Introduction
Breast cancer metastasis suppressor 1 (BRMS1) was
first discovered for its significant inhibition of metastasis in
breast cancer cells (1). Depending
on the cell types used, BRMS1 has been noted to inhibit multiple
steps in the invasion-metastasis cascade, including cell
communication (2,3), cell migration (4,5), cell
apoptosis (6,7), epithelial-mesenchymal transition (EMT)
(8), among others. Several
important BRMS1-interacting proteins have been identified,
providing possible clues to the molecular mechanisms of action of
BRMS1. For instance, BRMS1 participates in histone modification and
transcriptional regulation through interaction with the mSin3·HDAC
complex (9). BRMS1 also interacts
with the RelA/p65 subunit of NF-κB and promotes binding of HDAC1 to
RelA/p65, which suppresses the transcriptional activity of NF-κB
(6). More recently, BRMS1 has been
found to be posttranslationally regulated by CK2α via protein
interaction, which affects the nuclear exportation and degradation
of BRMS1 (10). Structural mapping
reveals that two coiled-coil motifs and the internal linker region
may be important for the different protein interactions of BRMS1
(9,11,12).
Fanconi anemia (FA) is a rare autosomal or X-linked
recessive inherited disease first described by Dr Guido Fanconi in
1927 (13). Although FA patients
mainly suffer from bone marrow failure, many of them also display
profound genome instability correlating with cancer predisposition
(14). A higher risk of head and
neck squamous cell carcinoma, leukemia, vulvar carcinoma, breast
and ovarian cancer has been described in different FA patients
(15). On the molecular level, the
FA pathway plays a role in resolving DNA damage, especially
interstrand crosslinks (ICLs) which covalently link the double
strands of the DNA. Removal of ICLs is particularly important for
cellular development, as ICLs strongly affect molecular processes
which require DNA unwinding and strand separation such as DNA
replication as well as transcription (16).
To date, 21 FA genes including FANCI have been
identified. FANCI was first characterized in 2007 by Smogorzewska
as a paralog of another FA gene, FANCD2. In response to DNA damage,
FANCI binds to FANCD2 to form a heterodimeric FANCI-FANCD2
(FANCI/D2) complex. The FANCI/D2 complex is then monoubiquitinated,
and downstream DNA repair proteins are further recruited to ICL
sites (17). Afterwards, ICLs are
removed so that genome stability can be guarded and cells can
survive from DNA damage. As an essential component of the FANCI/D2
complex, FANCI is not only required for the stability of FANCD2
(18), but is also required for
efficient FA core complex foci formation (19).
In the present study, we revealed an interactive
relationship between FANCI and BRMS1 through co-immunoprecipitation
assay for the first time. The association relationship prompted us
to ascertain whether BRMS1 has a role in the regulation of cell
sensitivity to DNA damage. Our results showed that depletion of
BRMS1 significantly diminished the monoubiquitination of
FANCI and FANCD2, leading to a reduced FANCD2 foci formation and
cell viability in response to ICL damage.
Materials and methods
Cell culture and transfection
293T and U2OS cells were purchased from the Cell
Bank of the Chinese Academy of Sciences (Shanghai, China). All
cells were cultured in Dulbecco's modified Eagle's medium (DMEM)
supplemented with 10% fetal bovine serum (FBS) in a 5%
CO2-humidified atmosphere at 37°C. All the cell culture
reagents were purchased from Gibco/Thermo Fisher Scientific, Inc.
(Waltham, MA, USA). Cells at 70% confluency were transfected with
Invitrogen™ Lipofectamine 3000 (Thermo Fisher Scientific, Inc.)
according to the manufacturer's instructions. In experiments
evaluating cell sensitivity to ICL damage, mitomycin C (MMC; Roche
Diagnostics, Indianapolis, IN, USA) was added to the cell culture
medium at the indicated dosages.
Immunoprecipitation
Cells were harvested with cell lysis buffer (Thermo
Fisher Scientific, Inc.) supplemented with protease inhibitor
cocktail (Thermo Fisher Scientific, Inc.) and the lysate was
centrifuged at 12,000 × g at 4°C for 15 min. The supernatant was
precleared with protein A/G beads (Thermo Fisher Scientific, Inc.)
and incubated with 1 µg specific primary antibodies at 4°C
overnight. The related antibodies included anti-Myc (cat. no.
05–724; Millipore, Bedford, MA, USA), anti-Flag (cat. no. F3165;
Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) and anti-FANCI (cat.
no. ab15344; Abcam, Cambridge, MA, USA). Afterwards, protein A/G
beads were added into the mixture and incubated at 4°C for at least
2 h. After washing four times, the beads were resuspended in
loading buffer and stored at −20°C before being subjected to
western blot analysis. Related recombinant plasmids used in the
co-immunoprecipitation include Myc-BRMS1, BRMS1 deletion mutants as
previously described (20) and
Flag-FANCI (a kind gift from Professor Jun Huang of Zhejiang
University, China).
Western blot analysis
Protein samples were collected with SDS lysis
buffer. Protein samples (15–30 µg) were separated by SDS-PAGE with
6 or 12% gel depending on specific experiment and then transferred
into PVDF membranes. After blocking in 5% fat-free milk for 1 h,
the membranes were incubated with specific primary antibodies at
4°C overnight. Afterwards, the membranes were washed and incubated
with secondary antibodies at room temperature for 1 h. Membranes
were visualized by enhanced chemiluminescence (ECL) kit (GE
Healthcare Life Sciences, Logan, UT, USA). The images are
representatives of several independent experiments with consistent
results and the densitometric values were quantified with Gene
Tools from Syngene software (Frederick, MD, USA). The related
antibodies included anti-Myc (1:3,000 dilution; cat. no. 05-724;
Millipore), anti-Flag (1:3,000 dilution; Sigma-Aldrich), anti-FANCI
(1:2,000 dilution; cat. no. ab15344; Abcam), anti-BRMS1 (1:3,000
dilution; cat. no. 16096-1-AP; Proteintech Group, Wuhan, China),
anti-FANCD2 (1:2,000 dilution; cat. no. ab2187; Abcam) and
peroxidase-conjugated goat anti-mouse (cat. no. IH-0031; DingGuo
Bio., Beijing, China)/rabbit (cat. no. IH-0011; DingGuo Bio.) IgG
diluted at 1:3,000 with 1% fat-free milk.
Quantitative real-time PCR
(qRT-PCR)
Total RNA was extracted from cultured cells using
Invitrogen™ Trizol (Thermo Fisher Scientific, Inc.) and 500 ng RNA
was applied for reverse transcription using reverse transcriptase
(Takara Biotechnology Co., Ltd., Dalian, China). Quantitative
real-time PCR analysis was performed using SYBR-Green Supermix kit
(Takara Biotechnology Co., Ltd.) with the CFX Connection detection
system (Bio-Rad Laboratories, Inc., Hercules, CA, USA). Reactions
with no cDNA template were performed as negative controls to rule
out contamination. Primers for BRMS1 and internal control
were previously described (7). Data
were analyzed as previously described (21).
Plasmid construction and selection of
stable transfectants
Recombinant pLenti-BRMS1-sgRNA plasmid was
previously described (20). U2OS
cells were transfected with pLenti-BRMS1-sgRNA or empty
vector before being subjected to selection with puromycin.
BRMS1-deficient clones and the control clones were selected
through anti-BRMS1 immunoblotting. Genomic DNA of these clones was
isolated to confirm BRMS1 mutation via Sanger
sequencing.
Immunofluorescence
U2OS cells were cultured in a 24-well plate with
coverslips inside and further treated with 1 µΜ MMC for 24 h. Cells
grown on coverslips were then washed, fixed with 4%
paraformaldehyde for 15 min and then permeabilized with 0.2% Triton
for 15 min. Cells were blocked in 5% BSA supplemented with 5%
donkey serum for 1 h, and then incubated with primary antibodies in
a wet container at 4°C overnight, followed by incubation with the
fluorescence-conjugated secondary antibodies at 37°C for 45 min.
Finally, cells were counterstained with Hoechst 33258
(Sigma-Aldrich) at 37°C for 20 min. The fluorescence images were
captured using an Axio Observer Z1 microscope (Carl Zeiss,
Oberkochen, Germany). Antibodies used in the immunofluorescence
included anti-FANCD2 (1:600 dilution; cat. no. ab2187; Abcam),
anti-Myc (1:200 dilution; cat. no. 05-724; Millipore), Alexa Fluor
488 goat anti-mouse IgG (1:500 dilution; cat. no. A-21202; Thermo
Fisher Scientific, Inc.) and Alexa 555 goat anti-rabbit IgG (1:300
dilution; cat. no. A-21430; Thermo Fisher Scientific, Inc.).
MMC sensitivity assay
Cells were seeded in 96-well plates at a density of
1,200 cells/well. After treatment with different concentrations of
MMC for 3 days, cell viability was calculated with MTS assay
(Promega, Madison, WI, USA) according to the manufacturer's
instructions. OD values were scanned by a microplate reader (BioTek
Instruments, Inc., Winooski, VT, USA).
Statistical analysis
Comparisons of quantitative data were analyzed by
Student's t-test. We considered two groups with a P-value <0.05
to be different, and with a P-value <0.01 to be significantly
different (labeled with * and **, respectively, in the
figures).
Results
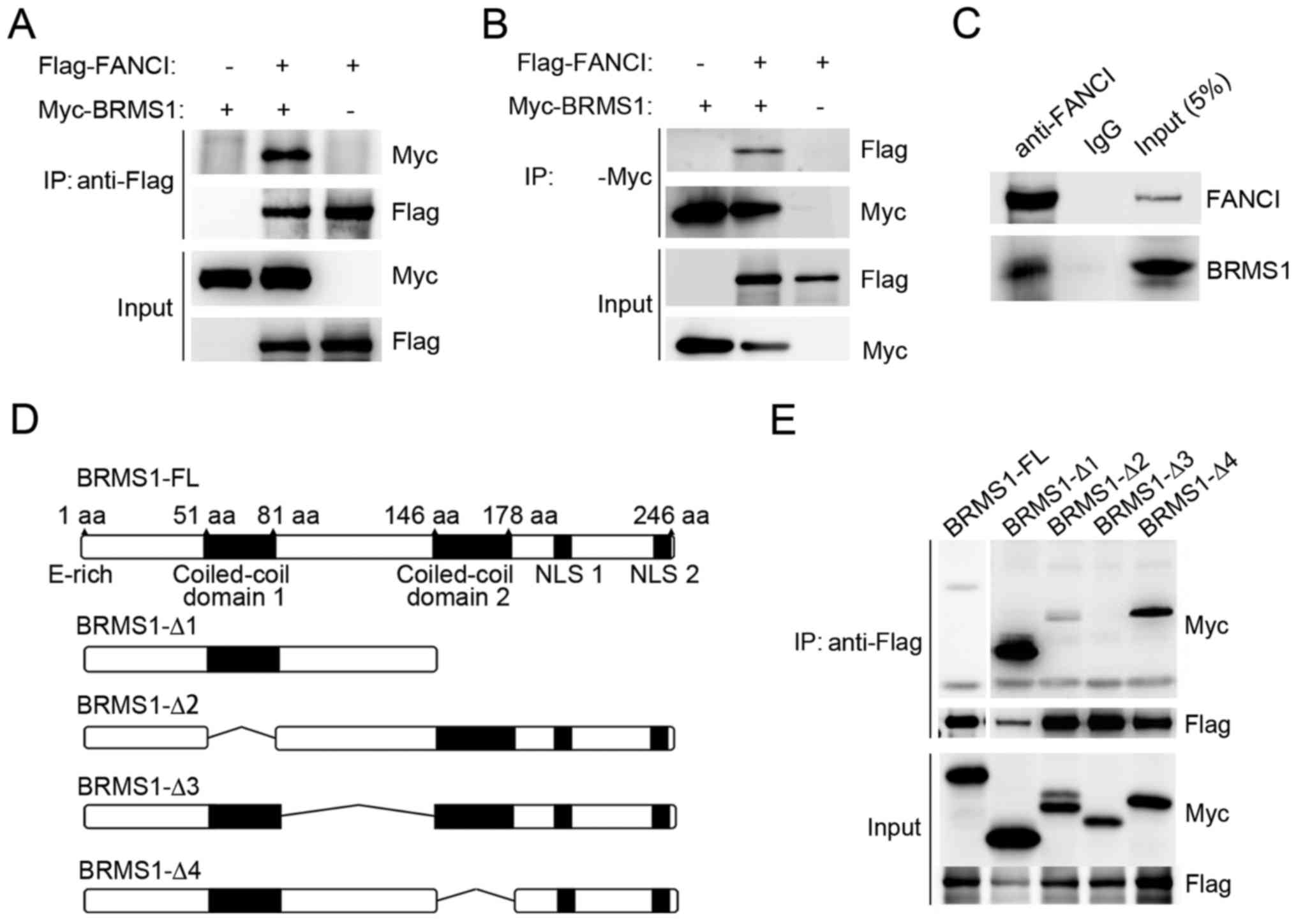
BRMS1 interacts with FANCI through its
linker region
Previously, we reported that BRMS1 is able to
interact with DBC1 through a large-scale tandem affinity
purification (20). In the present
study, another potential BRMS1-interacting protein, FANCI, was
confirmed by co-immunoprecipitation experiment. As shown in
Fig. 1A and B, when Myc-tagged
BRMS1 and Flag-tagged FANCI were co-expressed in 293T cells,
Flag-FANCI was detected in anti-Myc immunoprecipitates and vice
versa. No binding was detected in the control cells. In addition,
U2OS cells were utilized to detect the association between
endogenous FANCI and BRMS1. As shown in Fig. 1C, BRMS1 was readily
immunoprecipitated with anti-FANCI antibody, but not with the IgG
control. These data strongly suggest that BRMS1 is able to interact
with FANCI.
To identify the region of BRMS1 which is responsible
for BRMS1-FANCI interaction, a series of deletion mutants of BRMS1
were utilized as previously described (Fig. 1D) (20). As shown in Fig. 1E, among all the mutants, only
BRMS1-Δ3, which lost the linker region between two coiled-coil
motifs (residues 81–146) abolished the binding ability of BRMS1
with FANCI. By contrast, loss of either coiled-coil region
(BRMS1-Δ2, BRMS1-Δ4) or the C-terminal domain (BRMS1-Δ1) had no
effect on BRMS1-FANCI interaction.
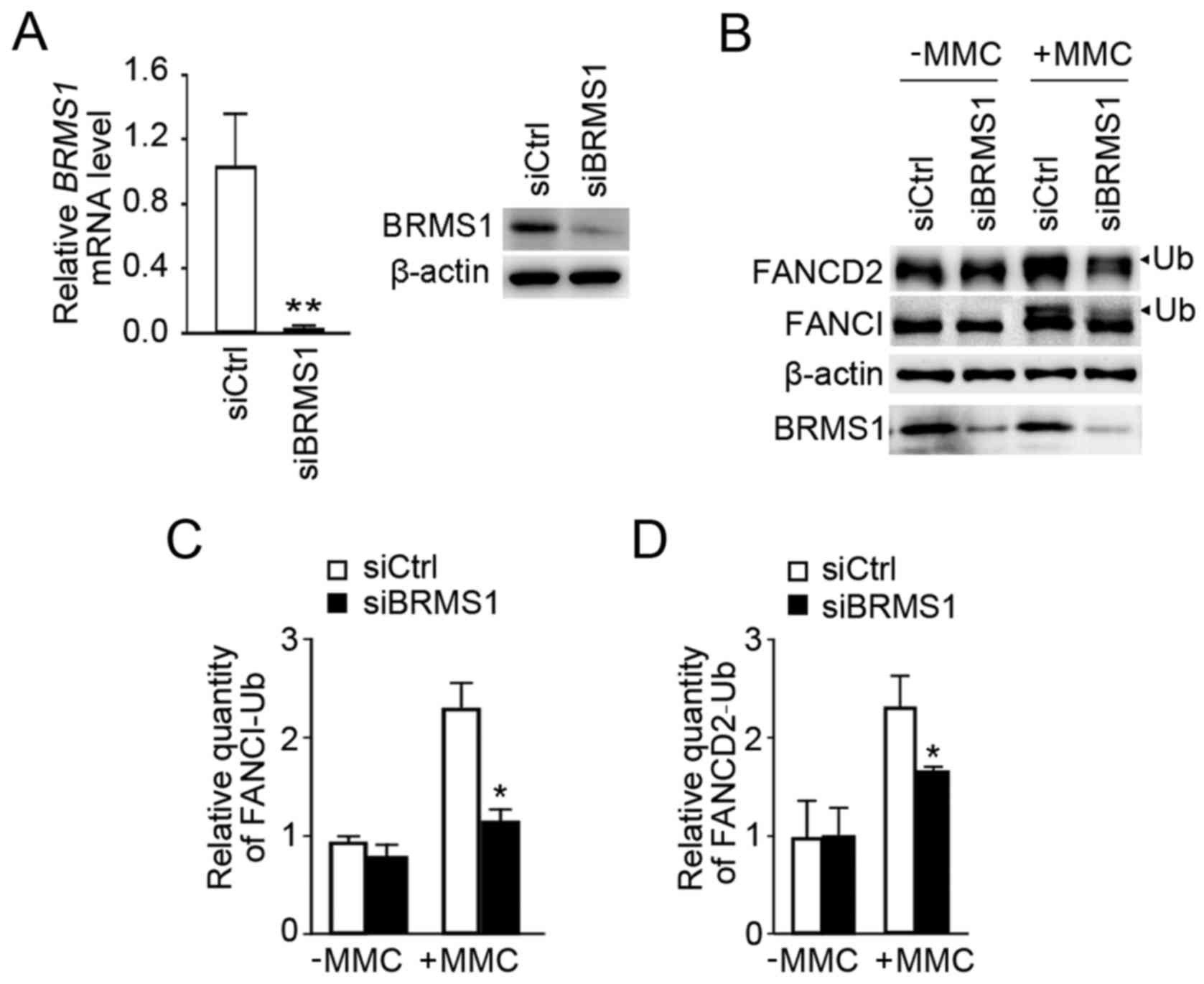
Knockdown of BRMS1 decreases the
monoubiquitination of FANCI/D2 upon DNA damage
An effective siRNA targeting BRMS1 was
utilized as previously described (7), and it successfully suppressed
endogenous BRMS1 expression in U2OS cells (siBRMS1) by comparison
with U2OS control cells (siCtrl) at both the mRNA and protein level
(Fig. 2A). MMC is widely used for
inducing DNA ICLs. As shown in Fig.
2B, although both siBRMS1 and siCtrl cells exhibited
monoubiquitiniation of FANCI/FANCD2 with MMC treatment, knockdown
of BRMS1 led to a significant reduction in FANCI/D2
monoubiquitination levels. Statistical analysis revealed that the
relative quantity of monoubiquitinated FANCI and FANCD2 declined by
52 and 28% separately (Fig. 2C and
D). These data initially indicated a potential role of BRMS1 in
regulating FANCI/D2 monoubiquitination in the response to
MMC-induced ICLs.
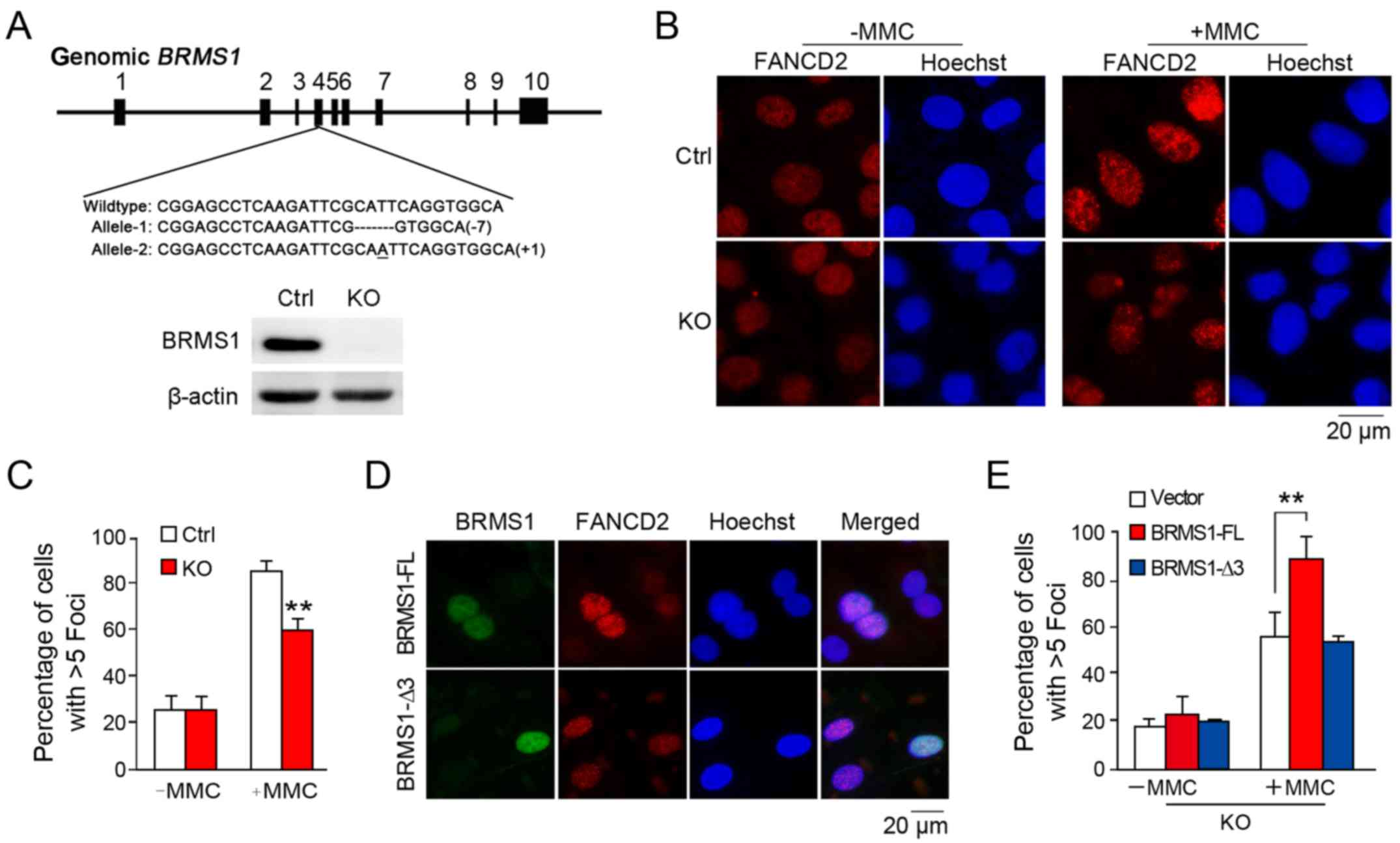
BRMS1 is involved in FANCD2 foci
formation upon DNA damage
When FANCI/D2 are monoubiquitinated, they gather to
the damage sites in the nucleus and further recruit downstream
exonucleases to repair DNA, leading to FANCD2 foci formation. To
answer whether BRMS1 is also involved in FANCD2 localization upon
DNA damage, we further generated a BRMS1-deficient cell
clone in the U2OS cell line through CRISPR/Cas9 method as
previously described (20). As
shown in Fig. 3A, two frame-shift
mutations of BRMS1 alleles were introduced into the
BRMS1-knockout U2OS clone (KO), leading to complete
depletion of BRMS1. As shown in Fig.
3B, while control cells exhibited markedly increased FANCD2
foci (85.3±3.9%) with MMC treatment, BRMS1-deficient cells
displayed hypersensitivity to DNA damage, leading to relatively
defective FANCD2 foci formation (59.3±4.7%) (Fig. 3C). This result is consistent with
that from the FANCI/D2 monoubiquitination analysis, since
modification of FANCI/D2 is pivotal to FANCD2 localization.
To further confirm our finding, a rescue assay was
designed. BRMS1-deficient cells were separately
reconstituted with full-length BRMS1 (BRMS1-FL) and the BRMS1-Δ3
mutant without FANCI-interacting ability before being subjected to
FANCD2 immunofluorescence staining. As shown in Fig. 3D, FANCD2 signals in the nuclear foci
were increased in cells with exogenous BRMS1 expression instead of
BRMS1-Δ3 expression. Statistically, only the full-length BRMS1, but
not the BRMS1-Δ3 mutant was able to rescue the diminished FANCD2
foci induced by BRMS1 depletion (Fig.
3E). These findings strongly suggest that BRMS1-FANCI
interaction may be essential for the regulatory effect of BRMS1 on
FANCD2 localization.
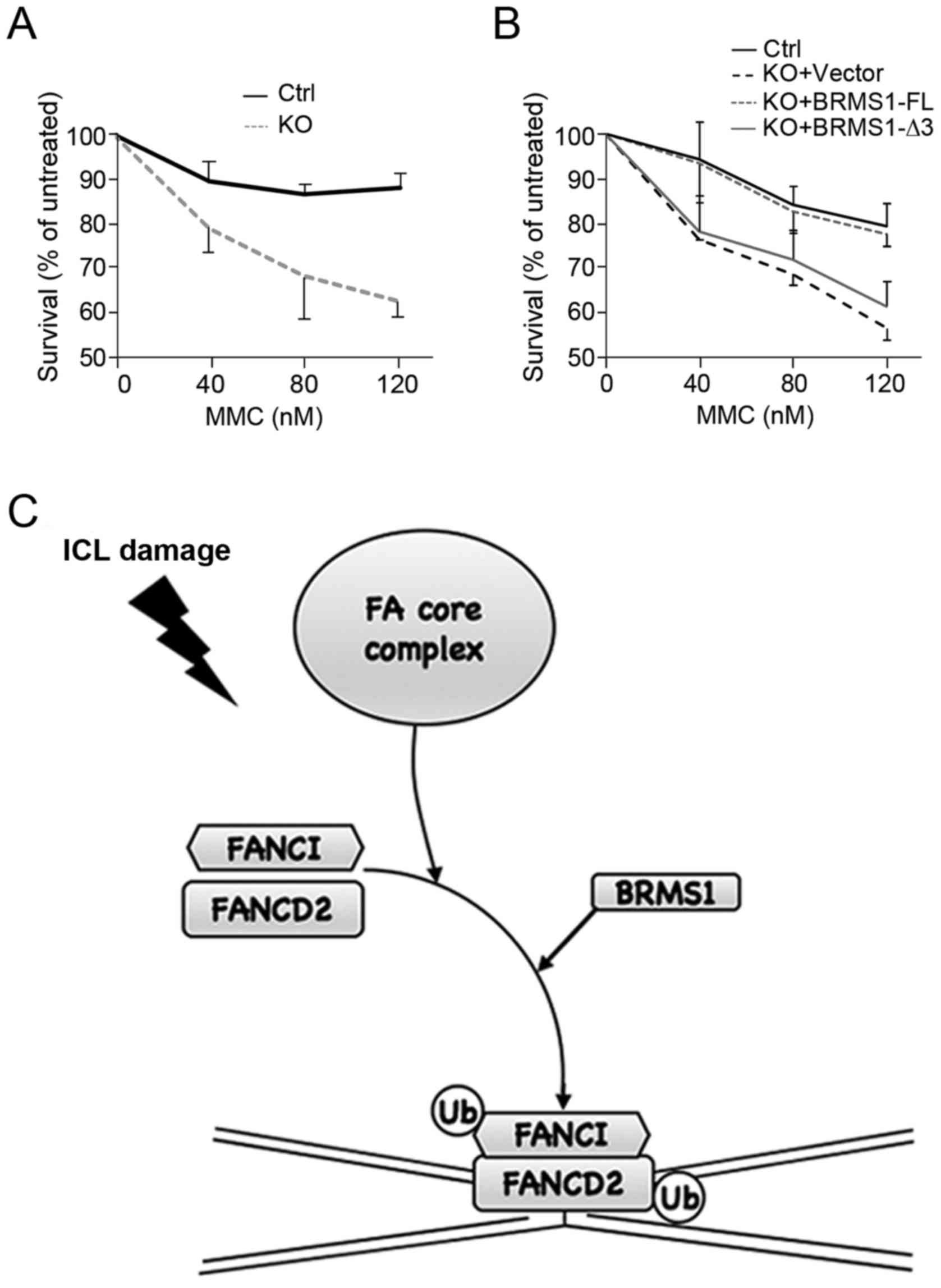
BRMS1 contributes to regulation of
cell sensitivity to MMC
Based on findings above, we further investigated
cell viability upon MMC treatment. As shown in Fig. 4A, U2OS control cells were resistant
to high concentrations of MMC, displaying relatively high
viability. By contrast, BRMS1-deficient cells (KO) were much
more sensitive to MMC in a dose-dependent manner, showing a more
than 20% decrease in the survival ratio with MMC treatment (120
nM). Moreover, cells reconstituted with BRMS1-FL, instead of
BRMS1-Δ3, displayed obvious recovery of cell viability (Fig. 4B). The reduced cell viability in
BRMS1-deficient cells not only corresponded to the decrease
in FANCD2 foci, but also provided another piece of evidence that
BRMS1-FANCI interaction is able to affect the downstream DNA repair
process of FA pathway.
Discussion
In the present study, FANCI was identified as a
novel protein associated with BRMS1 by tandem affinity purification
and co-immunoprecipitation. The linker region between two
coiled-coil motifs of BRMS1 may be responsible for the interaction,
and this domain was also reported to be the binding domain of BRMS1
with other proteins such as p300 and DBC1 (11,22).
Further functional studies revealed that depletion of BRMS1 led to
decreased FANCI/D2 monoubiquitination, FANCD2 foci formation and
cell viability with MMC treatment. Rescue experiments additionally
demonstrated that BRMS1-FANCI interaction is necessary for the
effect of BRMS1 on the Fanconi anemia (FA) pathway (Fig. 4C). Since proteins reported to
interact with BRMS1 are mostly involved in cellular signal
transduction and gene expression regulation, our findings bring new
insight into the potential function of BRMS1 in genome
maintenance.
In the context of the FA pathway, some other
proteins have been reported to affect monoubiquitination of the
FANCI/D2 complex through protein-protein interaction. For example,
RAD18 interacts with FANCD2 and regulates chromatin loading of the
FANCI/D2 complex. Depletion of RAD18 reduced the
monoubiquitination of FANCI/D2 and finally led to a delay in FANCD2
foci formation together with hypersensitivity of ICL damage
(23). In addition, UBL5 could
directly interact with FANCI and stabilize FANCI via modulating
pre-mRNA splicing of FANCI (24). UBL5 was also important for FANCI/D2
complex formation and monoubiquitination. In our study, while BRMS1
was found to regulate the monoubiquitination of FANCI/D2, slight
reduction in the FANCI/D2 protein level was also observed in
BRMS1-knockdown cells (Fig.
2B). Additional qRT-PCR was carried out and no obvious
difference was shown in FANCI/D2 mRNA levels after
interference of BRMS1 expression (data not shown). It has
been previously shown that FANCI is required for FANCD2 stability,
but not vice versa (18,25,26).
Moreover, E3 ligase function of BRMS1 induces polyubiquitination of
p300 and further proteasome-mediated protein degradation (11), providing another piece of evidence
that BRMS1 may affect protein modification and stability of its
interacting partners. Whether BRMS1-FANCI interaction may influence
the stability of the FANCI/D2 complex remains to be addressed in
our future work.
Genome maintenance systems can ensure genome
stability via detecting and resolving DNA damages, replication
errors, among others. Many tumor-suppressor genes are involved in
DNA damage response pathway, since their mutations can facilitate
cancer cells to accumulate additional mutations required for
transformation. Some of them also contribute to tumor metastasis
suppression, such as RAD9, PARP1, BRCA1/2, ATM, TP53, NM23,
among others. NM23 is the first identified tumor metastasis
suppressor gene (27). The
3′-5′exonuclease activity of NM23 in the DNA repair pathway was
demonstrated to be essential for metastasis suppression (28). NM23, instead of its mutant without
exonuclease activity, could inhibit invasive capacities of 1205LU
melanoma cells in vitro and suppress spontaneous metastasis
in vivo. The potential relationship between the FA pathway
and tumor metastasis can also be observed in the well-known tumor
suppressors, BRCA1/BRCA2, which are also called FANCS/FANCD1 in the
FA pathway (29). They both act in
the downstream of the FA pathway. BRCA2 interacts with RAD51 to
control its localization and assembly in the DNA damage site, while
BRCA1 interacts with the MRE11-RAD50-NBS1 complex implicated in
homologous recombination (30,31).
Mutations in BRCA1/BRCA2 could decrease the efficiency of the FA
pathway and induce genomic instability (27). Moreover, a recent study revealed
that the FA/BRCA pathway plays an important role in
chemoradiotherapy failure and distant metastasis of cervical cancer
(32). In our study, we raised the
hypothesis that BRMS1 may be another functional regulator of the FA
pathway which is also deeply involved in tumor metastasis. BRMS1
exhibits a strong metastatic suppressive effect in many types of
cancers by affecting different steps of the metastatic cascade.
Whether BRMS1-FANCI interaction also contributes to the metastatic
suppressive role of BRMS1 warrants further investigation.
Acknowledgements
We thank Professor Jun Huang (Zhejiang University,
China) for the kind gift of the pcDNA-FANCI.
Funding
The present study was supported by the National Key
Research and Development Program of China (2017YFC1001101).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YW, JD, YZ and SQ conceived and designed the
experiments. JD and XL performed the co-immunoprecipitation and the
western blot analysis. JD and YZ performed RNAi, stable
transfectants, immunofluorescence staining and cell viability
assay. YW, JD, YZ, XQ, XY and WX participated in the western blot
and data analysis. YW, YZ, JD and SQ wrote the paper. All authors
read, edited and approved the manuscript and agree to be
accountable for all aspects of the research in ensuring that the
accuracy or integrity of any part of the work are appropriately
investigated and resolved.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Phillips KK, Welch DR, Miele ME, Lee JH,
Wei LL and Weissman BE: Suppression of MDA-MB-435 breast carcinoma
cell metastasis following the introduction of human chromosome 11.
Cancer Res. 56:1222–1227. 1996.PubMed/NCBI
|
|
2
|
Bodenstine TM, Vaidya KS, Ismail A, Beck
BH, Cook LM, Diers AR, Landar A and Welch DR: Homotypic gap
junctional communication associated with metastasis suppression
increases with PKA activity and is unaffected by PI3K inhibition.
Cancer Res. 70:10002–10011. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Shevde LA, Samant RS, Goldberg SF,
Sikaneta T, Alessandrini A, Donahue HJ, Mauger DT and Welch DR:
Suppression of human melanoma metastasis by the metastasis
suppressor gene, BRMS1. Exp Cell Res. 273:229–239. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Roesley SN, Suryadinata R, Morrish E, Tan
AR, Issa SM, Oakhill JS, Bernard O, Welch DR and Šarčević B:
Cyclin-dependent kinase-mediated phosphorylation of breast cancer
metastasis suppressor 1 (BRMS1) affects cell migration. Cell Cycle.
15:137–151. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yang YL, Chen CZ, Jin LP, Ji QQ, Chen YZ,
Li Q, Zhang XH and Qu JM: Effect and mechanism of the metastasis
suppressor gene BRMS1 on the migration of breast cancer cells. Int
J Clin Exp Med. 6:908–916. 2013.PubMed/NCBI
|
|
6
|
Liu Y, Smith PW and Jones DR: Breast
cancer metastasis suppressor 1 functions as a corepressor by
enhancing histone deacetylase 1-mediated deacetylation of RelA/p65
and promoting apoptosis. Mol Cell Biol. 26:8683–8696. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wu Y, Jiang W, Wang Y, Wu J, Saiyin H,
Qiao X, Mei X, Guo B, Fang X, Zhang L, et al: Breast cancer
metastasis suppressor 1 regulates hepatocellular carcinoma cell
apoptosis via suppressing osteopontin expression. PLoS One.
7:e429762012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liu Y, Mayo MW, Xiao A, Hall EH, Amin EB,
Kadota K, Adusumilli PS and Jones DR: Loss of BRMS1 promotes a
mesenchymal phenotype through NF-κB-dependent regulation of Twist1.
Mol Cell Biol. 35:303–317. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Meehan WJ, Samant RS, Hopper JE, Carrozza
MJ, Shevde LA, Workman JL, Eckert KA, Verderame MF and Welch DR:
Breast cancer metastasis suppressor 1 (BRMS1) forms complexes with
retinoblastoma-binding protein 1 (RBP1) and the mSin3 histone
deacetylase complex and represses transcription. J Biol Chem.
279:1562–1569. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu Y, Amin EB, Mayo MW, Chudgar NP,
Bucciarelli PR, Kadota K, Adusumilli PS and Jones DR: CK2alpha'
drives lung cancer metastasis by targeting BRMS1 nuclear export and
degradation. Cancer Res. 76:2675–2686. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu Y, Mayo MW, Nagji AS, Hall EH, Shock
LS, Xiao A, Stelow EB and Jones DR: BRMS1 suppresses lung cancer
metastases through an E3 ligase function on histone
acetyltransferase p300. Cancer Res. 73:1308–1317. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Spinola-Amilibia M, Rivera J,
Ortiz-Lombardia M, Romero A, Neira JL and Bravo J: The structure of
BRMS1 nuclear export signal and SNX6 interacting region reveals a
hexamer formed by antiparallel coiled coils. J Mol Biol.
411:1114–1127. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lobitz S and Velleuer E: Guido fanconi
(1892–1979): A jack of all trades. Nat Rev Cancer. 6:893–898. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Auerbach AD: Fanconi anemia and its
diagnosis. Mutat Res. 668:4–10. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Stoepker C, Ameziane N, van der Lelij P,
Kooi IE, Oostra AB, Rooimans MA, van Mil SE, Brink A, Dietrich R,
Balk JA, et al: Defects in the Fanconi anemia pathway and chromatid
cohesion in head and neck cancer. Cancer Res. 75:3543–3553. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Scharer OD: DNA interstrand crosslinks:
Natural and drug-induced DNA adducts that induce unique cellular
responses. Chembiochem. 6:27–32. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ceccaldi R, Sarangi P and D'Andrea AD: The
Fanconi anaemia pathway: New players and new functions. Nat Rev Mol
Cell Biol. 17:337–349. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sims AE, Spiteri E, Sims RJ III, Arita AG,
Lach FP, Landers T, Wurm M, Freund M, Neveling K, Hanenberg H, et
al: FANCI is a second monoubiquitinated member of the Fanconi
anemia pathway. Nat Struct Mol Biol. 14:564–567. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Castella M, Jacquemont C, Thompson EL, Yeo
JE, Cheung RS, Huang JW, Sobeck A, Hendrickson EA and Taniguchi T:
FANCI regulates recruitment of the FA core complex at sites of DNA
damage independently of FANCD2. PLoS Genet. 11:e10055632015.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liu X, Ehmed E, Li B, Dou J, Qiao X, Jiang
W, Yang X, Qiao S and Wu Y: Breast cancer metastasis suppressor 1
modulates SIRT1-dependent p53 deacetylation through interacting
with DBC1. Am J Cancer Res. 6:1441–1449. 2016.PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCT method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shimamura A and Alter BP: Pathophysiology
and management of inherited bone marrow failure syndromes. Blood
Rev. 24:101–122. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Williams SA, Longerich S, Sung P, Vaziri C
and Kupfer GM: The E3 ubiquitin ligase RAD18 regulates
ubiquitylation and chromatin loading of FANCD2 and FANCI. Blood.
117:5078–5087. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Oka Y, Bekker-Jensen S and Mailand N:
Ubiquitin-like protein UBL5 promotes the functional integrity of
the Fanconi anemia pathway. EMBO J. 34:1385–1398. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Dorsman JC, Levitus M, Rockx D, Rooimans
MA, Oostra AB, Haitjema A, Bakker ST, Steltenpool J, Schuler D,
Mohan S, et al: Identification of the Fanconi anemia
complementation group I gene, FANCI. Cell Oncol. 29:211–218.
2007.PubMed/NCBI
|
|
26
|
Smogorzewska A, Matsuoka S, Vinciguerra P,
McDonald ER III, Hurov KE, Luo J, Ballif BA, Gygi SP, Hofmann K,
D'Andrea AD, et al: Identification of the FANCI protein, a
monoubiquitinated FANCD2 paralog required for DNA repair. Cell.
129:289–301. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tutt A, Gabriel A, Bertwistle D, Connor F,
Paterson H, Peacock J, Ross G and Ashworth A: Absence of Brca2
causes genome instability by chromosome breakage and loss
associated with centrosome amplification. Curr Biol. 9:1107–1110.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhang Q, McCorkle JR, Novak M, Yang M and
Kaetzel DM: Metastasis suppressor function of NM23-H1 requires its
3′-5′exonuclease activity. Int J Cancer. 128:40–50. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Castro E, Goh C, Olmos D, Saunders E,
Leongamornlert D, Tymrakiewicz M, Mahmud N, Dadaev T, Govindasami
K, Guy M, et al: Germline BRCA mutations are associated with higher
risk of nodal involvement, distant metastasis, and poor survival
outcomes in prostate cancer. J Clin Oncol. 31:1748–1757. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yu DS, Sonoda E, Takeda S, Huang CL,
Pellegrini L, Blundell TL and Venkitaraman AR: Dynamic control of
Rad51 recombinase by self-association and interaction with BRCA2.
Mol Cell. 12:1029–1041. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ree AH, Bratland A, Nome RV, Stokke T and
Fodstad Ø: Repression of mRNA for the PLK cell cycle gene after DNA
damage requires BRCA1. Oncogene. 22:8952–8955. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Balacescu O, Balacescu L, Tudoran O, Todor
N, Rus M, Buiga R, Susman S, Fetica B, Pop L, Maja L, et al: Gene
expression profiling reveals activation of the FA/BRCA pathway in
advanced squamous cervical cancer with intrinsic resistance and
therapy failure. BMC Cancer. 14:2462014. View Article : Google Scholar : PubMed/NCBI
|