Introduction
Pancreatic cancer (PC) is one of the most malignant
tumor types, with a 5-year overall survival rate of <6%
(1). Globally, pancreatic cancer is
the fourth most common cause of cancer-associated mortality. By
2030, pancreatic cancer may surpass breast cancer as the second
most deadly tumor in the US (2).
Patients with pancreatic cancer with no obvious clinical symptoms
are frequently not diagnosed until advanced tumor stages. In recent
years, with the development of molecular diagnoses and targeted
therapies, there has been a huge improvement in diagnosis and
treatment. However, pancreatic cancer, as a highly heterogeneous
disease, still lacks effective diagnostic and prognostic biomarkers
(3). To improve the rate of early
diagnosis and extend the survival time of patients with pancreatic
cancer, it is essential to identify effective tumor biomarkers for
early diagnosis and prognosis risk stratification. This will not
only help clinicians choose effective and individualized treatment
programs, it will also be conducive to future investigations into
the molecular mechanisms of the occurrence and development of
pancreatic cancer.
Due to the development of genomic and transcriptomic
sequencing techniques, an increasing number of non-coding RNAs
(ncRNAs), which differ from protein-coding RNAs, have been
identified. ncRNAs are divided into two categories according to
size: Short ncRNAs and long ncRNAs (lncRNAs). lncRNAs are defined
as >200 nucleotides in length (4). The Human Genome Project has indicated
that protein-coding genes account for <2% of the total sequence
of the genome (5). Our
understanding of lncRNAs is also very limited because the number of
ncRNAs, including lncRNAs, is huge. It has been reported that the
majority lncRNAs do not have gene regulatory functions; however,
there is growing evidence to support the idea that lncRNAs may
regulate gene expression at multiple levels, including genomic
imprinting and transcriptional and post-transcriptional regulation
(6). Chen et al (7) demonstrated that lncRNA small nucleolar
RNA host gene 20 promotes the proliferation and migration of lung
cancer cells by silencing p21 expression. Additionally the lncRNA
DPP10 antisense RNA 1 inhibits the metastasis of colorectal cancer
by upregulating TIMP metallopeptidase inhibitor 3, which suggests
that this lncRNA could be used as a new therapeutic target for
colorectal cancer (8). In recent
years, a novel mechanistic type of lncRNA, known as a competing
endogenous RNA (ceRNA), has been reported (6). lncRNAs of this class harbor microRNA
(miR) response elements and can compete with miRs to communicate
with mRNAs and to regulate the expression of genes (9). The study demonstrated that the lncRNA
colon cancer associated transcript 2 regulates miR-145 expression
by inhibiting its maturation in colon cancer cells (10). Another study reported that long
intergenic non-protein coding RNA 941 acted as a ceRNA for miR-34a,
leading to Snail1 upregulation and epithelial-mesenchymal
transition activation in hepatocarcinogenesis (11).
Regarding pancreatic cancer, it has been reported
that several lncRNAs, including HOX transcript antisense RNA
(12), HOXA distal transcript
antisense RNA (13), metastasis
associated lung adenocarcinoma transcript 1 (14,15)
and long intergenic non-protein coding RNA, regulator of
reprogramming (16), are
differentially expressed between pancreatic cancer tissues and
normal tissues. These findings indicate that lncRNAs may be
potential diagnostic or prognostic biomarkers for pancreatic
cancer. However, a single lncRNA biomarker is often limited in its
diagnostic and prognostic predictive value. Recent studies have
reported that several lncRNA signatures may improve prognosis
predictions for certain malignant tumors, including lymphoma
(17), colorectal (18), esophageal (19) and lung cancer (20). However, the prognostic role of
lncRNAs in pancreatic cancer has not been investigated. To
determine an effective signature of lncRNAs for predicting
pancreatic cancer survival, lncRNA expression profiles from a large
number of patients with pancreatic cancer were obtained and
analyzed by repurposing the publicly available Cancer Genome Atlas
(TCGA) database in this study.
Materials and methods
Data processing
The lncRNA expression data and corresponding
clinical data of pancreatic cancer were obtained from the TCGA
database (portal.gdc.cancer.gov/) using the search terms
‘Project = TCGA-PAAD’. The data included lncRNA expression profiles
of 178 cancer tissue samples and 4 normal tissue samples from 177
patients with pancreatic cancer. Analysis of differentially
expressed lncRNAs (DElncRNAs) between pancreatic tumor tissues and
normal tissues was performed using the edger package (bioconductor.org/packages/release/bioc/html/edgeR.html)
in R (software version 3.1.4; r-project.org)
(21). lncRNAs satisfying the
conditions log2|fold change (FC)|>1.0 and P<0.05
were considered to be differentially expressed. Unsupervised
hierarchical cluster analysis using the gplots package (cran.r-project.org/web/packages/gplots/index.html) in
R (software version 3.1.4; r-project.org) revealed
that normal tissues could be distinguished from pancreatic cancer
tissues based on the DElncRNA patterns. Unsupervised hierarchical
cluster analysis evaluated the degree of variation of DElncRNA
patterns from pancreatic cancer tissues and normal tissues.
Association of DElncRNAs with patient
prognosis
Patients with no prognostic information were
excluded and then combined the DElncRNA expression profiles with
the corresponding survival prognostic information. The prognostic
value of each DElncRNA was evaluated using a Kaplan-Meier curve and
the log-rank method. Patients were divided into the high expression
and low expression groups for each of the lncRNAs according to the
median value of the expression. The DElncRNAs that were
significantly associated with overall survival (OS) were identified
as potential prognostic lncRNA biomarkers.
Cox regression analysis and the
establishment of a prognostic risk score model
The potential prognostic lncRNAs obtained by
Kaplan-Meier survival analysis were included in a Cox regression
analysis. In the Cox regression analysis, it was attempted to
identify the best signature associated with patient survival by
using the least number of lncRNAs. Then, a prognostic risk model
consisting of multiple pancreatic cancer lncRNAs was established,
and a risk score was computed as follows:
Risk score=∑i=1N(Ei*Ci)
Risk score = (N, the number of prognostic lncRNAs;
Ei, the expression value of lncRNAi;
Ci, the estimated repression coefficient of
lncRNAi in the multivariable Cox regression analysis).
The risk score for each patient was calculated, and all patients
were divided into two groups (a high-risk group and a low-risk
group) according to the median risk score. The difference in
prognosis between the high-risk group and the low-risk group was
evaluated by the Kaplan-Meier method. Then, to evaluate the
prognostic risk score model for the 3- and 5-year survival rates of
patients with pancreatic cancer, the receiver operating
characteristic (ROC) curve was analyzed. The area under the ROC
curve (AUC) was calculated from the ROC curves.
Establishment of an lncRNA-mRNA co-expression
network and lncRNA functional predictions by bioinformatics
analysis. An lncRNA-mRNA co-expression network using the WCGNA
package in R. The co-expression relationships between the DElncRNAs
and DEprotein-coding genes were calculated using Pearson
correlation coefficients (the cut-off of Pearson correlation
coefficient was >0.6). Gene Ontology (GO) and Kyoto Encyclopedia
of Genes and Genomes (KEGG) enrichment analyses of the co-expressed
protein-coding genes with DElncRNAs were performed to predict the
biological function of DElncRNAs using the DAVID Bioinformatics
Tool version 6.8 (david.ncifcrf.gov/). Enrichment analysis was performed
using the functional annotation chart and was limited to GO terms
in the ‘Biological Process direct’ and KEGG pathways categories.
Functional annotations with a P<0.05 and an enrichment score of
>2 were considered significant.
Statistical analysis. T-test for continuous
variables and χ2 test for categorical variables were
performed to assess the relationship between the prognostic risk
score and clinical features. In the χ2 analysis, when
the total number of samples was <40 or the expected value of the
sample as <1, Fisher exact test was used. Univariate and
multivariate Cox regression analyses were performed to verify if
the predictive indicators of prognostic risk score calculated from
multiple lncRNAs were independent of other clinical features.
Statistical analyses were performed using IBM SPSS statistics
software program version 22.0 (IBM Corp., Armonk, NY, USA).
P<0.05 was considered to indicate a statistically significant
difference.
Results
Patient characteristic
In this study, expression profiles of 178 samples of
pancreatic cancer tissue and 4 samples of normal tissue were used
to identify DElncRNAs. These samples were from 177 patients with
pancreatic cancer included the following clinical and pathological
characteristics: Age, sex, tumor site, tumor size, grade, stage,
TNM status, chronic pancreatitis, smoking, alcohol consumption and
diabetes (Table I).
 | Table I.Patient demographics and
clinicopathological factors. |
Table I.
Patient demographics and
clinicopathological factors.
| Characteristic | Number of
patients |
|---|
| Age (years) |
|
|
<60/≥60 | 54/123 |
| Sex |
|
|
Male/female | 97/80 |
| Tumor site |
|
|
Head/other/NA | 138/28/11 |
| Tumor size |
|
|
<4/≥4 cm/NA | 96/68/13 |
| Grade |
|
|
1/2/3/4/x | 31/92/49/2/3 |
| Stage |
|
|
I/IIA/IIB/III/IV | 21/28/117/3/4 |
| T |
|
|
T1/T2/T3/T4/Tx | 8/24/141/3/1 |
| N |
|
|
N0/N1/Nx | 49/123/5 |
| M |
|
|
M0/M1/Mx | 79/4/94 |
| Chronic
pancreatitis |
|
|
None/yes/NA | 126/14/37 |
| Smoking
(years) |
|
|
<3/≥3/NA | 84/58/35 |
| Drinking |
|
|
None/yes/NA | 64/101/12 |
| Diabetes |
|
|
None/yes/NA | 108/37/32 |
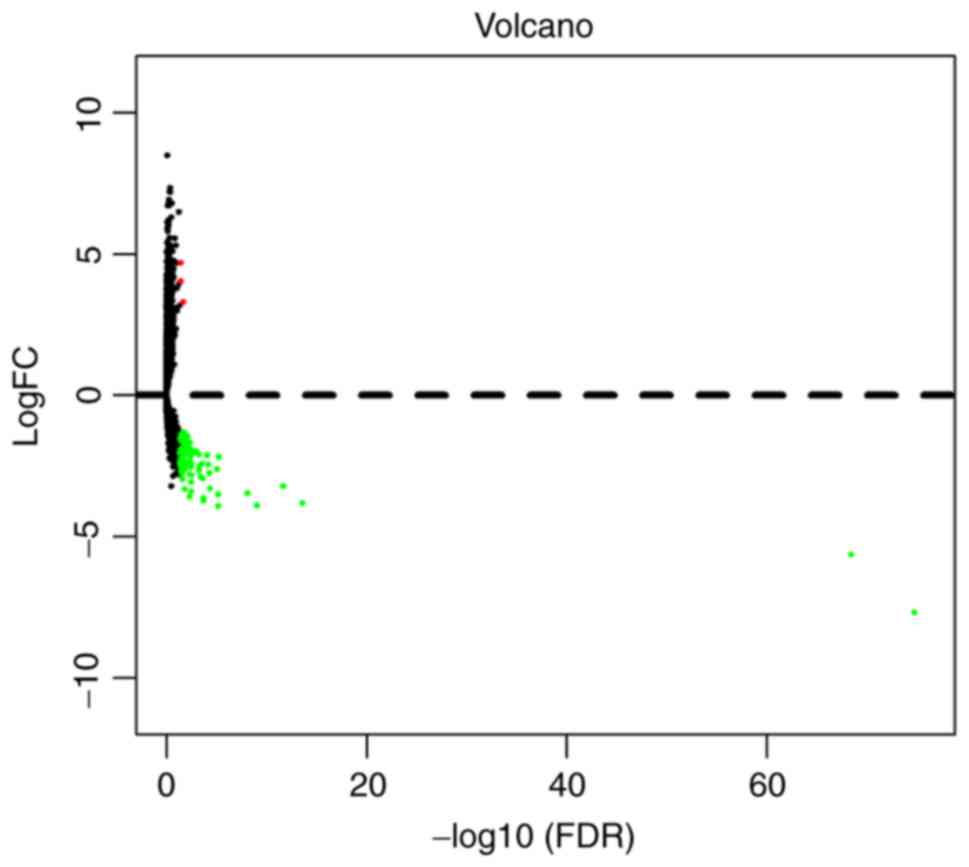
DElncRNAs between pancreatic tumor
tissues and normal tissues
According to the screening criteria (P<0.05 and
|log2FC|>2.0), a total of 92 DElncRNAs were
identified between pancreatic tumor tissues and normal tissues,
including 3 (3.3%) upregulated and 89 (96.7%) downregulated
lncRNAs. In order to visually display DElncRNA between the two
groups, the results were presented as a volcano plot (Fig. 1). Unsupervised hierarchical cluster
analysis showed that normal tissues could be distinguished from
pancreatic cancer tissues based on the DElncRNA patterns (Fig. 2).
Identification of potential lncRNA
biomarkers significantly associated with OS from DElncRNAs
Kaplan-Meier curves and the log-rank method were
used to identify potential lncRNA biomarkers significantly
associated with OS from the DElncRNAs. The results showed that
seven lncRNAs [AC008033.3, AC079015.1, MIR600 host gene (MIR600HG),
AP003086.1, FAM53B antisense RNA 1 (FAM53B-AS1), DNAH17 antisense
RNA 1 (DNAH17-AS1), AL137789.1; Table
II] were significantly associated with pancreatic cancer
prognosis. Of these, five were positively associated with OS
(AC008033.3, AC079015.1, MIR600HG, AP003086.1 and FAM53B-AS1;
Fig. 3A-E), and two were negatively
associated with OS (DNAH17-AS1 and AL137789.1, Fig. 3F and G).
 | Table II.Seven long non-coding RNAs
significantly associated with the overall survival of patients with
pancreatic cancer. |
Table II.
Seven long non-coding RNAs
significantly associated with the overall survival of patients with
pancreatic cancer.
| Gene symbol | Ensembl ID | Chromosome
(GRCh38) | Log fold
change | P-value | False discovery
rate | Adjusted
P-value |
|---|
| DNAH17-AS1 |
ENSG00000267432 | 17:
78,484,882–78,503,056 | −3.46188237 | 6.46871E-12 | 8.72E-09 | 0.00565 |
| AL137789.1 |
ENSG00000236911 | 1:
207,551,925–207,606,555 | −3.62282343 | 3.81106E-07 | 0.000198 | 0.03455 |
| AC079015.1 |
ENSG00000253988 | 8:
138,063,268–138,073,240 | −2.46900918 | 1.08231E-06 | 0.000438 | 0.01524 |
| FAM53B-AS1 |
ENSG00000233334 | 10:
124,703,625–124,714,217 | −2.07318081 | 2.6809E-05 | 0.00529 | 0.0453 |
| AP003086.1 |
ENSG00000251323 | 11:
78,423,982–78,429,836 | −1.73351271 | 7.38534E-05 | 0.012111 | 0.00115 |
| MIR600HG |
ENSG00000236901 | 9:
123,109,494–123,115,477 | −1.60319345 | 9.46888E-05 | 0.013928 | 0.00736 |
| AC008033.3 |
ENSG00000273824 | 12:
68,426,331–68,427,737 | −2.5734859 | 0.000352377 | 0.035634 | 0.00959 |
Establishment of a three-lncRNA
signature associated with pancreatic cancer patient survival
Three lncRNAs (MIR600HG, AL137789.1, AC079015.1)
were selected from seven potential prognostic lncRNAs for logistic
regression analysis according to the algorithm by trying to
incorporate fewer lncRNAs, while achieving the best fit for
patient's survival. Then, a formula was established using these
three-lncRNAs to assess prognostic risk as follows: Risk score =
(0.3073 × expression value of AL137789.1) + (−0.2044 × expression
value of AC079015.1) + (−0.7195 × expression value of MIR600HG).
The risk score for each patient was calculated and all patients
were divided into a high-risk group (88 cases) and a low-risk group
(89 cases) according to the cut-off median risk score (Fig. 4A and B). The OS of the high-risk
group was significantly shorter than that of the low-risk group
(median OS 1.33 vs. 3.65 years; P<0.0001). The Kaplan-Meier
survival curve is shown in Fig. 4C.
As the risk score rose, the expression of AL137789.1 increased, and
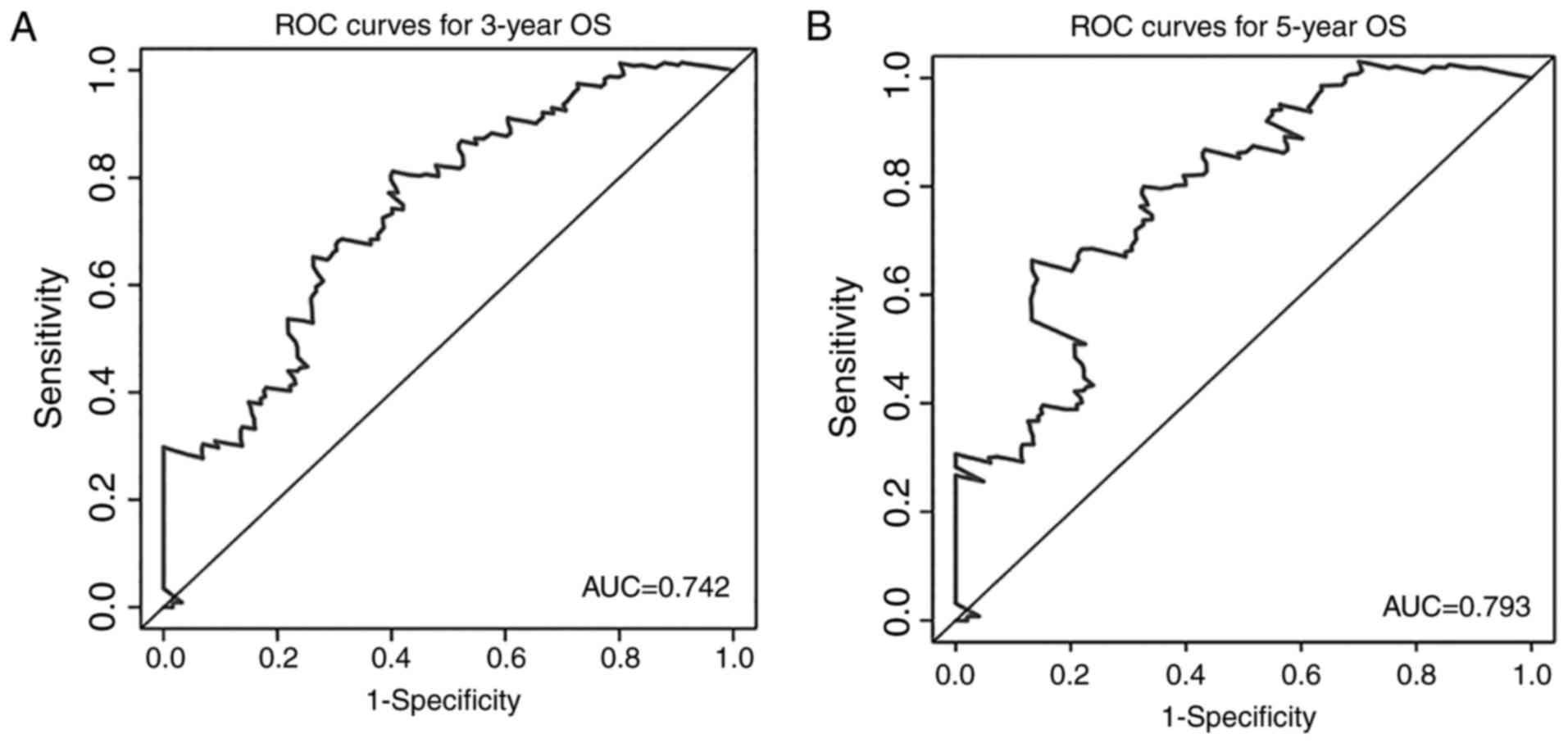
the expression of MIR600HG and AC079015.1 decreased (Fig. 4D). To evaluate the ability of the
risk score to predict 3- and 5-year survival rates, a
time-dependent ROC curve was used (Fig.
5). The results showed that the AUC for the lncRNA signature
prognostic model was 0.742 for an OS of 3 years and 0.793 for an OS
of 5 years. These results suggested an improvement in survival
predictions based on the expression of these three pancreatic
cancer lncRNAs.
Association between the three-lncRNA
signature risk score and clinicopathological characteristics
Significant differences in tumor size, American
Joint Committee on Cancer (AJCC) (22) stage, AJCC tumor status (T) and AJCC
node status (N) were identified between patients in with high and
low risk scores; however, there was no difference in age, sex,
tumor site, grade, AJCC metastasis status, chronic pancreatitis or
smoking between the patients with high and low risk scores
(Table III).
 | Table III.Association between the risk score of
three-long non-coding RNAs signature and clinicopathological
characteristics. |
Table III.
Association between the risk score of
three-long non-coding RNAs signature and clinicopathological
characteristics.
|
| Number of patients
(%) |
|
|
|---|
|
|
|
|
|
|---|
| Characteristic | Low risk | High risk | χ2 | P-value |
|---|
| Total patients | 89 (100) | 88 (100) |
|
|
| Age (years) |
|
| 0.077 | 0.782 |
|
<60 | 28 (31.5) | 26 (29.5) |
|
|
|
≥60 | 61 (68.5) | 62 (70.5) |
|
|
| Sex |
|
| 0.950 | 0.330 |
|
Male | 52 (58.4%) | 45 (51.1%) |
|
|
|
Female | 37 (41.6%) | 43 (48.9%) |
|
|
| Tumor site |
|
| 0.020 | 0.889 |
|
Head | 67 (75.3) | 71 (80.7) |
|
|
|
Others | 14 (15.7) | 14 (15.9) |
|
|
| NA | 8 (9.0) | 3 (3.4) |
|
|
| Tumor size
(cm) |
|
| 5.796 | 0.016 |
|
<4 | 54 (60.7) | 42 (47.7) |
|
|
| ≥4 | 27 (30.3) | 41 (46.6) |
|
|
| NA | 8 (9.0%) | 5(5.7%) |
|
|
| Grade |
|
| 0.005 | 0.945 |
|
G1 +
G2 | 62 (69.7) | 61 (69.3) |
|
|
|
G3 +
G4 | 26 (29.2) | 25 (28.4) |
|
|
|
GX | 1 (1.1) | 2 (2.3) |
|
|
| AJCC stage |
|
| 4.192 | 0.041 |
| I +
IIA | 31 (34.8) | 18 (20.5) |
|
|
| IIB +
III + IV | 56 (62.9) | 68 (77.3) |
|
|
| NA | 2 (2.2) | 2 (2.3) |
|
|
| AJCC T |
|
| 6.116 | 0.013 |
|
T1 +
T2 | 22 (24.7) | 10 (11.4) |
|
|
|
T3 +
T4 | 66 (74.1) | 78 (88.7) |
|
|
|
TX | 1 (1.1) | 0 (0.0) |
|
|
| AJCC N |
|
| 5.256 | 0.022 |
|
N0 | 31 (34.8) | 18 (20.5) |
|
|
|
N1 | 54 (60.7) | 69 (78.4) |
|
|
|
NX | 4 (4.5) | 1 (1.1) |
|
|
| AJCC M |
|
|
| 0.348 |
|
M0 | 42 (47.2) | 37 (42.0) |
|
|
|
M1 | 1(1.1) | 3 (3.4) |
|
|
|
MX | 46 (51.7) | 48 (54.5) |
|
|
| Chronic
pancreatitis |
|
| 0.013 | 0.910 |
|
None | 65 (73.0) | 61 (69.3) |
|
|
|
Yes | 7 (7.9) | 7 (8.0) |
|
|
| NA | 17 (19.1) | 20 (22.7) |
|
|
| Smoking
(years) |
|
| 1.049 | 0.306 |
|
<3 | 45 (50.6) | 39 (44.3) |
|
|
| ≥3 | 26 (29.2) | 32 (36.4) |
|
|
| NA | 18 (20.3) | 17 (19.3) |
|
|
| Alcohol
consumption |
|
| 0.004 | 0.951 |
|
None | 32 (36) | 32 (36.4) |
|
|
|
Yes | 51 (57.3) | 50 (56.8) |
|
|
| NA | 6 (6.7) | 6 (6.8) |
|
|
| Diabetes |
|
| 1.651 | 0.199 |
|
None | 51 (57.3) | 57 (64.8) |
|
|
|
Yes | 22 (24.7) | 15 (17.0) |
|
|
| NA | 16 (18.0) | 16 (18.2) |
|
|
Prognostic value of the three-lncRNA
signature is independent of other clinicopathological factors
Furthermore, univariate analyses were used to test
the prognostic value of the three-lncRNA signature on OS. The
results showed that risk score [hazard ratio (HR)=2.442;
P<0.001), tumor site (HR=0.343; P=0.004), grade (HR=1.584;
P=0.035), AJCC stage (HR=2.295: P=0.001), AJCC T (HR=1.774;
P=0.051), and AJCC N (HR=1.968; P=0.008) were associated with OS in
patients with pancreatic cancer (Table
IV). These significant indicators from univariate analyses were
subsequently included in multivariate Cox regression analyses
(because AJCC stage is not independent of the AJCC T and AJCC N
indicators, AJCC stage was excluded as an indicator). The results
of multivariate Cox regression analyses indicated that risk score
(HR=1.909; P=0.007) is an independent prognostic factor for
pancreatic cancer and that a high risk score is associated with a
poor prognosis (Table IV).
 | Table IV.Univariate and multivariate analyses
of the three-long non-coding RNA signature in the prediction of
pancreatic cancer overall survival. |
Table IV.
Univariate and multivariate analyses
of the three-long non-coding RNA signature in the prediction of
pancreatic cancer overall survival.
|
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|
|---|
|
| HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Risk score (high
vs. low) | 2.442
(1.576–3.784) | 0.000 | 1.909
(1.195–3.050) | 0.007 |
| Age, years (<60
vs. <60) | 1.291
(0.820–2.035) | 0.270 |
|
|
| Sex (female vs.
male) | 0.900
(0.597–1.357) | 0.616 |
|
|
| Tumor site (other
vs. head) | 0.343
(0.165–0.712) | 0.004 | 0.415
(0.194–0.885) | 0.023 |
| Tumor Size (≥4 cm
vs. <4 cm) | 1.016
(0.666–1.549) | 0.943 |
|
|
| Grade
(G3 + G4 vs. G1 +
G2) | 1.584
(1.032–2.432) | 0.035 | 1.643
(1.052–2.567) | 0.029 |
| AJCC stage (IIB +
III + IV vs. I + IIA) | 2.295
(1.383–3.808) | 0.001 |
|
|
| AJCC T
(T3 + T4 vs. T1 +
T2) | 1.774
(0.997–3.159) | 0.051 | 0.937
(0.496–1.769) | 0.841 |
| AJCC N
(N1 vs. N0) | 1.968
(1.191–3.252) | 0.008 | 1.596
(0.877–2.904) | 0.126 |
| AJCC M
(M1 vs. M0) | 0.571
(0.078–4.192) | 0.581 |
|
|
| Chronic
pancreatitis (yes vs. none) | 1.008
(0.460–2.208) | 0.983 |
|
|
| Smoking (3 years
vs. <3 years) | 0.941
(0.595–1.490) | 0.797 |
|
|
| Alcohol consumption
(yes vs. none) | 1.249
(0.805–3.159) | 0.322 |
|
|
| Diabetes (yes vs.
none) | 0.902
(0.518–1.569) | 0.714 |
|
|
Establishment of an lncRNA-mRNA
co-expression network and lncRNA functional predictions
To investigate the potential biological functions of
the DElncRNAs, a co-expression network was established between
DElncRNAs and DEprotein-coding genes (Fig. 6A). The expression of 153
protein-coding genes from 1,452 DEmRNAs was highly correlated with
18 DElncRNAs (Pearson correlation coefficient >0.60). lncRNA
AL137789.1, which is part of the three-lncRNA signature, was also
in this co-expression network. Subsequently, the co-expressed
protein-encoding genes were subjected to GO and KEGG pathway
enrichment analyses, with the background of the whole human genome.
GO functional annotation suggested that 153 protein-coding genes
were significantly enriched in 63 GO terms. The top 20 GO terms are
presented in Fig. 6B. KEGG
functional annotation suggested that protein-coding genes were
significantly enriched in 40 KEGG pathways. The top 20 pathways are
presented in Fig. 6C. The results
from GO and KEGG analyses were associated with immune responses
involving T, B and natural killer cells. Taken together, these
results suggested that lncRNAs may be involved in the tumor immune
response via interactions with protein-coding genes.
Discussion
The traditional view suggests that non-coding RNAs
do not affect gene expression. However, these non-coding RNAs
regulate the expression of genes at different levels, and it has
been reported that they can even encode proteins involved in
biological processes (23).
Increasing numbers of abnormally expressed lncRNAs have been
identified in a variety of tumors (6). These lncRNAs have complex biological
functions, including tumor promotion and tumor suppression, which
may be closely associated tumorigenesis and tumor progression.
It has been reported that the lncRNAs HOXA11
antisense RNA (24), DiGeorge
syndrome critical region gene 5 (25), maternally expressed 3 (26) and hepatocellular carcinoma
upregulated long non-coding RNA (27) promote metastasis and tumor cell
proliferation and are associated with poor prognosis in cancer
patients. However, the lncRNAs CPS1 intronic transcript 1 (28), cancer susceptibility 2 (29), long intergenic non-protein coding
RNA 589 (30) and growth arrest
specific 5 (31) can inhibit the
proliferation and metastasis of tumor cells, which indicates a
favorable survival. The role of certain lncRNAs, such as taurine
upregulated 1 (TUG1), in tumors is controversial. Studies have
reported that TUG1 contributes to tumor progression (32–34)
via its role in the promotion of cell proliferation, metastasis and
inhibition of cell apoptosis. Thus, it is thought to act as an
oncogene in various cancers, such as ovarian (35), thyroid (34) and colorectal cancer (36). However, other studies have reported
that the lncRNA TUG1 is a tumor suppressor (37,38). A
similar controversy occurred for the lncRNA prostate cancer
associated 3 (PCA3) (39,40). However, ncRNA accounts for 80% of
the whole genome, and the general understanding of the function of
lncRNAs is still superficial. Many lncRNAs have not been
functionally investigated.
In pancreatic cancer, several single lncRNA
biomarkers have been reported, including lncRNA PVT1 oncogene
(41), CCDC26 lncRNA (42) and long intergenic non-protein coding
RNA, p53 induced transcript (43).
The same controversy that occurred around PCA3 in prostate cancer
may also occur in pancreatic cancer. The expression of lncRNAs is
relatively low, thus it may be easy to introduce bias when using a
single lncRNA as a biomarker. Utilizing a combination of multiple
potential lncRNA biomarkers could improve accuracy. In this study,
a three-lncRNA signature that is associated with OS was established
by analyzing the RNA-sequencing data from 178 patients with
pancreatic cancer from the TCGA database.
The lncRNAs creating the three-lncRNA signature were
MIR600HG, AL137789.1 and AC079015.1. To the best of our knowledge,
these three lncRNAs have not been previously reported in any
cancer. The expression of all three lncRNAs was significantly lower
in tumor tissues than in normal tissues. Further, the expression of
these lncRNAs is closely associated with the OS of patients with
pancreatic cancer. Lower expression of AC079015.1 and MIR600HG was
associated with poorer OS, whereas lower expression of lncRNA
AL137789.1 was associated with favorable survival. A calculation
for prognosis risk score in patients with pancreatic cancer was
established according to the expression of the three lncRNAs. The
formula indicates that a higher risk score is associated with a
poorer prognosis. Multivariate analyses demonstrated that the
three-lncRNA signature was an independent prognostic predictor of
survival for patients with pancreatic cancer patient. Further, ROC
analyses showed that the AUC of the three-lncRNA signature was
0.742 for the prediction of 3-year OS and 0.793 for the prediction
of 5-year OS. These results demonstrate that these three lncRNAs
have a high accuracy in predicting the prognosis of patients with
pancreatic cancer. In analyses of the association between risk
score and clinicopathological characteristics, high risk scores
were significantly associated with larger tumor size and higher TNM
stage. This indicated that high risk is associated with tumor
progression from another perspective.
Thus far, the function of these three lncRNAs is
unknown. A lncRNA-mRNA co-expression network was used to predict
the function of lncRNAs by functional analysis of protein-coding
genes. The GO and KEGG enrichment results implied that the
DElncRNAs may be involved in tumorigenesis through effects on the
tumor immune response. Unfortunately, AL137789.1 was the only
lncRNA from three-lncRNA signature that was involved in the
co-expression network. AL137789.1 may increase the malignancy of
pancreatic cancer by affecting the tumor immune microenvironment,
despite the current lack of supporting evidence. A large number of
studies have demonstrated that tumor biological behavior is
regulated by the tumor immune microenvironment. In pancreatic
cancer, the tumor immune microenvironment is more complicated due
to the rich stromal environment of the pancreas. Tumor-infiltrating
lymphocytes and cytokines secreted from the stroma constitute the
main components of the tumor immune microenvironment, and have a
role in the immune regulation of tumors. However, lncRNAs may be
involved in certain tumor immunity processes and may aid in tumor
cell immune evasion (44,45). Notably, studies have reported that
lncRNAs are involved in the human immune system, including roles in
dendritic cells, T cells and macrophages (44,45).
Jiang et al (45) reported
that the lncRNA lnc-epidermal growth factor receptor stimulates
T-regulatory cell differentiation, thus promoting hepatocellular
carcinoma immune evasion. Another study reported that the lncRNA
IFNG antisense RNA 1, which is expressed by Th1 cells, specifically
decreases the differentiation of T-cells toward the Th2 phenotype
(46).
In summary, the current study established a
three-lncRNA signature associated with pancreatic cancer by
examining lncRNA expression profiles from the TCGA database.
Further analysis demonstrated that the three-lncRNA signature may
be an independent prognostic biomarker of survival of patients with
pancreatic cancer. Functional predictions revealed that one of
these three lncRNAs, AL137789.1, may be associated with the tumor
immune response.
Acknowledgements
Not applicable.
Funding
The present study was supported in part by the
Public Welfare Technology Research and Social Development Project
of the Department of Science and Technology of Zhejiang Province
(No. 2014C33139) and the Social Development Research and
Demonstration Application Project of the Bureau of Science and
Technology of Jiaxing City (No. 2016AY23057).
Availability of data and materials
The analyzed data sets generated during the study
are available from the corresponding author on reasonable
request.
Authors' contributions
BW and ZZ designed the experiments and analyzed the
data; YB and XW analyzed the data; KW, FC and JG performed lncRNA
functional predictions; JF and ZS performed survival analysis; all
authors read and approved the manuscript and agree to be
accountable for all aspects of the research in ensuring that the
accuracy or integrity of any part of the work are appropriately
investigated and resolved.
Ethics approval and consent to
participate
Ethical approval was obtained from the Second
Hospital of Jiaxing (Jiaxing, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Kamisawa T, Wood LD, Itoi T and Takaori K:
Pancreatic cancer. Lancet. 388:73–85. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Arkan MC: Cancer: Fat and the fate of
pancreatic tumours. Nature. 536:157–158. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Samuel N and Hudson TJ: The molecular and
cellular heterogeneity of pancreatic ductal adenocarcinoma. Nat Rev
Gastroenterol Hepatol. 9:77–87. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Birney E, Stamatoyannopoulos JA, Dutta A,
Guigó R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis
ET, Thurman RE, et al: Identification and analysis of functional
elements in 1% of the human genome by the ENCODE pilot project.
Nature. 447:799–816. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lander ES, Linton LM, Birren B, Nusbaum C,
Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al:
Initial sequencing and analysis of the human genome. Nature.
409:860–921. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Fatica A and Bozzoni I: Long non-coding
RNAs: New players in cell differentiation and development. Nat Rev
Genet. 15:7–21. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chen Z, Chen X, Chen P, Yu S, Nie F, Lu B,
Zhang T, Zhou Y, Chen Q, Wei C, et al: Long non-coding RNA SNHG20
promotes non-small cell lung cancer cell proliferation and
migration by epigenetically silencing of P21 expression. Cell Death
Dis. 8:e30922017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lin J, Tan X, Qiu L, Huang L, Zhou Y, Pan
Z, Liu R, Chen S, Geng R, Wu J and Huang W: Long noncoding RNA
BC032913 as a novel therapeutic target for colorectal cancer that
suppresses metastasis by upregulating TIMP3. Mol Ther Nucleic
Acids. 8:469–481. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tay Y, Rinn J and Pandolfi PP: The
multilayered complexity of ceRNA crosstalk and competition. Nature.
505:344–352. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yu Y, Nangia-Makker P, Farhana L and
Majumdar APN: A novel mechanism of lncRNA and miRNA interaction:
CCAT2 regulates miR-145 expression by suppressing its maturation
process in colon cancer cells. Mol Cancer. 16:1552017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yan X, Zhang D, Wu W, Wu S, Qian J, Hao Y,
Yan F, Zhu P, Wu J, Huang G, et al: Mesenchymal stem cells promote
hepatocarcinogenesis via lncRNA-MUF interaction with ANXA2 and
miR-34a. Cancer Res. 77:6704–6716. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yang SZ, Xu F, Zhou T, Zhao X, McDonald JM
and Chen Y: The long non-coding RNA HOTAIR enhances pancreatic
cancer resistance to TNF-related apoptosis-inducing ligand. J Biol
Chem. 292:10390–10397. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Fu Z, Chen C, Zhou Q, Wang Y, Zhao Y, Zhao
X, Li W, Zheng S, Ye H, Wang L, et al: LncRNA HOTTIP modulates
cancer stem cell properties in human pancreatic cancer by
regulating HOXA9. Cancer Lett. 410:68–81. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu JH, Chen G, Dang YW, Li CJ and Luo DZ:
Expression and prognostic significance of lncRNA MALAT1 in
pancreatic cancer tissues. Asian Pac J Cancer Prev. 15:2971–2977.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li L, Chen H, Gao Y, Wang YW, Zhang GQ,
Pan SH, Ji L, Kong R, Wang G, Jiia YH, et al: Long noncoding RNA
MALAT1 promotes aggressive pancreatic cancer proliferation and
metastasis via the stimulation of autophagy. Mol Cancer Ther.
15:2232–2243. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhan HX, Wang Y, Li C, Xu JW, Zhou B, Zhu
JK, Han HF, Wang L, Wang YS and Hu SY: LincRNA-ROR promotes
invasion, metastasis and tumor growth in pancreatic cancer through
activating ZEB1 pathway. Cancer Lett. 374:261–271. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sun J, Cheng L, Shi H, Zhang Z, Zhao H,
Wang Z and Zhou M: A potential panel of six-long non-coding RNA
signature to improve survival prediction of diffuse large-B-cell
lymphoma. Sci Rep. 6:278422016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hu Y, Chen HY, Yu CY, Xu J, Wang JL, Qian
J, Zhang X and Fang JY: A long non-coding RNA signature to improve
prognosis prediction of colorectal cancer. Oncotarget. 5:2230–2242.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li J, Chen Z, Tian L, Zhou C, He MY, Gao
Y, Wang S, Zhou F, Shi S, Feng X, et al: LncRNA profile study
reveals a three-lncRNA signature associated with the survival of
patients with oesophageal squamous cell carcinoma. Gut.
63:1700–1710. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhou M, Xu W, Yue X, Zhao H, Wang Z, Shi
H, Cheng L and Sun J: Relapse-related long non-coding RNA signature
to improve prognosis prediction of lung adenocarcinoma. Oncotarget.
7:29720–29738. 2016.PubMed/NCBI
|
|
21
|
Law CW, Alhamdoosh M, Su S, Smyth GK and
Ritchie ME: RNA-seq analysis is easy as 1-2-3 with limma, Glimma
and edgeR. F1000Res. 5:14082016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cuccurullo V and Mansi L: AJCC cancer
staging handbook: From the AJCC cancer staging manual (7th
edition). Eur J Nucl Med Mol Imaging. 38:408. 2011. View Article : Google Scholar
|
|
23
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liu Z, Chen Z, Fan R, Jiang B, Chen X,
Chen Q, Nie F, Lu K and Sun M: Over-expressed long noncoding RNA
HOXA11-AS promotes cell cycle progression and metastasis in gastric
cancer. Mol Cancer. 16:822017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Dong HX, Wang R, Jin XY, Zeng J and Pan J:
LncRNA DGCR5 promotes lung adenocarcinoma (LUAD) progression via
inhibiting hsa-mir-22-3p. J Cell Physiol. 233:4126–4136. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Gong X and Huang M: Long non-coding RNA
MEG3 promotes the proliferation of glioma cells through targeting
Wnt/β-catenin signal pathway. Cancer Gene Ther. 24:381–385. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chen S, Wu DD, Sang XB, Wang LL, Zong ZH,
Sun KX, Liu BL and Zhao Y: The lncRNA HULC functions as an oncogene
by targeting ATG7 and ITGB1 in epithelial ovarian carcinoma. Cell
Death Dis. 8:e31182017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhang W, Yuan W, Song J, Wang S and Gu X:
LncRNA CPS1-IT1 suppresses EMT and metastasis of colorectal cancer
by inhibiting hypoxia-induced autophagy through inactivation of
HIF-1α. Biochimie. 144:21–27. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang Y, Liu Z, Yao B, Li Q, Wang L, Wang
C, Dou C, Xu M, Liu Q and Tu K: Long non-coding RNA CASC2
suppresses epithelial-mesenchymal transition of hepatocellular
carcinoma cells through CASC2/miR-367/FBXW7 axis. Mol Cancer.
16:1232017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang J, Li Z, Liu L, Wang Q, Li S, Chen
D, Hu Z, Yu T, Ding J, Li J, et al: Long noncoding RNA TSLNC8 is a
tumor suppressor that inactivates the interleukin-6/STAT3 signaling
pathway. Hepatology. 67:171–187. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yang W, Hong L, Xu X, Wang Q, Huang J and
Jiang L: LncRNA GAS5 suppresses the tumorigenesis of cervical
cancer by downregulating miR-196a and miR-205. Tumour Biol.
39:10104283177113152017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li Z, Shen J, Chan MT and Wu WK: TUG1: A
pivotal oncogenic long non-coding RNA of human cancers. Cell
Prolif. 49:471–475. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Liu L, Chen X, Zhang Y, Hu Y, Shen X and
Zhu W: Long non-coding RNA TUG1 promotes endometrial cancer
development via inhibiting miR-299 and miR-34a-5p. Oncotarget.
8:31386–31394. 2017.PubMed/NCBI
|
|
34
|
Lei H, Gao Y and Xu X: LncRNA TUG1
influences papillary thyroid cancer cell proliferation, migration
and EMT formation through targeting miR-145. Acta Biochim Biophys
Sin. 49:588–597. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kuang D, Zhang X, Hua S, Dong W and Li Z:
Long non-coding RNA TUG1 regulates ovarian cancer proliferation and
metastasis via affecting epithelial-mesenchymal transition. Exp Mol
Pathol. 101:267–273. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Sun J, Ding C, Yang Z, Liu T, Zhang X,
Zhao C and Wang J: The long non-coding RNA TUG1 indicates a poor
prognosis for colorectal cancer and promotes metastasis by
affecting epithelial-mesenchymal transition. J Transl Med.
14:422016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhang EB, Yin DD, Sun M, Kong R, Liu XH,
You LH, Han L, Xia R, Wang KM, Yang JS, et al: P53-regulated long
non-coding RNA TUG1 affects cell proliferation in human non-small
cell lung cancer, partly through epigenetically regulating HOXB7
expression. Cell Death Dis. 5:e12432014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Li J, Zhang M, An G and Ma Q: LncRNA TUG1
acts as a tumor suppressor in human glioma by promoting cell
apoptosis. Exp Biol Med. 241:644–649. 2016. View Article : Google Scholar
|
|
39
|
Durand X, Xylinas E, Radulescu C,
Haus-Cheymol R, Moutereau S, Ploussard G, Forgues A, Robert G,
Vacherot F, Loric S, et al: The value of urinary prostate cancer
gene 3 (PCA3) scores in predicting pathological features at radical
prostatectomy. BJU Int. 110:43–49. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Augustin H, Mayrhofer K, Pummer K and
Mannweiler S: Relationship between prostate cancer gene 3 (PCA3)
and characteristics of tumor aggressiveness. Prostate. 73:203–210.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Xie Z, Chen X, Li J, Guo Y, Li H, Pan X,
Jiang J, Liu H and Wu B: Salivary HOTAIR and PVT1 as novel
biomarkers for early pancreatic cancer. Oncotarget. 7:25408–25419.
2016.PubMed/NCBI
|
|
42
|
Peng W and Jiang A: Long noncoding RNA
CCDC26 as a potential predictor biomarker contributes to
tumorigenesis in pancreatic cancer. Biomed Pharmacother.
83:712–717. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Li L, Zhang GQ, Chen H, Zhao ZJ, Chen HZ,
Liu H, Wang G, Jia YH, Pan SH, Kong R, et al: Plasma and tumor
levels of Linc-pint are diagnostic and prognostic biomarkers for
pancreatic cancer. Oncotarget. 7:71773–71781. 2016.PubMed/NCBI
|
|
44
|
Trimarchi T, Bilal E, Ntziachristos P,
Fabbri G, Dalla-Favera R, Tsirigos A and Aifantis I: Genome-wide
mapping and characterization of Notch-regulated long noncoding RNAs
in acute leukemia. Cell. 158:593–606. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wang P, Xue Y, Han Y, Lin L, Wu C, Xu S,
Jiang Z, Xu J, Liu Q and Cao X: The STAT3-binding long noncoding
RNA lnc-DC controls human dendritic cell differentiation. Science.
344:310–313. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Ranzani V, Rossetti G, Panzeri I, Arrigoni
A, Bonnal RJ, Curti S, Gruarin P, Provasi E, Sugliano E, Marconi M,
et al: The long intergenic noncoding RNA landscape of human
lymphocytes highlights the regulation of T cell differentiation by
linc-MAF-4. Nat Immunol. 16:318–325. 2015. View Article : Google Scholar : PubMed/NCBI
|