Introduction
Human esophageal squamous cell carcinoma (ESCC) has
a very high incidence, and has become one of the most common causes
of cancer-associated mortality worldwide (1,2).
Surgical resection and radiotherapy are the predominant therapeutic
interventions used to treat patients with ESCC, whereas neoadjuvant
chemotherapy and photodynamic therapy have been considered
promising alternative strategies (3). Despite advances in clinical and
experimental oncology methods, patients with ESCC often have a poor
prognosis (2–5). ESCC is associated with a poor outcome
due to its malignant biological characteristics, including
histopathological grade, and early metastasis and invasion
(6,7). The etiology of ESCC is a complex
process, and the exact molecular mechanism remains unclear.
Therefore, the identification of pathogenetic mechanisms that occur
during ESCC progression and metastasis is a priority in ESCC
research.
Human transforming growth factor-activated kinase
(TAK1)-binding protein 3 (TAB3) is essential for TAK1 activation
(8,9). TAK1 is an activator of nuclear factor
(NF)-κB, which serves critical roles in various cellular processes
that contribute to embryonic development, immunity, cell survival,
carcinogenesis, chemoresistance and others (10). These findings indicate that the
TAB3-TAK1-NF-κB pathway may be central to numerous cellular
processes. In addition, it has been reported that TAB3 is
overexpressed in several types of cancer, including breast, ovarian
and lung cancer, and that it is associated with tumor development
and invasion (11–13). However, the role of TAB3 in the
development of ESCC remains to be elucidated. The present study
aimed to explore the expression and function of TAB3 in ESCC cells.
The results indicated that TAB3 may enhance the proliferation and
invasion of ESCC cells; therefore, it may be of value with regards
experimental therapeutic strategies for the treatment of ESCC.
Materials and methods
Cell lines and cell culture
Human esophageal epithelial cells (HEECs), and the
TE-1, TE-10 and Eca-109 cell lines were purchased from the Shanghai
Institute of Cell Biology (Shanghai, China). The cells were
maintained in Dulbecco's modified Eagle's medium (DMEM; Gibco;
Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with
10% fetal bovine serum (FBS; Hangzhou Sijiqing Biological
Engineering Materials Co., Ltd., Hangzhou, China) at 37°C in a
humidified atmosphere containing 5% CO2.
Antibodies
The antibodies used in the present study were as
follows: Anti-TAB3 (cat. no. sc-166538, 1:500), anti-GAPDH (cat.
no. sc-166574, 1:1,000), anti-Ki-67 (cat. no. sc-23900, 1:100),
anti-cyclin D1 (cat. no. sc-450, 1:300), anti-proliferating cell
nuclear antigen (PCNA; cat. no. sc-25280, 1:1,000), anti-TAK1 (cat.
no. sc-7967, 1:300) and anti-p65 (cat. no. sc-71675, 1:100), all of
which were purchased from Santa Cruz Biotechnology, Inc. (Dallas,
TX, USA).
Tissue samples
Paired ESCC and adjacent non-cancerous tissues were
obtained from 80 patients who underwent surgery between June 2010
and September 2012 at the Department of Pathology of the Affiliated
Hospital of Nantong University (Nantong, China). No patient had
received chemotherapy or radiotherapy prior to surgery. Patients
with a family history of gastric/colorectal cancer were excluded.
All patients provided written informed consent for their tissue
samples to be used for scientific research. The present study was
approved by the Affiliated Hospital of Nantong University Ethics
Committee. The histological features of the specimens were
evaluated by a senior pathologist, according to the World Health
Organization classification criteria (14). All patients were followed up for
4.2–64.9 months. Patient follow-up was terminated on November 30,
2016. The main clinical and pathological characteristics of the
patients are presented in Table
I.
 | Table I.Expression of TAB3 in 80 human
esophageal squamous cell carcinoma tissues. |
Table I.
Expression of TAB3 in 80 human
esophageal squamous cell carcinoma tissues.
|
|
| TAB3 |
|
|---|
|
|
|
|
|
|---|
| Clinicopathological
parameters | Total | Low (n=42) | High (n=38) | P-value |
|---|
| Sex |
|
Male | 65 | 35 | 30 | 0.616 |
|
Female | 15 | 7 | 8 |
|
| Age (years) |
|
<60 | 26 | 17 | 9 | 0.109 |
|
≥60 | 54 | 25 | 29 |
|
| Lymph node
metastasis |
| No | 46 | 31 | 15 | 0.002a |
|
Yes | 34 | 11 | 23 |
|
| T stage |
| T1 | 9 | 11 | 2 | 0.019a |
| T2 | 17 | 13 | 10 |
|
| T3 | 64 | 18 | 26 |
|
| Pathological
grade |
|
Well | 25 | 15 | 10 | 0.001a |
|
Moderate | 34 | 23 | 11 |
|
|
Poor | 21 | 4 | 17 |
|
| Ki-67 |
|
Low | 32 | 22 | 10 | 0.017a |
|
High | 48 | 20 | 28 |
|
Immunohistochemistry (IHC)
For histological examination, all surgically excised
tissues were fixed with 10% formalin at 4°C for 24 h and embedded
in paraffin; subsequently, 4-µm specimen sections were prepared on
glass slides. The sections were deparaffinized in xylene and
rehydrated with graded alcohol washes, and antigen retrieval was
performed by heating the samples to 121°C for 3 min in 10 mM
citrate buffer (pH 6.0). Hydrogen peroxide (0.3%) was applied to
block endogenous peroxide activity for 20 min after cooling, in
order to block any nonspecific reactions. After rinsing in PBS (pH
7.2), the sections were incubated with rabbit anti-human TAB3
(1:100) for 2 h at room temperature. All slides were processed
using the peroxidase-antiperoxidase method (EnVision + Dual Link
system-horseradish peroxidase; Dako; Agilent Technologies, Inc.,
Santa Clara, CA, USA), according to the manufacturer's protocol.
After washing with PBS, the peroxidase reaction was visualized by
incubating the sections with DAB (0.1% phosphate buffer solution,
0.02% diaminobenzidine tetrahydrochloride and 3%
H2O2) at room temperature for 5 min. After
being rinsed in water, the sections were counterstained with
hematoxylin (0.5%) at room temperature for 1 min. Finally, the
sections were dehydrated with graded alcohol washes leicand cover
slips were added to the slides, which were observed under a
microscope (Leica DFC 300 FX; Leica Microsystems, Inc., Buffalo
Grove, IL, USA).
Evaluation of the results of
immunohistochemical staining
Three observers who were blind to the clinical and
follow-up data evaluated the staining results independently using a
multihead microscope. Sections were co-observed to reach a
consensus when the evaluations were divergent. For assessment of
TAB3, >1,000 cells from five high-power fields in each specimen
were randomly selected, and the staining was examined to determine
the mean percentage. The IHC staining was scored according to the
following method: Intensity of TAB3 staining was scored as 1
(negatively or poorly stained), 2 (moderately stained) or 3
(strongly stained). Percentage scores were assigned as 1 (0–49%), 2
(50–74%) or 3 (75–100%) for univariate analyses. After multiplying
the two scores, the specimens were divided into two groups
according to the scores (average=4.5): Score ≥4.5 was defined as
the high expression group and score <4.5 was defined as the low
expression group.
For Ki-67 detection, tissues were deparaffinized in
toluene, rehydrated in a graded series of ethanol solutions and
processed for immunohistochemistry. Briefly, after incubation with
0.3% hydrogen peroxide for 30 min at room temperature to quench
endogenous peroxidase, and heating for 15 min in 0.1 M citrate
buffer (pH 6.0) in a microwave oven for antigen retrieval, the
sections were blocked with normal serum (Vector Laboratories Inc.,
Burlingame, CA, USA) corresponding to the origin of the secondary
antibodies for 30 min at room temperature. The slides were
incubated overnight at 4°C with primary antibodies against Ki-67
(cat. no. sc-23900, 1:100; Santa Cruz Biotechnology, Inc.), After
washing with PBS, the slides were incubated with biotinylated
secondary antibodies (cat. no. SE131, 1:100; Beijing Solarbio
Science & Technology Co., Ltd., Beijing, China) for 30 min at
room temperature, followed by peroxidase-conjugated avidin-biotin
complex for 30 min at room temperature. Immunostaining was
visualized using 3,3′-diaminobenzidine as a chromogen. The slides
were then counterstained with hematoxylin. All sample sections were
observed under a microscope (Leica DFC 300 FX; Leica Microsystems,
Inc.). For determination of the Ki-67 proliferation index,
positively stained cells and nuclei were counted on a minimum of
six randomly selected fields from representative tumor sections.
The proliferative index was calculated as the number of
Ki-67-positive cells divided by the total number of cells counted.
In half of the samples, the staining was repeated three times to
avoid possible technical errors, and similar results were obtained
in these samples.
Western blotting
Cells were collected for immunoblotting analysis,
washed three times with ice-cold PBS and resuspended in 2X lysis
buffer (50 mM Tris-HCl, 120 mM NaCl, 0.5% Nonidet P-40, 100 mM NaF,
200 IM Na3VO4 and protease inhibitor
mixture). Cell lysates were centrifuged at 16,000 × g for 30 min at
4°C, and were then denatured at 100°C for 15 min. The total protein
concentration was determined using the Bio-Rad protein assay kit
(Bio-Rad Laboratories, Inc., Hercules, CA, USA). All protein
samples were stored at −20°C.
The proteins (30 µg/well) were resolved by 10%
SDS-PAGE and were then transferred onto polyvinylidene difluoride
membranes. The membranes were blocked with 5% non-fat milk in TBST
(20 mM Tris, 150 mM NaCl and 0.05% Tween-20) for 2 h at room
temperature, and were then incubated with primary antibodies for ≥6
h at room temperature. After being washed three times, the
membranes were incubated with horseradish peroxidase-conjugated
secondary human antibodies (cat. no. 31430, 1:5,000; Pierce; Thermo
Fisher Scientific, Inc.) for 2 h at room temperature, according to
the manufacturer's protocol. The bands were then detected by
enhanced chemiluminescence detection systems (Pierce; Thermo Fisher
Scientific, Inc.). The band intensity was measured using an ImageJ
(version 1.8.0; National Institutes of Health, Bethesda, MD,
USA).analysis system (National Institutes of Health).
Transient transfection
The TAB3 small interfering (si)RNA and control siRNA
were purchased from Shanghai GeneChem Co., Ltd. (Shanghai, China).
The TAB3-specific siRNA target sequence was
5′-GGTTGAAGTCTGAAGTTAA-3′ and the control-siRNA sequence was
5′-TTCTCCGAACGTGTCACGT-3′. The human ESCC cell line TE-1 was grown
in dishes until the cells reached 80% confluence, and were then
transfected with either control siRNA or TAB3-siRNA (50 nM) using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.), according to the manufacturers protocol. The
medium was replaced after 24 h with fresh medium for transfection.
The cells in the mock group were untreated. Cells were collected
for western blot analysis, wound healing assays, Transwell assays
and cell proliferation assays 36 h post-transfection.
Cell proliferation assays
Cell proliferation was determined using a commercial
Cell Counting Kit (CCK)-8 assay (Dojindo Molecular Technologies,
Inc., Kumamoto, Japan), according to the manufacturer's protocol.
Briefly, cells were plated into a 96-well plate at a density of
2×104 cells/well and were allowed to grow overnight.
CCK-8 reagent was then added to the wells for 2 h at 37°C, and the
absorbance was read at a wavelength of 490 nm using an automated
plate reader. The experiments were repeated at least three
times.
Cell cycle analysis
Serum starvation and a refeeding process were used
to synchronize the cell cycle. Briefly, TE-1 cells were incubated
in FBS-free DMEM for 72 h to synchronize the cells, after which the
medium was replaced with complete medium. Subsequently, the cells
were rapidly harvested after 48 h, fixed in 70% ethanol for ≥24 h
at −20°C, and incubated with 1 mg/ml RNase A (Invitrogen; Thermo
Fisher Scientific, Inc.) for 30 min at 37°C. Subsequently, the
cells were stained with propidium iodide (50 µg/ml) (BD
Biosciences, San Jose, CA, USA) in PBS with 0.5% Triton X-100, and
were analyzed using a BD FACScan flow cytometer (BD Biosciences)
with CellQuest acquisition and analysis software (version 1.8.0; BD
Biosciences, Franklin Lakes, NJ, USA).
Wound healing assay
Cells were grown in a monolayer in 6-well plates to
nearly 100% confluence. Subsequently, cells were serum-starved for
12 h post-transfection with control siRNA or TAB3 siRNA. The
monolayer was scratched with a 1-ml pipette tip, and the cells were
washed with PBS and cultured in 5% FBS-DMEM. A Leica inverted phase
contrast microscope (Leica DFC 300 FX; Leica Microsystems, Inc.)
was used to capture images of the cells under a 20× objective lens
at 0 and 24 h. The migration of cells was determined according to
the size of the wounded region.
Transwell migration/invasion
assay
Cells transfected with control siRNA or TAB3 siRNA
were starved overnight in DMEM with 0.1% FBS. Subsequently, the
cells were trypsinized and resuspended in DMEM containing 0.1%
bovine serum albumin (Gibco; Thermo Fisher Scientific, Inc.). Cells
(1×105) were added to the upper chambers of 24-well
Transwell plates (8 µm pore size; Corning Incorporation, Corning,
NY, USA), which were used to conduct migration and invasion assays.
The upper chambers of 24-well Transwell plates were not precoated
with Matrigel for the migration assays; however, to observe
invasive ability, the wells of the Transwell chamber were coated
with Matrigel. DMEM supplemented with 10% FBS was added to the
lower chambers. After overnight incubation at 37°C, the cells that
remained in the upper chamber were removed, whereas cells that had
migrated/invaded to the lower chamber were fixed with 10% methanol
for 20 min at 37°C and stained with 0.1% crystal violet for 30 min
at 37°C, in order to visualize their nuclei. The number of
migrated/invaded cells in five fields was counted under 200×
magnification (Leica DFC 300 FX; Leica Microsystems, Inc.), and the
mean number of cells per field was determined for each chamber. All
aforementioned experiments were conducted in triplicate and
repeated three times.
Co-immunoprecipitation assay
TE-1 cells were harvested and lysed in buffer (50
mmol/l Tris-HCl, pH 7.5; 150 mmol/l NaCl; 5 mmol/l EDTA; 1% NP-40,
0.5% deoxycholate; 0.1% SDS). A total of 30 µl supernatant was
collected as input. The remaining liquid was precleared with 30 µl
protein A/G agarose (Santa Cruz Biotechnology, Inc.) on a rocker at
4°C for 2 h. Subsequently, the precleared supernatant was separated
into two samples and incubated with 6 µl mouse monoclonal TAB3
antibody or TAK1 antibody (Santa Cruz Biotechnology, Inc.) or 0.3
µl mouse immunoglobulin G (cat. no. ESK7005-96T; Sangon Biotech
Co., Ltd., Shanghai, China) at 4°C overnight with gentle agitation.
The samples were then incubated with 30 µl protein A/G at 4°C for 2
h with gentle agitation. Finally, the precipitates were collected.
The immune complexes were analyzed by immunoblotting using specific
antibodies against TAB3 and TAK1.
Statistical analysis
All values are expressed as the means ± standard
error of the mean. Each experiment consisted of at least three
replicates per condition. Statistical analysis was performed using
SPSS software (version 19.0; IBM Corp., Armonk, NY, USA). TAB3
expression and clinicopathological features were analyzed by
Pearson's χ2 test. Spearman's correlation test was
performed to analyze the correlation between TAB3 and Ki-67
expression. For analysis of survival data, Kaplan-Meier curves were
constructed and log-rank tests were performed. Comparisons between
two groups were conducted using Student's t-test, whereas
comparisons between more than two groups were conducted using
analyses of variance; one-way analysis of variance followed by
Tukey's post hoc test or two-way analysis of variance followed by
Bonferroni's post hoc test were conducted. P<0.05 was considered
to indicate a statistically significant difference.
Results
TAB3 is overexpressed in ESCC
tissues
It has been reported that TAB3 is highly expressed
in various tumors, such as non-small cell lung cancer (NSCLC), and
breast and ovarian cancer (11–13).
However, to the best of our knowledge, its expression has not been
reported in ESCC. Therefore, immunohistochemical staining of 80
ESCC samples was performed to determine the expression of TAB3 in
ESCC tissues, and to confirm the association of TAB3 with ESCC
progression. As expected, TAB3 was significantly overexpressed in
ESCC tissues compared with the adjacent non-cancerous tissues in
most cases, and the immunoreactivity of TAB3 was predominantly
detected in the cytoplasm (Fig.
1A). Furthermore, the positive ratio of TAB3 was upregulated in
poorly differentiated ESCC tissues compared with
well-differentiated tissues, which was consistent with the Ki-67
index (Fig. 1B-D). As shown in
Fig. 1B and D, the Ki-67 index was
significantly elevated in poorly differentiated ESCC tissues; the
mean index increased from 5.1±2.2% in adjacent non-cancerous
tissues to 13.2±4.3% in well-differentiated tissues, 22.1±7.2% in
moderately differentiated tissues and 31.4±5.7% in poorly
differentiated tissues. Subsequently, the correlation between TAB3
and Ki-67 expression was analyzed. A moderate positive correlation
was determined between the expression status of TAB3 and that of
Ki-67. Spearman's correlation coefficient (γ2) for
TAB3-Ki-67 was 0.419 (P=0.001; Fig.
1E). To further demonstrate the association between TAB3
expression and clinicopathological parameters in ESCC, the data
were summarized in Table I. The
expression of TAB3 was associated with lymph node metastasis
(P=0.002), T stage (P=0.019), pathological grade (P=0.001) and
Ki-67 expression (P=0.017), whereas there was no association with
other prognostic factors.
TAB3 is overexpressed in ESCC cell
lines
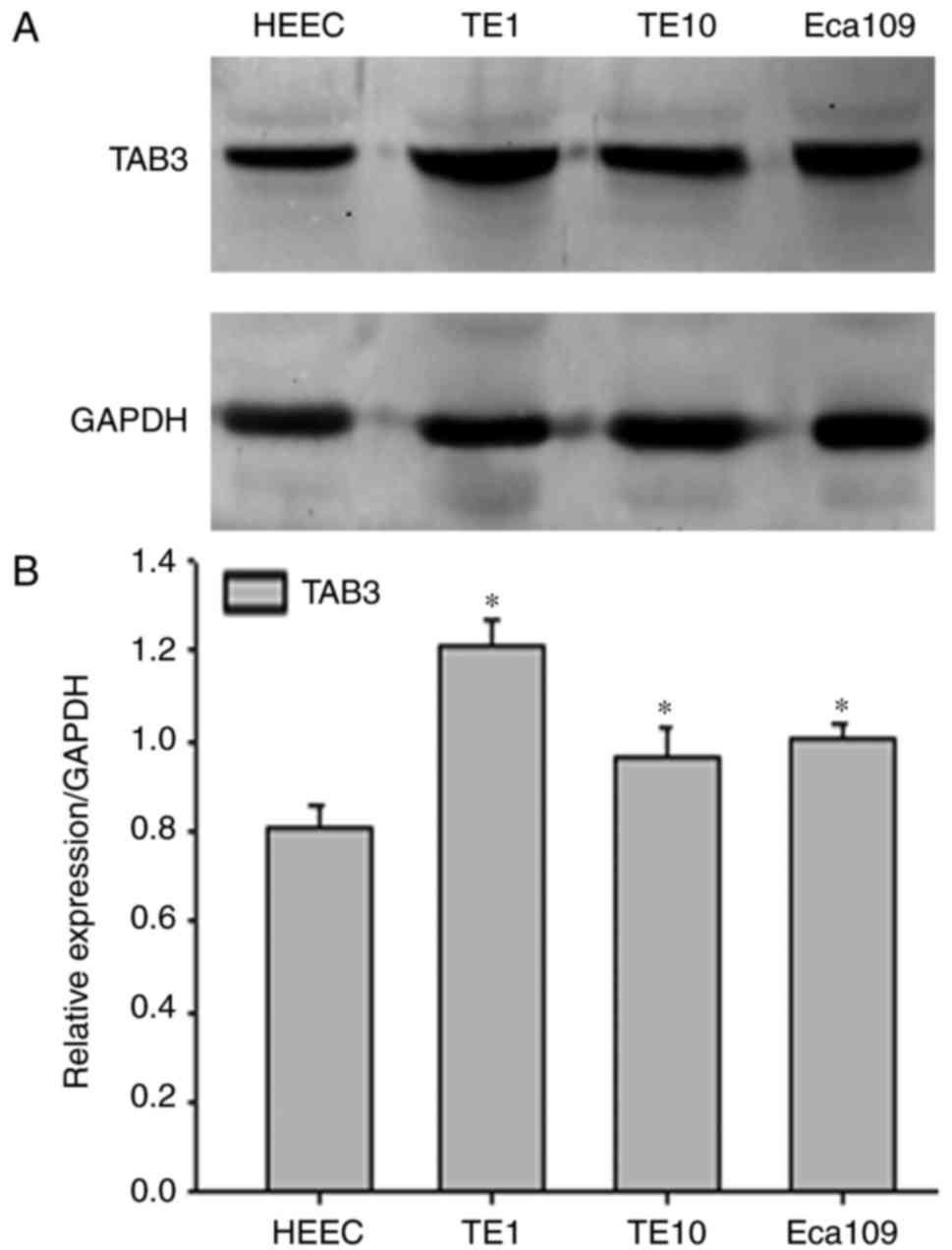
As shown in Fig. 1,
TAB3 may be significantly overexpressed in ESCC cancerous tissues
compared with in adjacent non-cancerous tissues in most cases. To
determine the possible role of TAB3, western blot analysis was used
to examine the protein expression levels of TAB3 HEECs and three
human ESCC cell lines (TE-1, TE-10 and Eca-109). As expected, the
expression levels of TAB3 were significantly higher in TE-1, TE-10
and Eca-109 cells compared with in HEEC cells (Fig. 2A and B). In addition, TE-1 cells
exhibited the highest protein levels among the three ESCC cell
lines; therefore, the TE-1 cell line was selected to conduct
subsequent experiments.
Association between TAB3 expression
and ESCC patient survival
The present study analyzed the association between
TAB3 expression and the survival status of 80 patients using
Kaplan-Meier survival curves. As shown in Fig. 3, high TAB3 expression (n=38) was
more significantly associated with poor survival compared with low
TAB3 expression (n=42). These results clearly indicated that TAB3
was associated with poor survival of patients.
Knockdown of TAB3 inhibits the
proliferation of ESCC cells
To further ascertain the role of TAB3 in ESCC cell
proliferation and invasion, TE-1 cells were transiently transfected
with TAB3 siRNA or control siRNA for 36 h. Western blotting was
used to confirm transfection efficiency. The results of western
blotting indicated that TAB3 silencing by TAB3 siRNA significantly
reduced the expression levels of TAB3 in TE-1 cells compared with
the control siRNA-transfected cells (Fig. 4A and B).
The effects of TAB3 on cell growth were subsequently
investigated. Western blot analysis demonstrated that following
TAB3 downregulation via siRNA transfection, the expression levels
of the cell proliferation marker PCNA and the cell cycle protein
cyclin D1 were concomitantly inhibited (Fig. 4C). In addition, the CCK-8 assay
demonstrated that knockdown of TAB3 significantly reduced cell
proliferation in a time-dependent manner (Fig. 4D). Furthermore, flow cytometric
analysis was conducted to reveal cell cycle distribution. The
percentage of TE-1 cells arrested in G0/G1
phase increased to 66.64% post-transfection with TAB3 siRNA,
whereas the percentage of cells in S phase decreased from 41.88 to
14.55%, thus suggesting that TAB3 may be able to promote the
G0/G1-S transition and thus promote cell
growth (Fig. 4E). These results
suggested that the expression of TAB3 may promote ESCC cell
proliferation.
Knockdown of TAB3 reduces the
migration and invasion of ESCC cells
Cancer cell motility is known to be indispensable
for cancer metastasis and invasion. Therefore, the present study
aimed to determine the effects of TAB3 on the migration and
invasion of ESCC cells. Wound healing and Transwell assays were
performed to assess the effects of TAB3 on cell migration and
invasion. As shown in Fig. 5A,
knockdown of TAB3 markedly decreased the rate of the wound healing
process. Subsequently, the Transwell migration assay was used to
confirm that TAB3 knockdown inhibited the migration of TE-1 cells
(Fig. 5B and C). The results
suggested that knockdown of TAB3 reduced the migration of TE-1
cells. Finally, Transwell invasion assays were conducted to
determine whether knockdown of TAB3 reduced the invasion of ESCC
cells. As shown in Fig. 5D and E,
knockdown of TAB3 significantly reduced the number of TE-1 cells
invading through the Matrigel-coated polycarbonate filter to the
lower chamber. These results indicated that TAB3 may increase the
migration and invasion of TE-1 cells.
Knockdown of TAB3 decreases the
expression of the NF-κB pathway in TE-1 cells
It has been reported that TAB3 binds to TAK1; the
compound is then phosphorylated, leading to activation of the NF-κB
pathway (15,16). The present study analyzed the
TAB3-TAK1-NF-κB pathway in ESCC cells. To further study the
mechanism of TAB3 in ESCC, co-immunoprecipitation was performed to
determine the relationship between TAB3 and TAK1; the results
revealed that TAB3 interacted with TAK1 in TE-1 cells (Fig. 6A). Subsequently, western blotting
was used to determine whether TAB3-TAK1 compounds could influence
the NF-κB pathway. The results demonstrated that silencing TAB3
with TAB3 siRNA decreased the expression of p65, which is a member
of the NF-κB family (Fig. 6B).
These results suggested that TAB3 may interact with TAK1 and
subsequently upregulate the NF-κB pathway.
Discussion
The present study demonstrated that TAB3 was
significantly upregulated in human ESCC tissues and cell lines, and
that TAB3 expression was associated with poor survival of patients.
In addition, knockdown of TAB3 expression by RNA interference
inhibited the proliferation and invasion of ESCC cells. Finally,
TAB3 was revealed to affect ESCC through activation of the
TAK1-NF-κB pathway. Therefore, TAB3 may be a promising therapeutic
target for the treatment of ESCC.
It has been reported that TAB3 is overexpressed in
several types of cancer, and that it is associated with tumor
development and invasion. Tao et al (12) used western blot analysis to
demonstrate that p38 mitogen-activated protein kinase (MAPK)
induces TAB3 and enhances TAB3-mediated NF-κB activation in breast
cancer. Furthermore, it was revealed that the expression of TAB3
increases cell migration and invasion by activating NF-κB, and TAB3
expression is significantly correlated with poor outcomes of
patients (12). Another study
demonstrated that in ovarian cancer, TAB3 is significantly
overexpressed, enhancing cancer cell proliferation and reducing
chemical sensitivity (13). Our
previous study revealed that in NSCLC, TAB3 overexpression promotes
cell proliferation and mediates chemoresistance to cisplatin via
the NF-κB pathway (11). TAB3 is
involved in several types of tumor; however, to the best of our
knowledge, its effect on ESCC remains to be studied. Therefore, the
present study is the first to observe that the expression of TAB3
may be markedly upregulated in ESCC cells, thus suggesting that
TAB3 may serve a role in the development and progression of
ESCC.
The cell cycle is associated with cell
transformation and carcinogenesis (17–19).
Boonstra and Moes demonstrated that the MAPK and phosphoinositide
3-kinase pathways have an essential role in early G1
phase (18), and cyclin D1 is a key
target of proliferative signals in G1 phase (20). The present study demonstrated that
following knockdown of TAB3 with siRNA, the expression levels of
PCNA and cyclin D1 were concomitantly inhibited; therefore, TAB3
may be involved in carcinogenesis and the development of ESCC. To
the best of our knowledge, the effects of TAB3 on other cell cycle
control molecules, such as cyclin-dependent kinase (cdk)4, cdk6,
p21CIP1, p27KIP1 and p53, have not been reported. The present study
did not analyze these molecules; therefore, further studies may
elucidate the effect of TAB3 on more cell cycle control
molecules.
Increasing evidence has revealed that the NF-κB
pathway serves a key role in promoting ESCC progression by
modulating cell cycle and invasion processes (21–24).
Activation of the NF-κB pathway induces epithelial-mesenchymal
transition in cells (25,26). Furthermore, it has been reported
that activation of both the activator protein 1 (AP-1) and the
NF-κB transcription factors may increase the invasive properties of
ESCC cells (27,28).
TAB3 was initially identified as the human ortholog
of Xenopus TAB3 in the NF-κB pathway and as a hypothetical
oncogene (29). Both AP-1 and NF-κB
transcription factors can be activated by overexpression of TAB3
(29). Numerous studies have
implicated TAB3 as a central factor in cell survival, proliferation
and metastasis, which are actions that are mediated by activating
the NF-κB pathway (11,12,16).
TAB3 and its homolog TAK1-binding protein 2 (TAB2) are adaptor
molecules linking tumor necrosis factor (TNF)-associated factor 6,
tumor necrosis factor α and interleukin-1, which are TNF-associated
factor molecules. Both TAB2 and TAB3 can activate TAK1. Previous
reports have demonstrated that inhibition of TAK1 may be an
effective target for the induction of cancer cell death (9,29). The
present study also revealed that knockdown of TAB3 significantly
inhibited proliferation and invasion of ESCC cells via the NF-κB
pathway. Based on these findings, it may be concluded that TAB3
participates in NF-κB pathway signaling in ESCC. However, another
way to determine whether TAB3 activates the NF-κB pathway is to
examine phosphorylated-p65 expression using nuclear extracts. As a
limitation of the present study, our future studies aim to examine
phosphorylated-p65.
In conclusion, the present study demonstrated that
high TAB3 expression was markedly associated with poor survival.
Furthermore, knockdown of TAB3 expression inhibited the
proliferation and invasion of ESCC cells by suppressing the NF-κB
pathway, which may represent a novel therapeutic target for the
treatment of ESCC. However, it has been suggested that knockdown of
TAB3 leads to chemoresistance in ovarian cancer and NSCLC (11,13).
Radiotherapy and chemotherapy are important therapeutic strategies
in the treatment of ESCC. Therefore, future studies, potentially
using TAB3 knockout mice to investigate the function of TAB3 in
tumor radiotherapy and chemotherapy in vivo, may further
elucidate the functional significance of this gene in the NF-κB
signaling pathway.
Acknowledgements
The authors would like to thank Dr Jian Zhao and Dr
Dan Dan for their valuable input.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant nos. 31670857 and
31700737), the Natural Science Foundation of Jiangsu Province
(grant no. BK20161152), the Key Scientific Research Program of Wuxi
Municipal Health Bureau (grant no. Z201509), the Program for
Innovative Research of Wuxi (grant no. CXTD004), and the Young
Talents Subsidy Project of Wuxi Municipal Commission of Health and
Family Planning (grant no. QNRC092).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JZ performed the histological examination of the
tissues and was a major contributor in writing the manuscript. LG
performed the wound healing assay and Transwell migration/invasion
assays. YG performed the western blotting and
co-immunoprecipitation assay. WX performed the cell proliferation
assays. DS, QL and WM analyzed the possible mechanism by which TAB3
participated in tumorigenesis of ESCC. FW performed the cell cycle
analysis. PL and JC designed the present study. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
All patients provided written informed consent for
their tissue samples to be used for scientific research. The
present study was approved by the Affiliated Hospital of Nantong
University Ethics Committee (Nantong, China).
Patient consent for publication
All patients that participated in this study
provided informed consent for publication.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yano T, Muto M, Minashi K, Iwasaki J,
Kojima T, Fuse N, Doi T, Kaneko K and Ohtsu A: Photodynamic therapy
as salvage treatment for local failure after chemoradiotherapy in
patients with esophageal squamous cell carcinoma: A phase II study.
Int J Cancer. 131:1228–1234. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhang L, Wu YD, Li P, Tu J, Niu YL, Xu CM
and Zhang ST: Effects of cyclooxygenase-2 on human esophageal
squamous cell carcinoma. World J Gastroenterol. 17:4572–4580. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lagergren J, Smyth E, Cunningham D and
Lagergren P: Oesophageal cancer. Lancet. 390:2383–2396. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Rustgi AK and El-Serag HB: Esophageal
carcinoma. N Engl J Med. 371:2499–2509. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Enzinger PC and Mayer RJ: Esophageal
cancer. N Engl J Med. 349:2241–2252. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Takaesu G, Kishida S, Hiyama A, Yamaguchi
K, Shibuya H, Irie K, Ninomiya-Tsuji J and Matsumoto K: TAB2, a
novel adaptor protein, mediates activation of TAK1 MAPKKK by
linking TAK1 to TRAF6 in the IL-1 signal transduction pathway. Mol
Cell. 5:649–658. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Besse A, Lamothe B, Campos AD, Webster WK,
Maddineni U, Lin SC, Wu H and Darnay BG: TAK1-dependent signaling
requires functional interaction with TAB2/TAB3. J Biol Chem.
282:3918–3928. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hu Y, Liu JP, Zhu Y and Lu NH: The
importance of toll-like receptors in NF-κB signaling pathway
activation by helicobacter pylori infection and the regulators of
this response. Helicobacter. 21:428–440. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen J, Gu J, Feng J, Liu Y, Xue Q, Ni T,
Wang Z, Jia L, Mao G and Ji L: TAB3 overexpression promotes cell
proliferation in non-small cell lung cancer and mediates
chemoresistance to CDDP in A549 cells via the NF-κB pathway. Tumour
Biol. 37:3851–3861. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tao T, He Z, Shao Z and Lu H: TAB3
O-GlcNAcylation promotes metastasis of triple negative breast
cancer. Oncotarget. 7:22807–22818. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chen Y, Wang X, Duan C, Chen J, Su M, Jin
Y, Deng Y, Wang D, Chen C, Zhou L, et al: Loss of TAB3 expression
by shRNA exhibits suppressive bioactivity and increased chemical
sensitivity of ovarian cancer cell lines via the NF-κB pathway.
Cell Prolif. 49:657–668. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sobin LH, Gospodarowicz MK and Wittekind
C: TNM classification of malignant tumours. John Wiley & Sons;
2011
|
|
15
|
Roh YS, Song J and Seki E: TAK1 regulates
hepatic cell survival and carcinogenesis. J Gastroenterol.
49:185–194. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Luo C, Yuan R, Chen L, Zhou W, Shen W, Qiu
Y and Shao J, Yan J and Shao J: TAB3 upregulates Survivin
expression to promote colorectal cancer invasion and metastasis by
binding to the TAK1-TRAF6 complex. Oncotarget. 8:106565–106576.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Malumbres M and Carnero A: Cell cycle
deregulation: A common motif in cancer. Prog Cell Cycle Res.
5:5–18. 2003.PubMed/NCBI
|
|
18
|
Boonstra J and Moes MJ: Signal
transduction and actin in the regulation of G1-phase progression.
Crit Rev Eukaryot Gene Expr. 15:255–276. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hulleman E and Boonstra J: Regulation of
G1 phase progression by growth factors and the extracellular
matrix. Cell Mol Life Sci. 58:80–93. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tashima Y, Hamada H, Okamoto M and Hanai
T: Prediction of key factor controlling G1/S phase in the mammalian
cell cycle using system analysis. J Biosci Bioeng. 106:368–374.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gan J, Ke X, Jiang J, Dong H, Yao Z, Lin
Y, Lin W, Wu X, Yan S, Zhuang Y, et al: Growth hormone-releasing
hormone receptor antagonists inhibit human gastric cancer through
downregulation of PAK1-STAT3/NF-kappaB signaling. Proc Natl Acad
Sci USA. 113:14745–14750. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Jung JU, Ravi S, Lee DW, McFadden K,
Kamradt ML, Toussaint LG and Sitcheran R: NIK/MAP3K14 regulates
mitochondrial dynamics and trafficking to promote cell invasion.
Curr Biol. 26:3288–3302. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gan X, Chen B, Shen Z, Liu Y, Li H, Xie X,
Xu X, Li H, Huang Z and Chen J: High GPX1 expression promotes
esophageal squamous cell carcinoma invasion, migration,
proliferation and cisplatin-resistance but can be reduced by
vitamin D. Int J Clin Exp Med. 7:2530–2540. 2014.PubMed/NCBI
|
|
24
|
Han Y, Guo XH, Zheng QF, Zhu YL, Fan YY
and Zhang XY: Down-regulation of platelet-derived growth factor-D
expression blockades NF-κB pathway to inhibit cell proliferation
and invasion as well as induce apoptosis in esophageal squamous
cell carcinoma. Mol Biol Rep. 40:2473–2483. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gao S, Sun Y, Zhang X, Hu L, Liu Y, Chua
CY, Phillips LM, Ren H, Fleming JB, Wang H, et al: IGFBP2 activates
the NF-κB pathway to drive epithelial-mesenchymal transition and
invasive character in pancreatic ductal adenocarcinoma. Cancer Res.
76:6543–6554. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Pires BR, Mencalha AL, Ferreira GM, de
Souza WF, Morgado-Díaz JA, Maia AM, Corrêa S and Abdelhay ES:
NF-kappaB is involved in the regulation of EMT genes in breast
cancer cells. PLoS One. 12:e01696222017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Shin WS, Hong Y, Lee HW and Lee ST:
Catalytically defective receptor protein tyrosine kinase PTK7
enhances invasive phenotype by inducing MMP-9 through activation of
AP-1 and NF-κB in esophageal squamous cell carcinoma cells.
Oncotarget. 7:73242–73256. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhou J, Zheng S, Liu T, Liu Q, Chen Y, Tan
D, Ma R and Lu X: MCP2 activates NF-κB signaling pathway promoting
the migration and invasion of ESCC cells. Cell Biol Int.
42:365–372. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Jin G, Klika A, Callahan M, Faga B, Danzig
J, Jiang Z, Li X, Stark GR, Harrington J and Sherf B:
Identification of a human NF-kappaB-activating protein, TAB3. Proc
Natl Acad Sci USA. 101:2028–2033. 2004. View Article : Google Scholar : PubMed/NCBI
|