Introduction
Cancer is the leading cause of death among children
aged 1–14 years in the United States, and neuroblastoma (NB)
accounts for 6% of those patients (1). NB is a common solid extracranial tumor
in children that is diagnosed as a metastatic disease, most often
metastasizing to the bone, bone marrow and lymph nodes (2,3) which
destroys the bodys immune system. Despite the latest methods of
chemotherapy and surgery, this type of tumor continues to make the
prognosis very dismal for children presenting with advanced stages
of the disease. Only 18–30% of patients with this type of cancer
survive (4). Even worse, the
mortality and recurrence rates of patients with high-risk (HR) NB
is >60% (5), which is a heavy
burden on both the patients and their families. An NB cell is a
poorly differentiated type of cancer cell; therefore, it contains
the functional and molecular characteristics of normal stem cells,
which enables it to evade growth suppressors (6) that would increase patient survival.
These characteristics provide NB with the ability to resist chemo-
and radiotherapy treatments (7). In
general, the overexpression of special cell-survival pathways and a
deficiency in normal cell senescence or apoptosis control
contribute to NB tumorigenesis and malignant transformation;
therefore, the breakdown or hindrance in the connection to
cell-survival pathways may decrease the potential for NB malignancy
and provide several avenues to explore new NB therapies.
The Ras/Raf/MEK/ERK cascade pathway has been
reported to play important roles in cancer cell growth and NB
survival (8–10). Active mutations and/or
overexpression of various components within this pathway (EGFR, Ras
and Raf) are frequently observed in NB (11,12),
especially in relapsing NB (13,14).
As such, researchers have focused on the Raf/MEK/ERK pathway in
intense studies to identify new target-based approaches for cancer
treatment to obtain more favorable results (15–17).
Unfortunately, the disease in a majority of NB patients who receive
treatment with Ras, Raf and MEK inhibitors becomes resistant after
~6–8 months and the drugs exhibit significantly reduced clinical
efficacy. In view of this, the ERK1/2 pathway may be one of the
best targets for NB treatment (17).
Previous studies have revealed that DCF1 plays a
vital role in various cell processes, including regulation of
neural stem-cell proliferation and differentiation (18,19),
glioma cell apoptosis (20) and
dendritic spine formation (21) and
maintaining energy balance (22).
Xie et al (23) demonstrated
that DCF1 was significantly alerted among different grades of
tumors and that DCF1 overexpression led to apoptosis by causing the
cell mitochondria to dysfunction.
In the present study, we confirmed that the DCF1
inhibited cell viability and motility, promoted the apoptosis of
Neuro-2a (N2a) and SK-N-SH cells, and identified the molecular
mechanism by which DCF1 exerts tumor-suppressive effects on NB
cells by inhibiting the ERK1/2 signaling pathway to promote
apoptosis.
Materials and methods
All experiments were approved and performed in
accordance with the guidelines and regulations of the Shanghai
University Ethics Committee (Shanghai, China).
Plasmid construction
Human dcf1 was amplified from the 293T
cellular cDNA library using polymerase chain reaction (PCR) with
KOD-Plus-Neo DNA polymerase (cat. no. KOD-401; Toyobo Shanghai
Biotech Co., Ltd., Shanghai, China) and cloned into the XhoI
and SmaI restriction sites of the pCAGGS-EGFP plasmid and
into the HindIII and XhoI restriction sites of the
pcDNA3.1-myc-his plasmid using following primers: sense,
5′-CGGAATTCATGGCGGCGCCGAAGGGGAG-3′ and antisense,
5′-CGGGATCCGTAAAATTTCAGAATGAGCA-3′. The small interfering RNA
plasmid of DCF1 (psi-DCF1) was preserved in our laboratory.
Cell culture
The N2a mouse brain neuroma cells were purchased
from the Cell Bank of the Type Culture Collection of the Chinese
Academy of Sciences (Shanghai, China). N2a cells were cultured
according to the methods used in a previous study (24). In brief, N2a cells were grown in an
incubator containing Minimum Essential Medium (MEM) (cat. no.
32561094; Thermo Fisher Scientific, Shanghai, China) supplemented
with GlutaMAX, 10% fetal bovine serum (FBS) (cat. no. 10100147;
Thermo Fisher Scientific) and 100 U/ml penicillin/streptomycin
(cat. no. 10378016; Thermo Fisher Scientific) in 5% CO2
at 37°C. SK-N-SH cells were purchased from FU Heng Biology (cat.
no. FH0164; FuHeng Cell Center, Shanghai, China). SK-N-SH cells
were grown in an incubator containing Dulbeccos modified Eagles
medium (DMEM) supplemented with 10% FBS and 100 U/ml
penicillin/streptomycin in 5% CO2 at 37°C. The culture
medium was changed every 1–2 days. For all experiments, the cells
were grown to ~90% confluence and subjected to no more than 20 cell
passages. The cells were harvested and seeded at a density of
2×105 cells/well in 12-well culture plates and used for
fluorescence correlation spectroscopy (FCS), western blotting, cell
migration assay, Matrigel invasion assay and immunofluorescence
after being transfected with plasmid pCAGGS-DCF1-EGFP and
pCAGGS-EGFP or pcDNA3.1 and pcDNA3.1-DCF1.
Cell transfection
Cells were plated on 12- or 24-well plates at a
density of 2×105 cells/well or 1×105
cells/well and cultured overnight at 37°C in an atmosphere of 5%
CO2 for DNA transfection of up to 70% confluence.
Lipofectamine 2000 (cat. no. 11668019; Thermo Fisher Scientific) or
calcium phosphate (cat. no. K278001; Thermo Fisher Scientific) was
used according to the manufacturers protocols to transfect l
µg/well plasmid into the cells.
Cell proliferation
Cell proliferation after DCF1 transfection was
assessed using the Cell Counting Kit-8 (CCK-8; product no. C0038;
Beyotime Institute of Biotechnology, Shanghai, China). Briefly, the
cells were seeded on 96-well plates for 18–20 h and transfected
with plasmid pcDNA3.1-myc-his or pcDNA3.1-DCF1-myc-his for the
appropriate time needed for proliferation. A mixture of 10 µl of
CCK-8 and 90 µl of serum-free DMEM culture medium solution was
added to each well at 12-h intervals. After being incubated with
CCK-8 solution for 1 h, the absorbance at 450 nm was measured using
a microplate reader (Bio-Rad Laboratories, Hercules, CA, USA). The
ability for the cells to proliferate was characterized by optical
absorption, and each experiment was conducted five times.
Wound healing assay
Cell wound healing assays were performed in 6-well
tissue culture plates. Cells at 30–40% confluence were transfected
with pcDNA3.1-myc-his or pcDNA3.1-DCF1-myc-his. After 24 h, cell
confluence reached ~75% and scratches in the wells were made using
1-ml pipette tips. The wells were then washed twice with full
medium to remove the suspended cells and allowed to grow for an
additional 48 h. Images were captured using a Nikon TI-S inverted
microscope (Nikon Corp., Tokyo, Japan) at 12-h intervals. The
migration gap distance was measured using Image-Pro Plus software
(Media Cybernetics, Rockville, MD, USA) and the % of wound closure
was calculated as follows:
[(At=0h-At=∆h)/At=0h] ×100%,
where, At=0h is the area of the wound measured
immediately after scratching, and At=∆h is
the area of the wound measured at 12, 24, 36 and 48 h after
scratching. When the samples were compared with the control cells,
the differences were considered significant if P<0.05, P<0.01
and P<0.001.
Cell invasion
The effect of DCF1 overexpression on the cells was
assessed using the BioCoat™ Matrigel™ Invasion Chamber (cat. no.
354480; BD Biosciences, San Jose, CA, USA). N2a and SK-N-SH cells
were transfected with pCAGGS-EGFP and pCAGGS-DCF1-EGFP for 48 h,
after which the cells were harvested and adjusted to a density of
5×104 cells/ml. After incubation in the invasion chamber
for 22 h in a humidified atmosphere of 5% CO2, the
non-invaded cells were scrubbed with a cotton swab, and the
remaining transfected cells were stained with 1% crystal violet and
observed under a Nikon Ti-S fluorescence microscope (Nikon Corp.)
at an ×100 magnification. Each experiment was performed in
triplicate.
Cell cycle detection using flow
cytometry
After transfection with pcDNA3.1-myc-his or
pcDNA3.1-DCF1-myc-his for 48 h, the cells were harvested using
trypsin [0.25% in phosphate-buffered solution (PBS) solution
without ethylenediaminetetraacetic acid (EDTA)]. Next, the cells
were washed twice with 2% bovine serum albumin (BSA)-PBS and
resuspended in 200 µl cold PBS, to which precooled 100% ethanol was
slowly added dropwise, and the solution was gently mixed to reach
70% ethanol. The mixture was fixed overnight at −20°C, after which
5 µl RNase (10 mg/ml) was added for 30 min at 37°C to digest the
RNA. To prepare for flow cytometry, the nuclei were stained in the
dark with 100 µg/ml propidium iodide (PI) for 30 min (Beckman
Coulter, Inc., Brea, CA, USA).
Cell apoptosis detection by flow
cytometry
After transfection with pcDNA3.1-myc-his or
pcDNA3.1-DCF1-myc-his for 48 h, the cells were harvested using
trypsin (0.25% in PBS solution without EDTA), washed twice with
2%-BSA-PBS, and resuspended in 100 µl binding buffer combined with
5 µl Annexin V-FITC and gently mixed. Next, 10 µl 7-AAD was added
to stain the cells according to the manufacturer's protocol for the
Annexin V Apoptosis Detection kit with 7-AAD (BioLegend, San Diego,
CA, USA). Samples were stained for 15 min at room temperature (RT),
after which 400 µl binding buffer was added to prepare the samples
for flow cytometry detection (Beckman Coulter, Inc.).
Cell tumorigenicity in nude mice
BALB/c athymic nude mice (Shanghai SLAC Laboratory
Animal Co., Ltd., Shanghai, China; 6–8 weeks of age, male) were
used, and each experimental group consisted of 4 mice (the control
group mice were infected subcutaneously with cells that were
transfected with pCAGGS-EGFP, and the experimental group mice were
infected subcutaneously with cells that were transfected with
pCAGGS-DCF1-EGFP). All animal procedures were carried out in
accordance with the guidelines and regulations of the Shanghai
University Ethics Committee. Briefly, N2A cells were transfected
with pCAGGS-EGFP (control) or pCAGGS-DCF1-EGFP for 48 h and
harvested using trypsin digestion. Single cell suspensions were
prepared, after which 2×106 cells were injected
subcutaneously into the right flanks of mice. The mice were raised
in the same environment with a controlled temperature (23±1°C) and
humidity (40–45%) on a 12-h light/dark cycle (08:00-20:00) with
enough food and water provided and were observed every week. The
tumor volume was calculated as the length × width2 ×
0.52 (25).
Immunofluorescence
Graft tumors removed from the nude mice were fixed
in precooled 4% paraformaldehyde in 0.1 M PBS (pH 7.4). After
fixing, the tumors were washed with PBS, dehydrated with 20 and 30%
sucrose, frozen in optimal cutting temperature (O.C.T.) compound
(Sakura Finetek US Inc., Torrance, CA, USA) and sliced into 20-µm
sections using a cryostat (Thermo Fisher Scientific, Inc., Waltham,
MA, USA). Sections were permeabilized with 0.1% Triton X-100 for 10
min and then washed three times with PBS. The slices were blocked
with 5% BSA in PBS for 60 min and incubated with primary antibodies
Tuj1 (dilution 1:500; cat. no. ab18207; Abcam Biotechnology,
Cambridge, UK) and Nestin (dilution 1:100; cat. no. sc-23927; Santa
Cruz Biotechnology, Inc., Santa Cruz, CA, USA) overnight at 4°C.
After washing three times with PBS, the slices were incubated with
secondary antibodies [cat. no. ZF-0513; Alexa Fluor®
594-conjugated goat anti-mouse IgG (H+L); cat. no. ZF-0511; Alexa
Fluor® 488-conjugated goat anti-mouse IgG (H+L); Beijing
Zhongshan Golden Bridge Biotechnology, Beijing, China] for 60 min
at RT. Finally, the nucleus was stained with
4,6-diamidino-2-phenylindole (DAPI) for 10 min, and fluorescence
was detected using the Zeiss LSM 710 confocal microscope (Carl
Zeiss, Oberkochen, Germany).
Western blotting
Protein lysates were boiled and subjected to
different densities (4–15%) of sodium dodecyl sulfate (SDS) then
transferred onto a nitrocellulose membrane. The membranes were
blocked with 5% BSA or non-fat milk for 1 h and incubated with the
following primary antibodies overnight at 4°C: Raf1 (dilution
1:1,000; cat. no. A0223; ABclonal, Wuhan, China), Ras (dilution
1:1,000; cat. no. A12212; ABclonal), Mek1/2 (dilution 1:1,000; cat.
no. A11122; ABclonal), p-Erk1/2 (dilution 1:1,000; cat. no. 9101;
Cell Signaling Technology, Danvers, MA, USA), Erk1/2 (dilution
1:1,000; cat. no. 137F5; Cell Signaling Technology), cell cyclin A2
(CCNA2; dilution 1:1,500; cat. no. A7632; ABclonal), cell cyclin
kinase 2 (CDK2; dilution 1:1,000; cat. no. A10810; ABclonal); Bax
(dilution 1:1,000; cat. no. sc-20067; Santa Cruz Biotechnology,
Inc.), BID (dilution 1:1,000; cat. no. sc-373939; Santa Cruz
Biotechnology, Inc.), Mcl-1 (dilution 1:1,000; cat. no. sc-53951;
Santa Cruz Biotechnology, Inc.), caspase-3 (dilution 1:1,000; cat.
no. A2156; ABclonal), survivin (dilution 1:800; cat. no. D221289;
Sangon Biotech Co., Ltd., Shanghai, China), Bcl-2 (dilution
1:1,000; cat. no. sc-56015; Santa Cruz Biotechnology, Inc.),
glyceraldehyde 3-phosphate dehydrogenase (GAPDH; dilution 1:1,000;
cat. no. sc-32233; Santa Cruz Biotechnology, Inc.) and α-tubulin
(dilution 1:5,000; cat. no. 2144; Cell Signaling Technology, Inc.).
The membranes were washed thrice for 5 min in a mixture of
Tris-buffered saline and Polysorbate 20 (TBST), incubated with KPL
Dylight 800 goat anti-rabbit immunoglobulin (Ig)G (dilution
1:1,000; cat. no. 5230-0412; KPL, Gaithersburg, MD, USA) or DyLight
700 goat anti-mouse immunoglobulin (Ig)G secondary antibodies
(dilution 1:1,000; cat. no. 072-06-18-06; KPL) for 1 h, and washed
again thrice for 5 min in TBST. Immunoblotting bands were observed
and quantified using the LI-COR Odyssey infrared imaging system
(simultaneous two-color targeted analysis) and software (LI-COR
Biosciences, Lincoln, NE, USA).
Statistical analyses
All data were expressed as the mean ± SEM.
Statistical analysis was performed with Independent Student's
t-test or one-way ANOVA was used to evaluate the statistical
comparison of two and multiple groups, respectively. Tukey's post
hoc test was used after the one-way ANOVA. Statistical significance
was considered for P<0.05, P<0.01, P<0.001,
P<0.0001.
Results
Overexpression of DCF1 inhibits the
viability and motility of N2a and SK-N-SH cells
To investigate the effect of DCF1 on NB cell growth,
N2a cells and SK-N-SH cells were transfected with pcDNA3.1-myc-his
or pcDNA3.1-DCF1-myc-his. The result of the CCK-8 assay revealed
that DCF1 significantly decreased the cell proliferation rate of
N2a (Fig. 1A) and SK-N-SH cells
(Fig. 1B). Our previous study
indicated that DCF1 inhibited the motility of a glioma cell line
(23). In the present study, we
conducted invasion and migration assays to detect whether DCF1
inhibited the motility of NB in vitro. We observed that the
overexpression of DCF1 could attenuate the invasion (Fig. 1C and D) and migration (Fig. 1E and F) abilities of N2a and SK-N-SH
cells compared with cells transfected with pcDNA3.1-myc-his.
Therefore, we concluded that DCF1 inhibited the viability and
motility of NB cells.
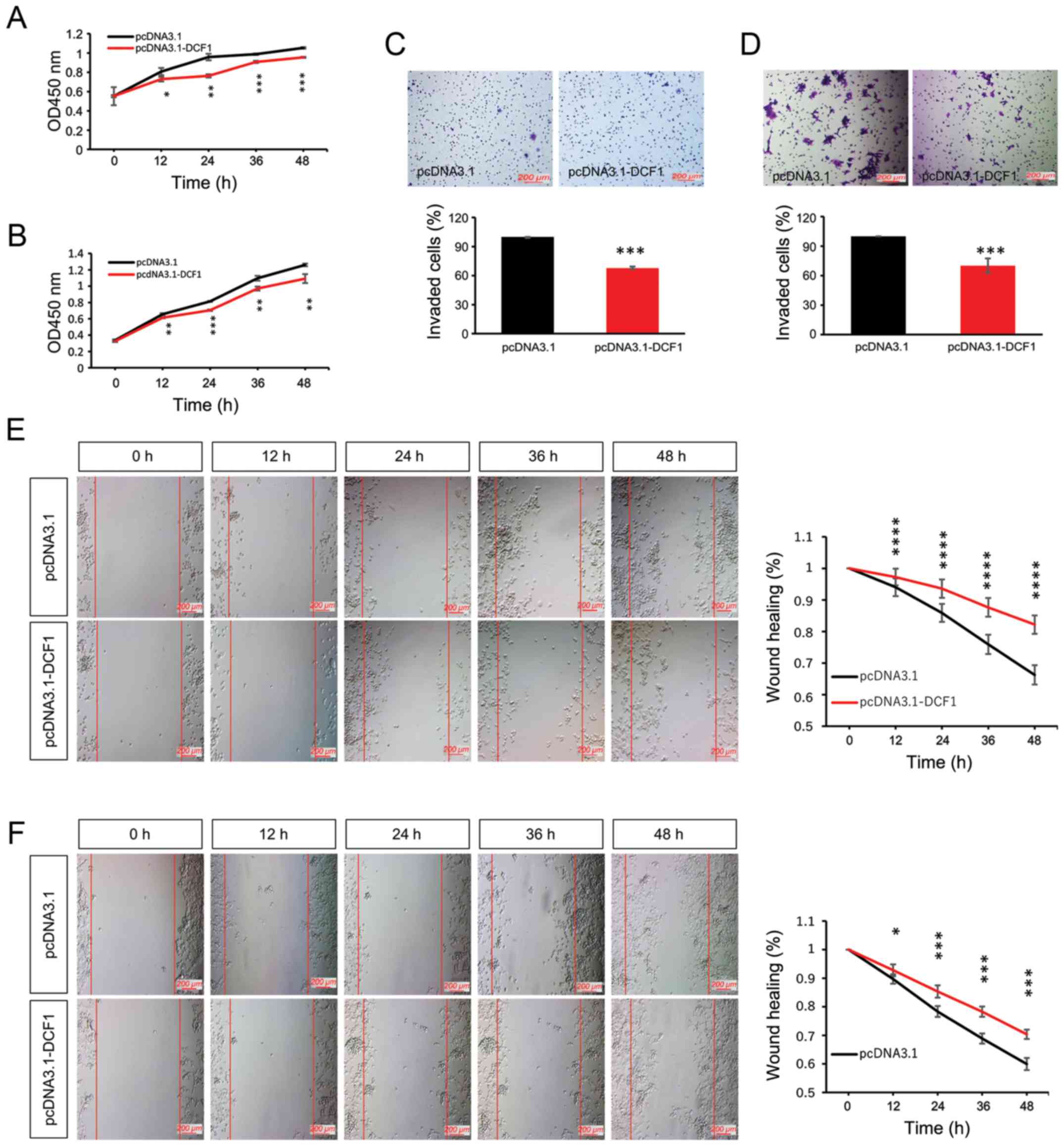 | Figure 1.Overexpression of DCF1 inhibits
proliferation and motility of NB cells in vitro. N2a and
SK-N-SH cells were transfected with pcDNA3.1 and pcDNA3.1-DCF1,
respectively. (A) A CCK-8 assay was performed to investigate the
effect of DCF1 on the proliferation of N2a cells at different
time-points, n=5. (B) A CCK-8 assay was performed to investigate
the effect of DCF1 on the proliferation of SK-N-SH cells at
different time-points, n=5. DCF1 inhibited the proliferation rate.
(C) A Transwell assay was used to evaluate the invasion of N2a
cells, n=4. (D) A Transwell assay was used to evaluate the invasion
of SK-N-SH cells, n=4. DCF1 decreased the percentage of invasive
cells. (E) The migration of N2a cells was evaluated by wound
healing assay, n=3, quadruplicate. (F) The migration of N2a cells
was evaluated by wound healing assay, n=3, quadruplicate. DCF1
significantly inhibited the migration rate of NB cells in
vitro. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001.
NB, neuroblastoma; DCF1, dendritic cell factor 1; N2a, Neuro-2a;
CCK-8, Cell Counting Kit-8. |
DCF1 has no effect on the cell cycle
and cell differentiation of N2a cells in vitro
Since NBs and their derived cell lines maintain the
potential to terminally differentiate, and since cell-cycle arrest
is tightly coupled to cell differentiation and affects the
viability of NB cells, we examined whether DCF1 induced
antiproliferative effects through cell-cycle arrest and cell
differentiation. To assess this hypothesis, N2a cells were
transfected with either pCAGGS-EGFP or pCAGGS-DCF1-EGFP and
subsequently processed for fluorescence-activated cell sorting and
immunoblotting analysis. The results of cytofluorimetric analysis
revealed that the cell cycle was not arrested with DCF1 (Fig. 2A). CCNA2, a cell-cycle protein,
controls both the G1/S and G2/M transition phases of the cell cycle
and functions by forming specific serine-threonine protein kinase
holoenzyme complexes with cyclin-dependent protein kinases, CDK1 or
CDK2 (26). CCNA2 was also
investigated using immunoblotting. No significant differences were
observed (Fig. 2B), which was
consistent with the results of the flow cytometric analysis. Next,
to examine whether DCF1 induced NB cell differentiation, we
monitored the levels of neuronal lineage markers SRY-box 2 (Sox2),
Nestin, glial fibrillary acidic protein (GFAP) and neuron-specific
Class III β-tubulin (Tuj1). The expression levels of these markers
were not altered after the cells were transfected with DCF1
(Fig. 2C). These results indicated
that DCF1 had no effect on the cell cycle or the differentiation of
N2a cells.
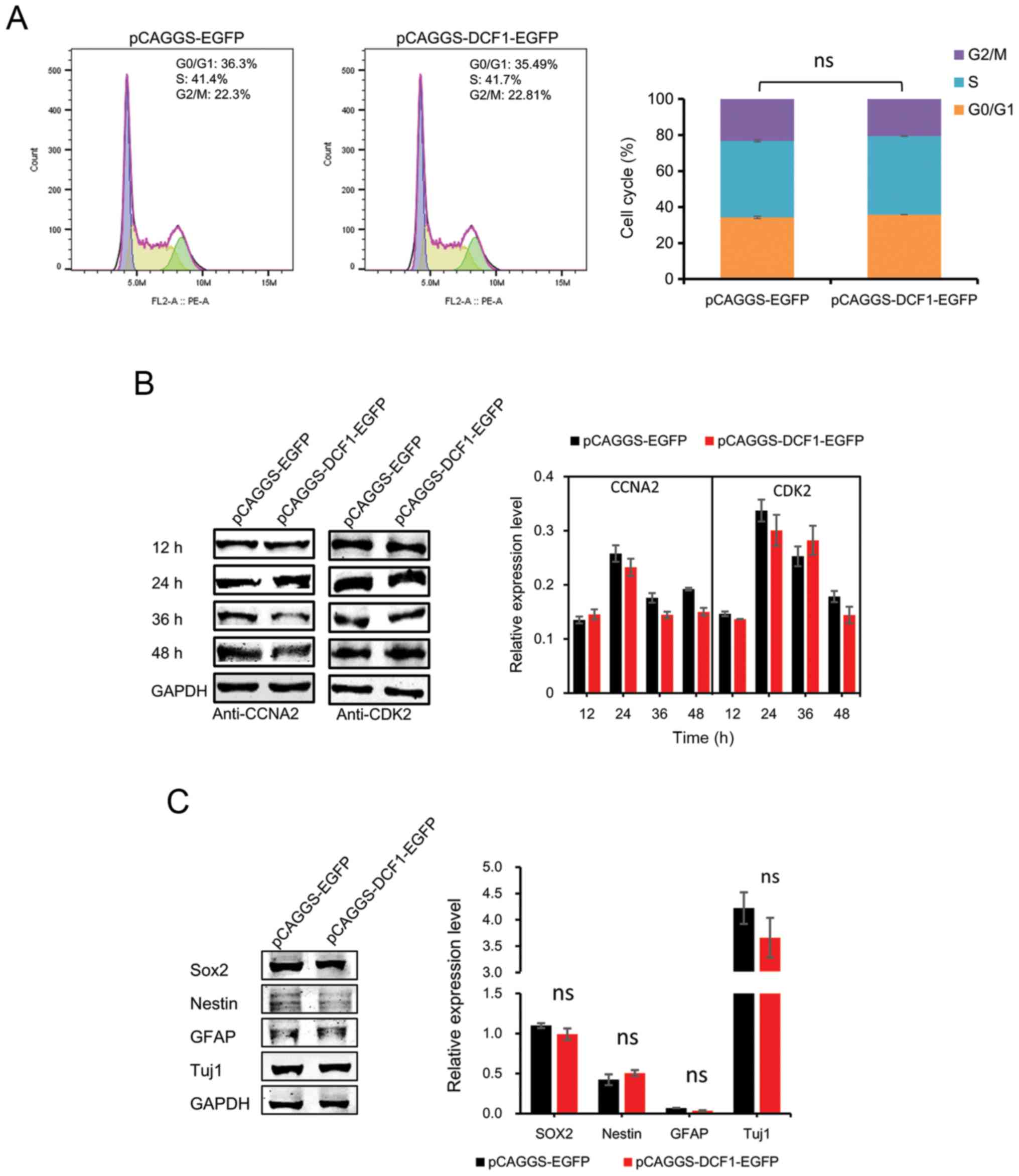 | Figure 2.Overexpression of DCF1 has no effect
on the cell cycle and cell differentiation of N2a cells in
vitro. N2a cells were transfected with pCAGGS-EGFP or
pCAGGS-DCF1-EGFP, respectively. (A) N2a cells were analyzed by flow
cytometry and quantified DNA distributions by ModFit program, n=6.
(B) Cell cycle regulators CCNA2 and CDK2 were examined by western
blotting and normalized to GAPDH, n=4. (C) Forty-eight hours after
transfection, western blot analysis of neuronal differentiation
markers, Sox2, Nestin, GFAP and Tuj1 in N2a cells was performed,
n=4. DCF1, dendritic cell factor 1; N2a, Neuro-2a; CCNA2, cell
cyclin A2; CDK2, cell cyclin kinase 2; Sox2, SRY-box 2; GFAP, glial
fibrillary acidic protein; Tuj1, neuron-specific Class III
β-tubulin. |
DCF1 promotes apoptosis of
neuroblastoma cells
In view of the fact that DCF1 had no effect on the
cell cycle or cell differentiation, we then examined apoptosis,
another factor that affects cell viability. After the cells were
transfected with pcDNA3.1-myc-his and pcDNA3.1-DCF1-myc-his,
respectively, for 48 h, apoptosis in neuroblastoma cells was
investigated and analyzed using fluorescence-activated cell
detection according to the manufacturers manual. The results of
flow cytometric analysis revealed that the average proportion of
apoptotic N2a cells containing pcDNA3.1-DCF1-myc-his (66.67±0.58%)
was significantly higher than those containing pcDNA3.1-myc-his
(24.79±1.90%) (Fig. 3A), and that
the average proportion of apoptotic SK-N-SH cells transfected with
pcDNA3.1-DCF1-myc-his (59.54±6.6%) was higher than those
transfected with pcDNA3.1-myc-his (30.09±3.6%) (Fig. 3B). These results indicated that DCF1
had a huge impact on the process of apoptosis in both N2a and
SK-N-SH cells. DAPI staining also revealed an increasing number of
N2a cells that experienced programmed cell death after being
transfected with DCF1. The cells with pCAGGS-DCF1-EGFP exhibited
more shrinkage in the nucleus than those with pCAGGS-EGFP (Fig. 3C).
 | Figure 3.DCF1 promotes NB cells into
apoptosis. (A) N2a cells were transfected with pcDNA3.1-myc-his or
pcDNA3.1-DCF1-myc-his, respectively, after transfection with dcf1
for 48 h. The effect of DCF1 on apoptosis of N2a was analyzed by
Annexin V-APC/7-AAD kit with flow cytometry (upper panel).
Quantification confirmed a significant increase in the percentage
of late apoptosis (P=0.000365), while no significant difference in
early apoptosis was observed compared with the control group, n=6
(lower panel). (B) SK-N-SH cells were transfected with
pcDNA3.1-myc-his or pcDNA3.1-DCF1-myc-his, respectively, after
transfection with DCF1 for 48 h. The effect of DCF1 on apoptosis of
N2a was analyzed by Annexin V-APC/7-AAD kit with flow cytometry
(upper panel). Quantification confirmed a significant increase in
the percentage of early (P=0.00162) and late apoptosis
(P=0.000352), n=3 (lower panel). (C) N2a cells transfected with
N2-DCF1-EGFP revealed shrinking morphology and condensed chromosome
in nuclei, indicated by white arrows. (D) Western blotting revealed
that pro-apoptotic protein (Bax) was upregulated, anti-apoptotic
proteins Bcl-2 and survivin were downregulated and apoptotic
executor caspase-3 was activated, n=4. (E) Western blotting
revealed that pro-apoptotic proteins (Bax and BID) were
upregulated, anti-apoptotic proteins Bcl-2, Mcl-1 and survivin were
downregulated and apoptotic executor caspase-3 was activated, n=4.
*P<0.05, **P<0.01, ***P<0.001, ****P<0.0001. DCF1,
dendritic cell factor 1; NB, neuroblastoma; N2a, Neuro-2a. |
To examine the molecular mechanism by which DCF1
induces apoptosis, we evaluated the changes in the expression of
apoptotic proteins, including Bax and BID, both pro-apoptotic
proteins and Bcl-2, Mcl-1 and survivin (27,28),
anti-apoptotic proteins that become prominently expressed in
transformed cell lines and in all of the most common human cancers,
such as those of the lung, colon, pancreas, prostate and breast
(28). Notably, survivin is a
direct inhibitor of caspase-3 and caspase-7 (29) and obstructs the process of
apoptosis. Western blot analysis revealed that overexpressed DCF1
led to enhancement of Bax, a decrease of anti-apoptotic proteins
Bcl-2 and survivin, and activated apoptotic executor caspase-3
(Fig. 3D). The pro-apoptotic
proteins Bax and BID were significantly increased; and the
anti-apoptotic proteins Bcl-2, Mcl-1 and survivin were
significantly decreased in SK-N-SH cells (Fig. 3E). We also observed that the
apoptosis executor caspase-3 was increased and activated with DCF1
in N2A cells (Fig. 3D) and SK-N-SH
cells (Fig. 3E). These data
revealed that DCF1 induced apoptosis through a caspase-dependent
pathway in neuroblastoma cells.
DCF1 inhibits the growth of NB cells
in vivo
The in vitro studies indicated that DCF1 may
act as a tumor suppressor in NB cells by promoting apoptosis. In
the present study, we investigated whether DCF1 could inhibit NB
tumorigenesis in vivo. N2a cells (2×106)
transfected with either pCAGGS-EGFP or pCAGGS-DCF1-EGFP were
subcutaneously injected into the right flank of nude mice (8 male
6-week old mice were used in conducting the cell tumorigenicity
experiment; all mice were randomly assigned into two groups, each
group consisted of 4 mice) to investigate the relationship between
DCF1 and tumorigenesis. As revealed in Fig. 4A, the volume of the tumor with DCF1
was smaller (0.65±0.08 cm3) than that from the negative
control (2.09±0.25 cm3).
Considering that DCF1 overexpression leads to the
maintenance of neural stem cells (NSCs) in an undifferentiated
state (18) and the potential for
NB cells to terminally differentiate, the expression levels of
Nestin and Tuj1 were assessed by immunofluorescence in the tissue
slices from the nude mice transplanted with N2a cells to examine
whether DCF1 induced differentiation in vivo. The results
revealed that there was no significant difference between Nestin
and Tuj1 in DCF1-expressing graft tumors and the control graft
tumor (Fig. 4B). Therefore, it was
clear that DCF1 inhibited the growth of tumors in vivo but
failed to induce differentiation in NB cells.
ERK1/2 signaling pathway is the target
of DCF1 in NB cells
To gain deeper insight into the mechanisms by which
DCF1 controls cell viability, motility and apoptosis in N2a cells
and SK-N-SH cells, we focused on the mitogen-activated protein
kinase (MAPK) cascade pathway since previous studies have indicated
that the activation or inhibition of the classical MAPK pathway (or
the ERK1/2 signaling pathway) is crucial for controlling tumor cell
proliferation, migration, invasion and survival (30–35).
We first evaluated the phosphorylation level of ERK1/2 using
western blotting. The result revealed that the phosphorylation
level of ERK was significantly decreased (Fig. 5A). Notably, downregulating DCF1 by
small interfering RNA increased the phosphorylation level of ERK1/2
(Fig. 5B), which suggested that
DCF1 inhibited the activation of the ERK1/2 signaling pathway to
regulate the apoptosis of NB cells. Subsequently, we detected the
upstream regulator of ERK1/2, including Ras, Raf1 and MEK1/2 using
immunoblotting. The results revealed that DCF1 significantly
decreased the protein expression levels of Ras, Raf1 and MEK1/2
(Fig. 5A), while downregulated
expression of DCF1 enhanced the expression of the proteins involved
in the ERK1/2 signaling pathway, which indicated the activation of
the ERK1/2 pathway (Fig. 5B). These
results demonstrated that DCF1 regulated the viability and motility
of N2a cells by inhibiting the ERK signaling pathway
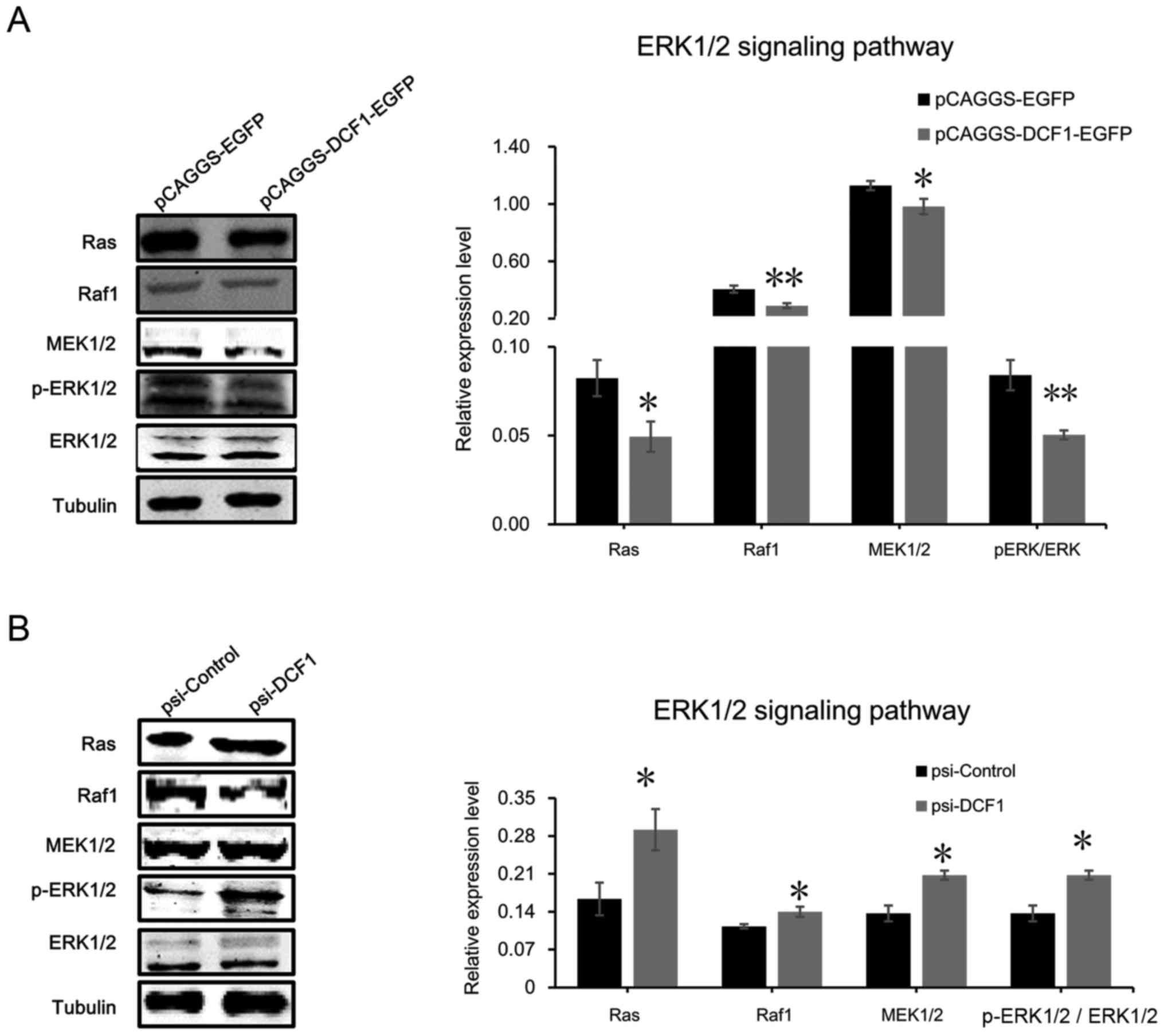 | Figure 5.DCF1 targets the ERK1/2 signaling
pathway in NB. (A) After transfection of DCF1 for 48 h, N2a cell
lysis was detected with ERK1/2 signaling pathway-associated
proteins, Ras, Raf1, MEK1/2, p-ERK1/2 and ERK1/2, by western
blotting (left panel) and quantification revealed that the
expression level of Ras, Raf1, MEK1/2, ERK1/2 and p-ERK1/2 was
significantly decreased (right panel), which indicated that
overexpressed DCF1 inhibited the ERK1/2 pathway, n=4. (B) After
transfection of psi-DCF1 to interfere with the expression of DCF1,
N2a cell lysis was detected with ERK1/2 signaling
pathway-associated proteins, Ras, Raf1, MEK1/2, p-ERK1/2 and
ERK1/2, by western blotting (left panel) and quantification
revealed that the expression level of Ras, Raf1, MEK1/2, ERK1/2 and
p-ERK1/2 was significantly increased (right panel), which indicated
that downregulated DCF1 activated the ERK1/2 pathway, n=4.
*P<0.05, **P<0.01. DCF1, dendritic cell factor 1; NB,
neuroblastoma; N2a, Neuro-2a. |
Discussion
NB is a lethal cancer of the brain, and despite
great endeavors to improve treatment methods and to understand the
molecular mechanisms underlying this disease, the results remain
far from our expectations; therefore, it is important and critical
to find a treatment method to eradicate or inhibit NB. In the
present study, for the first time, the critical role of DCF1 was
demonstrated with regard to the inhibition of proliferation,
motility, invasion and apoptosis of N2a and SK-N-SH cells. We also
examined the expression levels of pro-apoptotic proteins Bax and
BID, and anti-apoptotic proteins Bcl-2, Mcl-1, survivin and
caspase-3. As anticipated, Bcl-2, Mcl-1 and survivin were
downregulated, and Bax and BID were upregulated. Finally, DCF1
activated the apoptotic executor caspase-3 and suppressed the
growth, but not the differentiation, of tumorigenesis in
vivo. Although previous studies have shown that DCF1 can
inhibit tumors, the mechanism by which DCF1 functions in cancers
has not been fully investigated. In the present, we identified the
ERK signaling pathway as a target of DCF1. The Ras/Raf/MEK/ERK
cascade played a critical role in the regulation of cell-cycle
progression and cell survival, and the aberrant activation of genes
within the cascade was involved in the development and progression
of nearly one-third of all human cancers (36–39).
BRAF or RAS (KRAS, HRAS, or NRAS)
mutations could be detected in nearly all tumor cases. RAS
mutations are found in >50% of all pancreatic cancers, and
BRAF mutations, particularly those involved with the Valine
600 codon, are found in very high percentages in several cancers,
such as hairy cell leukemia, melanomas and Langerhans cell
histiocytosis (40). Considering
that these mutations exhibited wide distribution and played a
critical role in several malignancies, great efforts are being made
to develop drugs specifically targeting members of this pathway.
Unfortunately, although considerable clinical efficacy has emerged
by treating cancer patients with Ras, Raf and MEK inhibitors, the
acquired resistance from this therapy is one negative side effect.
Most cases of resistance were considered ERK dependent, which
implies that inhibiting the activity of Ras, Raf, MEK or ERK1/2
could still be an effective method by which to activate the
downstream substrate or target genes. Based on this, it has been
suggested that ERK1/2 may be an optimal target to conquer the
acquired resistance to Ras, Raf and MEK inhibitors (17); therefore, it is now the target in
anticancer therapy and it is the subject of more intense studies
(41). Our data revealed that DCF1
overexpression reduced the expression or inhibited the activity of
Ras, Raf1 and MEK to inhibit activation of the ERK1/2 pathway;
therefore, we propose that DCF1 should be regarded as a potential
target for treating NB.
DCF1 plays an important role in controlling tumor
viability, promoting tumor cell apoptosis, and inhibiting
tumorigenesis in vivo, and the data aforementioned
demonstrated that DCF1 could be a potential new inhibitor for
components of the ERK1/2 signaling pathway and helped our
understanding of the molecular mechanism of NB.
Acknowledgements
Not applicable.
Funding
The present study was funded by the National Natural
Science Foundation of China (nos. 81271253 and 81471162), the
Science and Technology Commission of Shanghai (no. 14JC1402400) and
the Key Innovation Project of Shanghai Municipal Education
Commission (grant no. 14ZZ090).
Availability of data and materials
All data used in this study are included in this
published article.
Authors contributions
TW conceived the study and revised the manuscript.
GL designed and conducted the experiments, prepared the figures and
wrote the manuscript. RF designed the experiments and analyzed the
data. YS cultured the Neuro-2A and SK-N-SH cancer cells and
conducted the transfection experiment. LZ fed the BALB/c athymic
nude mice and conducted tumor graft in nude mice. YW performed the
cellular tests and the plasmid construction; YC participated in the
IHC related experiments and in western blotting. All authors read
and approved the manuscript and agree to be accountable for all
aspects of the research in ensuring that the accuracy or integrity
of any part of the work are appropriately investigated and
resolved.
Ethics approval and consent to
participate
All animal procedures were carried out in accordance
with the guidelines and regulations of the Shanghai University
Ethics Committee.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
Statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Maris JM, Hogarty MD, Bagatell R and Cohn
SL: Neuroblastoma. Lancet. 369:2106–2120. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
DuBois SG, Kalika Y, Lukens JN, Brodeur
GM, Seeger RC, Atkinson JB, Haase GM, Black CT, Perez C, Shimada H,
et al: Metastatic sites in stage IV and IVS neuroblastoma correlate
with age, tumor biology, and survival. J Pediatr Hematol Oncol.
21:181–189. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cotterill SJ, Parker L, More L and Craft
AW: Neuroblastoma: Changing incidence and survival in young people
aged 0–24 years. A report from the North of England Young Persons'
Malignant Disease Registry. Med Pediatr Oncol. 36:231–234. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cohn SL, Pearson AD, London WB, Monclair
T, Ambros PF, Brodeur GM, Faldum A, Hero B, Iehara T, Machin D, et
al: ; INRG Task Force: The International Neuroblastoma Risk Group
(INRG) classification system: An INRG Task Force report. J Clin
Oncol. 27:289–297. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yang XH, Tang F, Shin J and Cunningham JM:
A c-Myc-regulated stem cell-like signature in high-risk
neuroblastoma: A systematic discovery (Target neuroblastoma
ESC-like signature). Sci Rep. 7:412017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
DeNardo BD, Holloway MP, Ji Q, Nguyen KT,
Cheng Y, Valentine MB, Salomon A and Altura RA: Quantitative
phosphoproteomic analysis identifies activation of the RET and
IGF-1R/IR signaling pathways in neuroblastoma. PLoS One.
8:e825132013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Martinez MA, Ubeda A, Cid MA and Trillo
MA: The proliferative response of NB69 human neuroblastoma cells to
a 50 Hz magnetic field is mediated by ERK1/2 signaling. Cell
Physiol Biochem. 29:675–686. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Vieira GC, Chockalingam S, Melegh Z,
Greenhough A, Malik S, Szemes M, Park JH, Kaidi A, Zhou L,
Catchpoole D, et al: LGR5 regulates pro-survival MEK/ERK and
proliferative Wnt/β-catenin signalling in neuroblastoma.
Oncotarget. 6:40053–40067. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Vujic I, Posch C, Sanlorenzo M, Yen AJ,
Tsumura A, Kwong A, Feichtenschlager V, Lai K, Arneson DV,
Rappersberger K, et al: Mutant NRASQ61 shares signaling
similarities across various cancer types-potential implications for
future therapies. Oncotarget. 5:7936–7944. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Pugh TJ, Morozova O, Attiyeh EF,
Asgharzadeh S, Wei JS, Auclair D, Carter SL, Cibulskis K, Hanna M,
Kiezun A, et al: The genetic landscape of high-risk neuroblastoma.
Nat Genet. 45:279–284. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Eleveld TF, Oldridge DA, Bernard V, Koster
J, Colmet Daage L, Diskin SJ, Schild L, Bentahar NB, Bellini A,
Chicard M, et al: Relapsed neuroblastomas show frequent RAS-MAPK
pathway mutations. Nat Genet. 47:864–871. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Schramm A, Köster J, Assenov Y, Althoff K,
Peifer M, Mahlow E, Odersky A, Beisser D, Ernst C, Henssen AG, et
al: Mutational dynamics between primary and relapse neuroblastomas.
Nat Genet. 47:872–877. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Roberts PJ and Der CJ: Targeting the
Raf-MEK-ERK mitogen-activated protein kinase cascade for the
treatment of cancer. Oncogene. 26:3291–3310. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Montagut C and Settleman J: Targeting the
RAF-MEK-ERK pathway in cancer therapy. Cancer Lett. 283:125–134.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yu Z, Ye S, Hu G, Lv M, Tu Z, Zhou K and
Li Q: The RAF-MEK-ERK pathway: Targeting ERK to overcome obstacles
to effective cancer therapy. Future Med Chem. 7:269–289. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang L, Wang J, Wu Y, Wu J, Pang S, Pan R
and Wen T: A novel function of dcf1 during the differentiation of
neural stem cells in vitro. Cell Mol Neurobiol. 28:887–894. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li X, Feng R, Huang C, Wang H, Wang J,
Zhang Z, Yan H and Wen T: MicroRNA-351 regulates TMEM 59 (DCF1)
expression and mediates neural stem cell morphogenesis. RNA Biol.
9:292–301. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xie Y, Li Q, Yang Q, Yang M, Zhang Z, Zhu
L, Yan H, Feng R, Zhang S, Huang C, et al: Overexpression of DCF1
inhibits glioma through destruction of mitochondria and activation
of apoptosis pathway. Sci Rep. 4:37022014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liu Q, Feng R, Chen Y, Luo G, Yan H, Chen
L, Lin R, Ding Y and Wen T: Dcf1 triggers dendritic spine formation
and facilitates memory acquisition. Mol Neurobiol. 55:763–775.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liu Q, Chen Y, Li Q, Wu L and Wen T: Dcf1
regulates neuropeptide expression and maintains energy balance.
Neurosci Lett. 650:1–7. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xie Y, Li Q, Yang Q, Yang M, Zhang Z, Zhu
L, Yan H, Feng R, Zhang S, Huang C, et al: Overexpression of DCF1
inhibits glioma through destruction of mitochondria and activation
of apoptosis pathway. Sci Rep. 4:pp. 37022014, https://doi.org/10.1038/srep03702
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kim MS, Yu JM, Kim HJ, Kim HB, Kim ST,
Jang SK, Choi YW, Lee DI and Joo SS: Ginsenoside Re and Rd enhance
the expression of cholinergic markers and neuronal differentiation
in Neuro-2a cells. Biol Pharm Bull. 37:826–833. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Weibel S, Hofmann E, Basse-Luesebrink TC,
Donat U, Seubert C, Adelfinger M, Gnamlin P, Kober C, Frentzen A,
Gentschev I, et al: Treatment of malignant effusion by oncolytic
virotherapy in an experimental subcutaneous xenograft model of lung
cancer. J Transl Med. 11:1062013.https://doi.org/10.1186/1479-5876-11-106
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Pagano M, Pepperkok R, Verde F, Ansorge W
and Draetta G: Cyclin A is required at two points in the human cell
cycle. EMBO J. 11:961–971. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chipuk JE, Fisher JC, Dillon CP, Kriwacki
RW, Kuwana T and Green DR: Mechanism of apoptosis induction by
inhibition of the anti-apoptotic BCL-2 proteins. Proc Natl Acad Sci
USA. 105:20327–20332. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ambrosini G, Adida C and Altieri DC: A
novel anti-apoptosis gene, survivin, expressed in cancer and
lymphoma. Nat Med. 3:917–921. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Shin S, Sung BJ, Cho YS, Kim HJ, Ha NC,
Hwang JI, Chung CW, Jung YK and Oh BH: An anti-apoptotic protein
human survivin is a direct inhibitor of caspase-3 and-7.
Biochemistry. 40:1117–1123. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lavoie JN, L'Allemain G, Brunet A, Müller
R and Pouysségur J: Cyclin D1 expression is regulated positively by
the p42/p44MAPK and negatively by the p38/HOGMAPK pathway. J Biol
Chem. 271:20608–20616. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Caunt CJ, Sale MJ, Smith PD and Cook SJ:
ME K1 and MEK2 inhibitors and cancer therapy: The long and winding
road. Nat Rev Cancer. 15:577–592. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Liu Y, Zhang X, Yang B, Zhuang H, Guo H,
Wei W, Li Y, Chen R, Li Y and Zhang N: Demethylation-induced
overexpression of Shc3 drives c-Raf-independent activation of
MEK/ERK in HCC. Cancer Res. 78:2219–2232. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yang C, Zheng W and Du W: CXCR3A
contributes to the invasion and metastasis of gastric cancer cells.
Oncol Rep. 36:1686–1692. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hou H, Ge C, Sun H, Li H, Li J and Tian H:
Tunicamycin inhibits cell proliferation and migration in
hepatocellular carcinoma through suppression of CD44s and the
ERK1/2 pathway. Cancer Sci. 109:1088–1100. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhao H, Liu B and Li J: LIM and SH3
protein 1 knockdown suppresses proliferation and metastasis of
colorectal carcinoma cells via inhibition of the mitogen-activated
protein kinase signaling pathway. Oncol Lett. 15:6839–6844.
2018.PubMed/NCBI
|
|
36
|
Ferreira-Silva GA, Lages CC, Sartorelli P,
Hasegawa FR, Soares MG and Ionta M: Casearin D inhibits ERK
phosphorylation and induces downregulation of cyclin D1 in HepG2
cells. Toxicol In Vitro. 38:27–32. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Xu X, Wang Q, He Y, Ding L, Zhong F, Ou Y,
Shen Y, Liu H and He S: ADP-ribosylation factor 1 (ARF1) takes part
in cell proliferation and cell adhesion-mediated drug resistance
(CAM-DR). Ann Hematol. 96:847–858. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yan Y, Wang L, He J, Liu P, Lv X and Zhang
Y, Xu X, Zhang L and Zhang Y: Synergy with interferon-lambda 3 and
sorafenib suppresses hepatocellular carcinoma proliferation. Biomed
Pharmacother. 88:395–402. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Uehling DE and Harris PA: Recent progress
on MAP kinase pathway inhibitors. Bioorg Med Chem Lett.
25:4047–4056. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wu PK and Park JI: MEK1/2 Inhibitors:
Molecular activity and resistance mechanisms. Semin Oncol.
42:849–862. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Serini S and Calviello G: Modulation of
Ras/ERK and phosphoinositide signaling by long-chain n-3 PUFA in
breast cancer and their potential complementary role in combination
with targeted drugs. Nutrients. 9:1852017.doi:10.3390/nu9030185.
View Article : Google Scholar :
|



















