in
vivo. BMC Cancer. 17:1892017. View Article : Google Scholar : PubMed/NCBI
|