Introduction
Lymphoma is a malignant tumor that occurs in the
lymphoid and hematologic systems, such as lymph nodes, spleen,
thymus and extranodal lymphatic tissues and organs. Lymphomas are
associated with immune cell malignant transformation, and thus are
categorized as immune system malignant tumors (1,2). Among
the immune system malignant tumors, non-Hodgkin lymphoma (NHL) is
the most commonly diagnosed type of lymphoma (3). Diffuse large B cell lymphoma (DLBCL)
is the most prevalent type of B cell-derived non-Hodgkin lymphoma
(B-NHL). DLBCL is also the most common NHL in adults (4,5). DLBCL
is an invasive lymphoma with a low rate of cure and poor
prognosis.
Increased activation of the extracellular signal
regulated kinase (ERK)/mitogen activated protein kinase (MAPK)
signal transduction pathway is closely related to the occurrence,
development and metastasis of multiple cancers. MAPK kinase 1
(MEK1) is a double-specific protein kinase which can act upstream
of ERK protein and phosphorylate the tyrosine/threonine (Tyr/Thr)
residues of ERK protein, thereby activating the ERK/MAPK signaling
pathway. A number of studies have shown that an abnormal increase
in MEK1 expression or functional activity is related to the
occurrence, progression, drug resistance and poor prognosis of
numerous types of tumors, such as pancreatic, ovarian and lung
cancers (6–8). Moreover, it has been shown that the
expression or function enhancement of MEK1 plays an important role
in lymphomas (9). MicroRNAs
(miRNAs) are a type of small molecule non-coding RNAs with a length
of 22–25 nucleotides that were initially discovered in eukaryotes.
miRNAs can be adjusted to match the target gene mRNA 3′-end
untranslated region (3′-UTR) with complete or incomplete
complementation, and can regulate the expression of >30% of
human target genes by degradation or inhibition of target gene mRNA
translation. Depending on the targeting-specific genes, miRNAs are
involved in the pathogenesis of tumors acting as oncogenes or
tumor-suppressor genes (10–13).
Previous studies have shown that the expression of miR-101 in
colorectal (14), esophageal
(15) and ovarian cancers (16) is abnormally reduced, suggesting that
miR-101 may play a role as a tumor-suppressor gene in
tumorigenesis. It has also been reported that the abnormal decrease
in miR-101 expression is associated with the incidence of
lymphomas, suggesting a possible tumor-suppressor effect (17,18).
Bioinformatic analysis showed a complementary binding site between
miR-101 and the 3′-UTR of MEK1 mRNA. Therefore, in the present
study we determined the role of miR-101 in regulating the
expression of MEK1, the activity of the ERK/MAPK pathway, and
lymphoma cell proliferation and apoptosis.
Materials and methods
Main reagents and materials
Normal human B lymphoblastoid cell lines (HCC1954 BL
and NCI-BL2009) were purchased from the American Type Culture
Collection (ATCC; Manassas, VA, USA). 293T cells were purchased
from Hunan Fenghui Biotechnology Co., Ltd. (Hunan, China). Human
DLBCL cell lines (SU-DHL-4 and Farage) were purchased from ATCC.
Gibco™ RPMI-1640 medium, fetal bovine serum (FBS), penicillin and
streptomycin were purchased from Thermo Fisher Scientific, Inc.
(Waltham, MA, USA). The RNA extraction reagent, TRIzol, and the
transfection reagent, Lipofectamine 2000, were purchased from
Thermo Fisher Scientific, Inc. (Invitrogen™; Waltham, MA, USA). The
PrimeScript RT reagent kit and SYBR Fast qPCR Mix were purchased
from Takara (Dalian, China). Rabbit anti-human MEK1 (cat. no.
12671), p-MEK1 (cat. no. 98195) and Bcl-2 (cat. no. 4223)
antibodies were purchased from Cell Signaling Technology, Inc.
(Beverly, MA, USA). Mouse anti-human ERK1/2 (cat. no. ab36991),
p-ERK1/2 (cat. no. ab126423) and PCNA antibodies (cat. no. ab70472)
were purchased from Abcam (Cambridge, MA, USA). The Annexin V/PI
apoptosis detection kit and Cell Cycle detection kit were purchased
from Biyuntian (Jiangsu, China). siRNA-MEK1 and siRNA-NC were
purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). A
cell proliferation detection kit (Click-iT® EdU Alexa
Fluor 488 Flow Cytometry Assay kit) was obtained from Molecular
Probes (Eugene, OR, USA). The Dual-Glo® Luciferase Assay
System and pMIR luciferase reporter gene plasmid were provided by
Promega (Madison, WI, USA).
Clinical data
A total of 72 patients diagnosed with DLBCL who
received treatment at our hospital between September 2011 and April
2015 and had complete follow-up data were enrolled, including 38
males and 34 females (age range, 19–77 years; median age, 58 years;
and average age, 50.7 years). Patients who did not undergo previous
radiotherapy and chemotherapy were included, and patients who
underwent antitumor treatment pre-operatively were excluded. The
tumor tissue specimens were collected intra-operatively,
immediately frozen in liquid nitrogen, and stored at −80°C. The
tissue specimens were staged according to the Ann Arbor staging
scheme as follows: I, invasion of a lymph node or an extranodal
organ or part; II, diaphragmatic side, involving two or more lymph
nodes or external limitations infringing on an extranodal organ or
part; III, violation of both surfaces of the diaphragm, the
junction, or additional limitations to infringe upon an extranodal
organ or part or the spleen; and IV, diffuse invasion or
disseminated extranodal organ or part of one or more. Among the
participants, 42 were stages I–II and 30 were stages III–IV. The
distribution of patients according to the Lymphoma International
Prognostic Index (IPI) was as follows: 0–1, n=25; 2, n=22; 3, n=16;
and 4–5, n=9. There were 28 patients with B symptoms, and 44
patients had no B symptoms.
The lymphatic tissue samples from 30 patients with
reactive hyperplasia of lymph nodes were designated as the control
group. All of the patients signed informed consent, which was
approved by the Ethics Committee of Zhangzhou Affiliated Hospital
of Fujian Medical University (protocol no. 2010032).
Cell culture
HCC1954 BL, NCI-BL2009, SU-DHL-4, and Farage cells
were cultured in RPMI-1640 medium containing 10% FBS and 1%
penicillin-streptomycin. The cells were passaged at a 1:4 ratio,
and cells with suitable growth in the logarithmic phase were used
in all experiments.
Construction of the recombinant
plasmid with double luciferase gene report
The whole genome DNA of 293T cells was used as a
template, and PCR amplification contained the MEK1 3′-UTR region of
the miR-101 binding site. SacI and HindIII enzyme
cutting sites and protective bases were added to the 5′-terminal of
the upstream and downstream primers, respectively. The primer
sequences for MEK1 were as follows: Sense,
5′-CGAGCTCAAGCAACAAAGAGCGAGTCCCGA0-3′ and antisense,
5′-CCAAGCTTGCAAAGCATGCTTCACATGCACT-3′ (SacI and
HindIII enzyme sites are underlined; the former is a
protective base). The amplified product was the 10–465 bp range of
the 3′-UTR region of MEK1. The total PCR amplification reaction
volume consisted of the following: 2X KOD buffer 10 µl; d NTP (2
mM) 4 µl; primer F (10 µM) 0.5 µl; primer R (10 µM) 0.5 µl; KOD FX
0.4 µl; template DNA 2 µl; and ddH2O 3.6 µl. The cycling
conditions were as follows: 95°C for 10 min, followed by 30 cycles
at 94°C for 30 sec; 58°C for 30 sec; and 72°C for 10 min, with 4°C
infinity. After PCR amplification, SacI and HindIII
double enzymes were used to cut the PCR products and pMIR Noblets.
The conditions for the enzyme digestion were 37°C for 4 h. The
purified product was recovered by 1.5% agarose gel electrophoresis,
and the purified PCR products were incubated at 16°C for overnight
connection. The products were transformed into DH5α competent
cells, inoculated on plates containing penicillin, and incubated at
37°C overnight. Then, a single positive clone was selected and
incubated in Escherichia coli culture medium at 37°C
overnight to extract the plasmids. The sequence was identified, and
designated as pMIR-MEK1-WT. Using the wild-type plasmid as a
template, miR-101 and MEK1 3′-UTR binding region mutations for
meaningless sequence design of MEK1 point mutation primers were as
follows: forward, 5′-CTCTTGTGTTAATAAATATTACTGTCT-3′ and reverse,
5′-AAGTGTCATATATTAAAAATGACAAGA-3′ (mutant sequences are
underlined). The detailed construction steps were the same as the
wild-type, and the constructed mutant plasmid was designated as
pMIR-MEK1-MUT.
Dual-luciferase reporter gene
assay
The 293T (1×105) cells were inoculated in
24-well plates. After being cultured in DMEM containing 10% FBS for
24 h, the cells were divided into 4 transfection groups:
pMIR-MEK1-WT+miR-101 mimic group; pMIR-MEK1-WT+miR-NC group;
pMIR-MEK1-MUT+miR-101 mimic group; and pMIR-MEK1-MUT+miR-NC group.
Each group had 3 duplicate pores. The specific transfection
additions were as follows: 0.8 µg of gene recombinant plasmid; 0.02
µg of pRL-SV40 plasmid; and 20 pmol miR-101 mimic or miR-NC in 50
µl of Opti-MEM. The mixture was then stored for 5 min at room
temperature. Lipofectamine™ 2000 (2 µl) was added to 50 µl of
Opti-MEM, and was placed at room temperature for 5 min after gentle
mixing. The two mixtures were combined after mixing for 20 min at
room temperature. The 4 groups of cells were added to 24-hole
plates. DMEM fresh medium was replaced by culture medium at 37°C
for 6 h, and the cultures were continued for 48 h. A Dual-Glo
Luciferase Assay System kit was used to detect luciferase activity,
as follows: 100 µl of Passive Lysis Buffer cell lysate was added to
each hole of a 24-well plate; the plates were slowly shaken at room
temperature for 15 min; 20 µl of cell lysate was added to 100 µl of
luciferase assay reagent II (LAR II), mixed, and the fluorescence
of the firefly luciferase activity was detected. Fluorescein (100
µl) was added to the detection kit. The Renilla luciferase
activity of firefly luciferase activity to Renilla
luciferase activity/relative activity value was analyzed.
Cell transfection and grouping
The day before transfection, SU-DHL-4 and Farage
cells were cultured in 10-cm culture dishes, ensuring that the
transfection cell density was 50–60% on the day of fusion, and the
transfected cells were divided into the following 4 groups: miR-NC
group; miR-101 mimic group; siRNA-NC group; and siRNA-MEK1 group.
The miR-NC, miR-101 mimic, siRNA-NC and siRNA-MEK1 group cells were
diluted with Opti-MEM and incubated at room temperature for 5 min.
Each transfected nucleotide was then mixed with Lipofectamine 2000
and gently inverted by mixing, and incubated for 30 min at room
temperature. The cell culture medium was removed and washed twice
in PBS to remove serum, and then replaced with Opti-MEM serum-free
cell culture medium. The transfection complexes were added to the
culture medium and mixed. After culturing for 6 h, they were
replaced with RPMI-1640 medium containing 10% FBS and 1%
penicillin. After being cultured for 72 h, they were trypsinized.
Cells were harvested for gene, protein expression, and flow
cytometric assays.
qRT-PCR detection of gene
expression
One milliliter of TRIzol was added to every 20 mg of
tissue or every 3×106 cells. After full lysis, the
tissues or cells were added to chloroform for extraction. After
stratification, the RNA supernatant was transferred to a new EP
tube. After washing with isopropanol and 70% ethanol, DEPC water
was dissolved to obtain RNA. The RNA was reversed transcribed into
cDNA using PrimeScript RT reagent kit with cDNA as the template and
qPCR detecting gene expression. The reverse transcription reaction
system contained 0.5 µl (50 µM), 0.5 µl of random 6 mers (100 µM),
0.5 µl of PrimeScript RT Enzyme Mix, 1.0 µg of RNA, 2 µl of 5X
PrimeScript Buffer, and RNase-free H2O2 was
added to a total volume of 10 µl. The reverse transcription
conditions were 37°C for 15 min and 85°C for 5 sec. The PCR
reaction system contained SYBR Fast qPCR Mix (10.0 µl), 0.8 µl of
forward primer (10 µM), 0.8 µl of reverse primer (10 µM), 2.0 µl of
cDNA, and 6.4 µl of RNase-free dH2O. The PCR reaction
consisted of 95°C pre-denaturation for 10 min, followed by 40
cycles at 95°C for 5 sec, 60°C for 20 sec, and extension at 72°C
for 15 sec. Real-time PCR was performed on Bio-Rad CFX96/CFX
Connect™ (Bio-Rad Laboratories, Inc., Hercules, CA, USA) to test
the relative expression.
Western blot analysis
Total protein was extracted by RIPA from cells or
tissues. A total of 40 µg of protein was separated by 8–10% sodium
lauryl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and 5%
concentration gel for 3 h and transferred to nitrocellulose
membranes. Next, the membranes were blocked with 5% skim milk at
room temperature for 60 min and incubated with the primary antibody
at 4°C overnight (MEK1, MMP-2, anti ERK1/2, ERK1/2, Bcl-2 and
β-actin; the dilution ratios were 1:2,000, 1:1,000, 1:2,000,
1:1,000, 1:2,000, 1:10,000 and PBST, respectively). After
thrice-washing, the membranes were incubated with horseradish
peroxidase (HRP)-labeled secondary antibody (1:20,000) for 60 min,
then thrice-washed. Finally, protein expression was detected by
enhanced chemiluminescence (ECL). Data were analyzed with ImageJ
software k1.45 (National Institutes of Health, Bethesda, MD,
USA).
Flow cytometry to detect cell
proliferation
Each transfection group of cells was resuspended in
complete medium. After a 120-min incubation with 10 µM EdU, the
cells were continuously cultured for 48 h, and then the cells were
collected after trypsinization. The cells were washed during 250 ×
g centrifugation 1 time with PBS containing 1% BSA, and then 100 µl
of Click-iT fixative was added for 15 min and washed during 250 × g
centrifugation 1 time with PBS containing 1% BSA. The cells were
added to 100 µl of 1X Click-iT saponin-based permeabilization for
15 min at room temperature and 500 µl of PBS, CuSO4,
Alexa Fluor 488, and buffer was added before incubation in the dark
at room temperature for 30 min. The cells were added to 3 ml of 1X
Click-iT saponin-based permeabilization and washed during 250 × g
centrifugation 1 time. The cells were then added to 500 µl of 1X
Click-iT saponin-based permeabilization and wash reagent and tested
on a Beckman Kurt FC 500 MCL/MPL flow cytometry device (Beckman
Coulter, Inc., Brea, CA, USA) to evaluate cell proliferation.
Flow cytometry to detect cell
apoptosis
The transfected cells were digested by trypsin and
collected after 250 × g centrifugation, then washed 2 times in PBS.
Cells were incubated in 5 µl of Annexin V-FITC and 5 µl of PI in
the dark for 10 min. Next, the cells were resuspended in 400 µl of
binding buffer and tested on a Beckmann FC 500 MCL/MPL flow
cytometry device to evaluate cell apoptosis.
Statistical analysis
All data analyses were performed using SPSS 18.0
software (SPSS, Inc., Chicago, IL, USA). The measurement data are
depicted as the mean ± standard deviation and were compared by
t-tests. For two-group comparisons, a Chi-square test was used, and
when making comparisons among multiple groups, one way ANOVA test
was used to determine the relationship between the level of miR-101
expression and the clinical characteristics of the patients. The
Mann-Whitney U test was used to compare the level of miR-101 and
MEK1 mRNA expression in the lymphoid tissues of the two groups. The
survival curve was drawn with the Kaplan-Meier method, and the
survival rate was compared using a log-rank test. A P<0.05 was
considered statistically significant.
Results
miR-101 targeted regulation of MEK1
expression
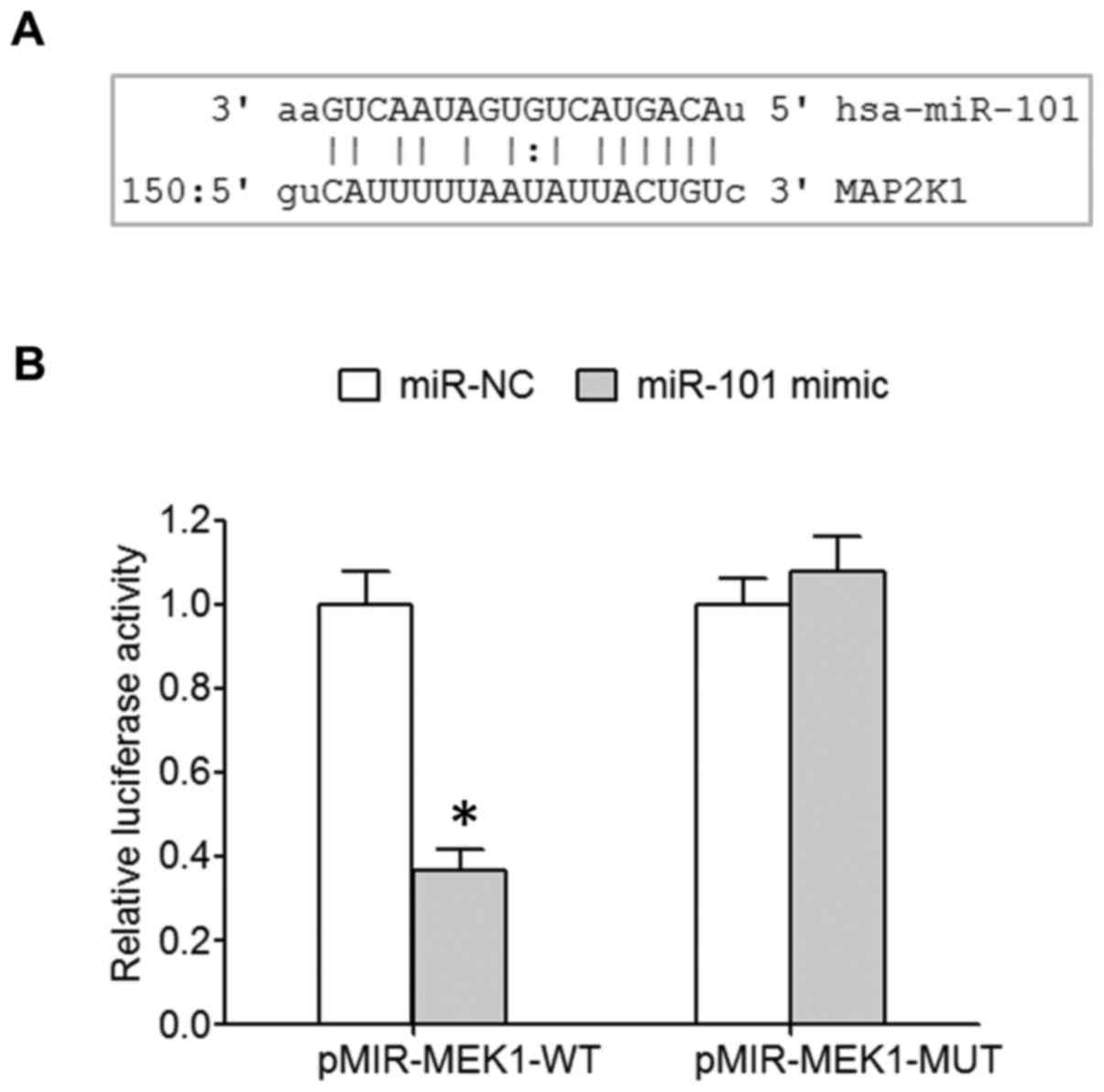
MicroRNA.org online prediction showed
the targeted binding site between miR-101 and 3′-UTR of MEK1 mRNA
(Fig. 1A). A dual luciferase assay
revealed that miR-101 mimic transfection significantly decreased
the relative luciferase activity of 293T cells transfected with
wild-type pMIR-MEK1-WT, while the relative luciferase activity of
293T cells transfected with mutant pMIR-MEK1-MUT was not affected
(Fig. 1B), indicating the
regulatory relationship between miR-101 and MEK1.
miR-101 is reduced, while MEK1 is
increased in DLBCL tissues
Based on quantitative RT-PCR (qRT-PCR), it was shown
that the expression of miR-101 in tumor tissues from DLBCL patients
was significantly decreased (Mann-Whitney U value=367, P<0.05)
when compared with proliferative lymph node tissues in the control
group (Fig. 2A), while the
expression of MEK1 mRNA was elevated in the tumor tissues from
DLBCL patients compared with the control group (Mann-Whitney U=269,
P<0.05; Fig. 2A). Western blot
analysis showed that the expression levels of MEK1 and p-MEK1
protein were significantly increased in the DLBCL tissues when
compared with levels in the control tissues; representative
illustrations are shown in Fig. 2B.
Based on the median miR-101 expression, DLBCL patients were divided
into a miR-101 high-expression group and a miR-101 low-expression
group. The relationship between clinical features and the level of
miR-101 expression was analyzed. According to χ2 test
analysis, the number of patients with high expression of miR-101
was higher in patients with clinical stage I–II and the number of
patients with low expression of miR-101 was significantly higher in
patients with clinical stage III–IV (P=0.017). The proportion of
patients with an IPI score >3 points was higher in the low
miR-101 expression group and the proportion of patients with an IPI
score <3 was significantly higher in the high miR-101 expression
group (P=0.026), yet patient age, gender, no B symptoms, and
miR-101 expression had no apparent relationship (P>0.05;
Table I). Survival analysis showed
that the survival and prognosis of patients with high expression of
miR-101 were significantly better than the patients with low
miR-101 expression (log-rank test χ2=5.684, P=0.017).
Survival and prognosis of patients with low expression MEK1 mRNA
were better than patients with high expression of MEK1 mRNA
(log-rank test χ2=5.360, P=0.021; Fig. 2C).
 | Table I.Relationship between the expression
of miR-101 and clinical characteristics of the DLBCL cases. |
Table I.
Relationship between the expression
of miR-101 and clinical characteristics of the DLBCL cases.
|
|
| miR-101 expression
level |
|
|
|---|
|
|
|
|
|
|
|---|
| Clinical
features | n | High | Low | χ2 | P-value |
|---|
| Age (years) |
|
|
| 1.399 | 0.237 |
|
≤60 | 33 | 19 | 14 |
|
|
|
>60 | 39 | 17 | 22 |
|
|
| Sex |
|
|
| 0.892 | 0.345 |
|
Male | 38 | 21 | 17 |
|
|
|
Female | 34 | 15 | 19 |
|
|
| Clinical stage |
|
|
| 5.714 | 0.017 |
|
I–II | 42 | 26 | 16 |
|
|
|
III–IV | 30 | 10 | 20 |
|
|
| IPI score |
|
|
| 4.963 | 0.026 |
|
0–2 | 47 | 28 | 19 |
|
|
|
3–5 | 25 | 8 | 17 |
|
|
| B symptoms |
|
|
| 2.104 | 0.147 |
|
Present | 28 | 17 | 11 |
|
|
|
Absent | 44 | 19 | 25 |
|
|
Overexpression of miR-101
downregulates ERK/MAPK pathway activity
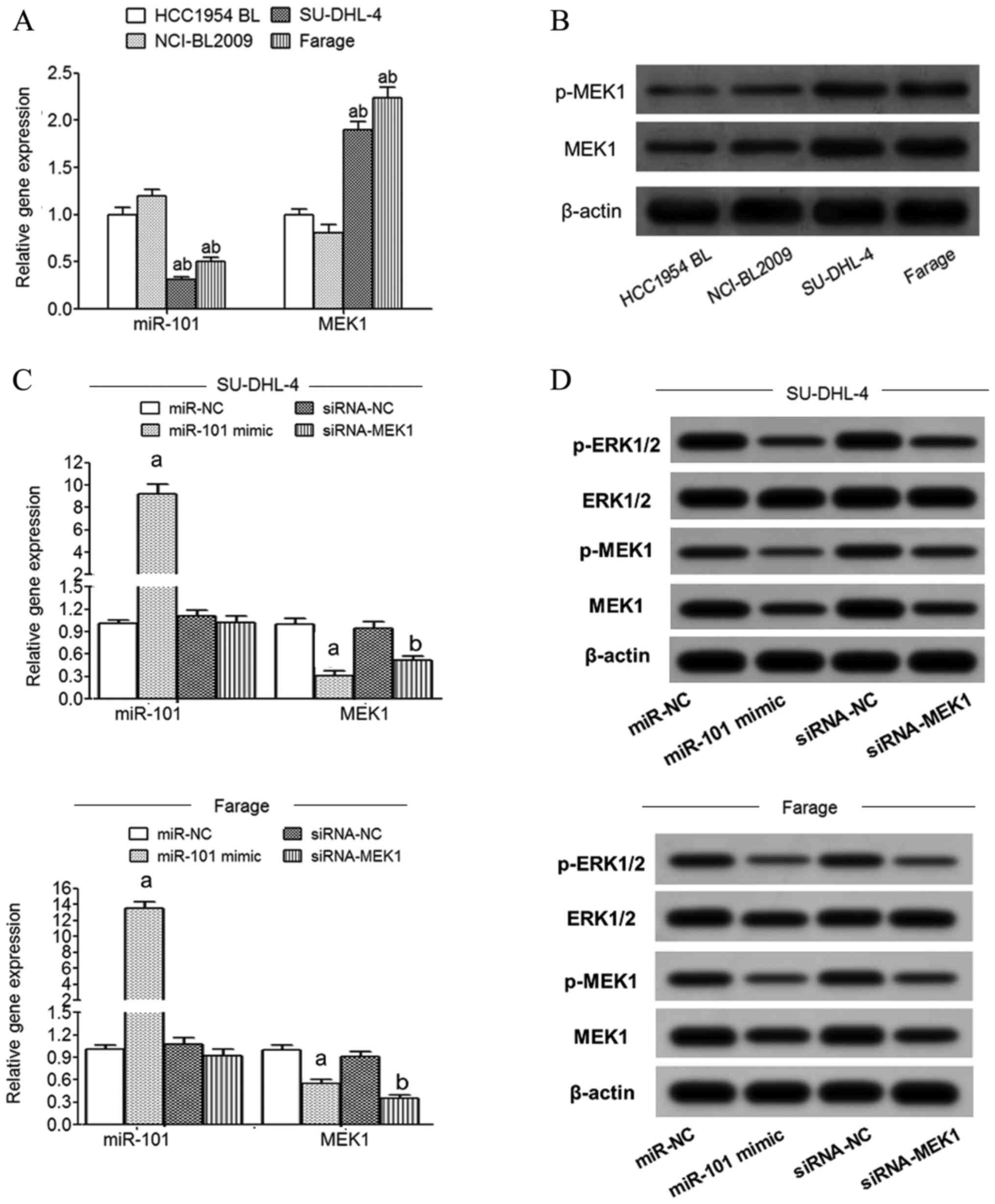
qRT-PCR detection showed that compared with normal
lymphoblastoid cells (HCC1954 BL and NCI-BL2009), the expression of
miR-101 in SU-DHL-4 and Farage cells was significantly decreased
and MEK1 mRNA expression was significantly increased (Fig. 3A). Western blot detection showed
that compared with normal lymphoblastoid cells (HCC1954 BL and
NCI-BL2009), the expression of MEK1 and p-MEK1 protein in SU-DHL-4
and Farage cells was significantly increased (Fig. 3B). The expression of miR-101 in
SU-DHL-4 and Farage cells was significantly increased in the
miR-101 mimic transfection group compared with the miR-NC group
while MEK1 mRNA expression was significantly decreased (Fig. 3C). Compared with the siRNA-NC group,
the expression of MEK1 mRNA in the SU-DHL-4 and Farage cells of the
siRNA-MEK1 transfected group was significantly decreased (Fig. 3C). Western blot analysis showed that
transfection of miR-101 mimic or siRNA-MEK1 significantly reduced
MEK1, p-MEK1 and p-ERK1/2 protein expression in the SU-DHL-4 and
Farage cells (Fig. 3D).
Overexpression of miR-101 inhibits the
proliferation of lymphoma cells
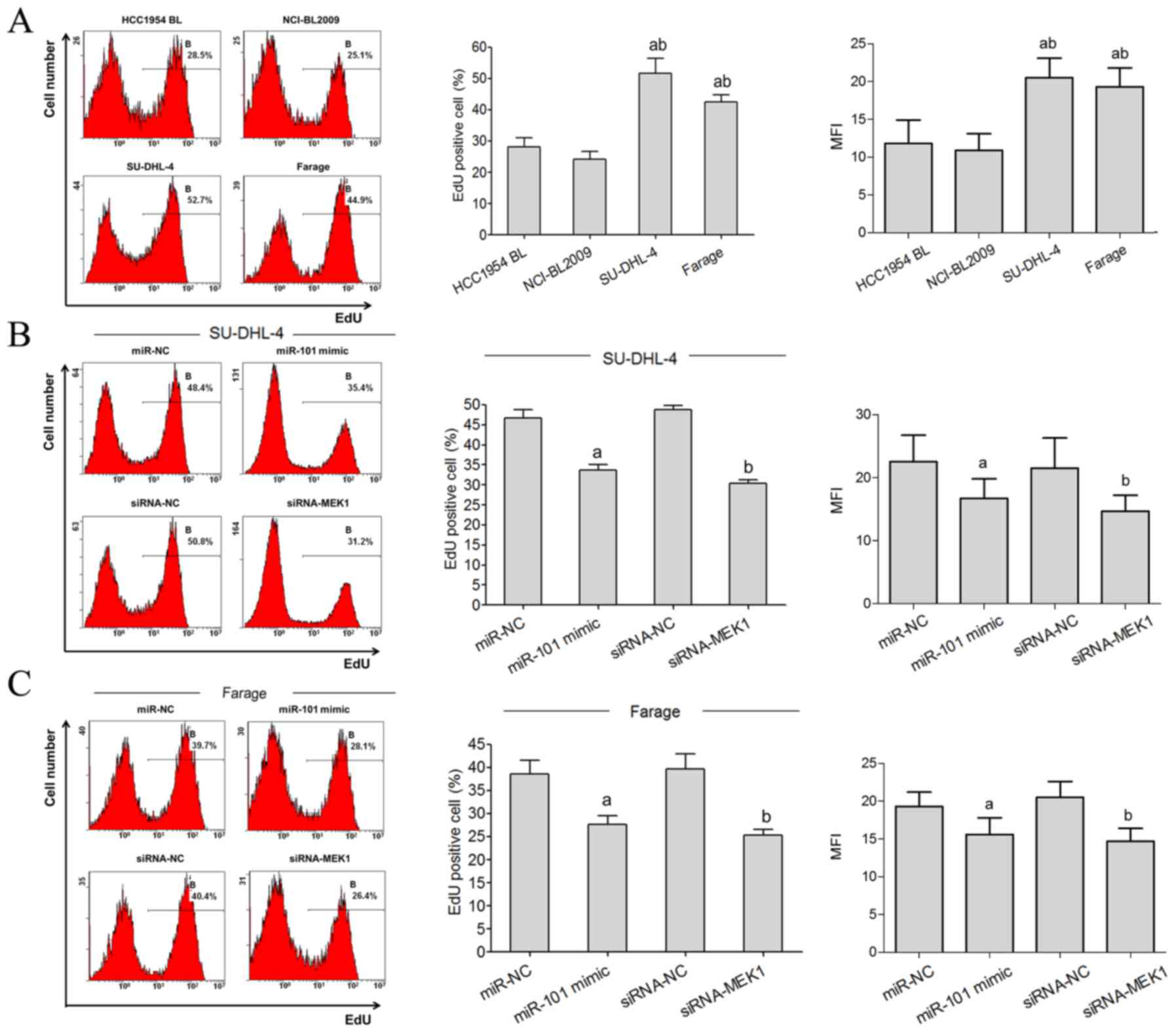
Flow cytometric analysis showed that the
proliferative rate of SU-DHL-4 and Farage cells was significantly
higher than the HCC1954 BL and NCI-BL2009 cells (Fig. 4A). The EdU staining flow test showed
that the EdU-positive rate of SU-DHL-4 and Farage cells in the
miR-101 mimic transfected group was significantly lower than the
miR-NC group. Compared with the siRNA-NC group, the EdU-positive
rates of SU-DHL-4 and Farage cells in the siRNA-MEK1 transfected
group were significantly lower (Fig. 4B
and C).
Overexpression of miR-101 promotes the
apoptosis of lymphoma cells
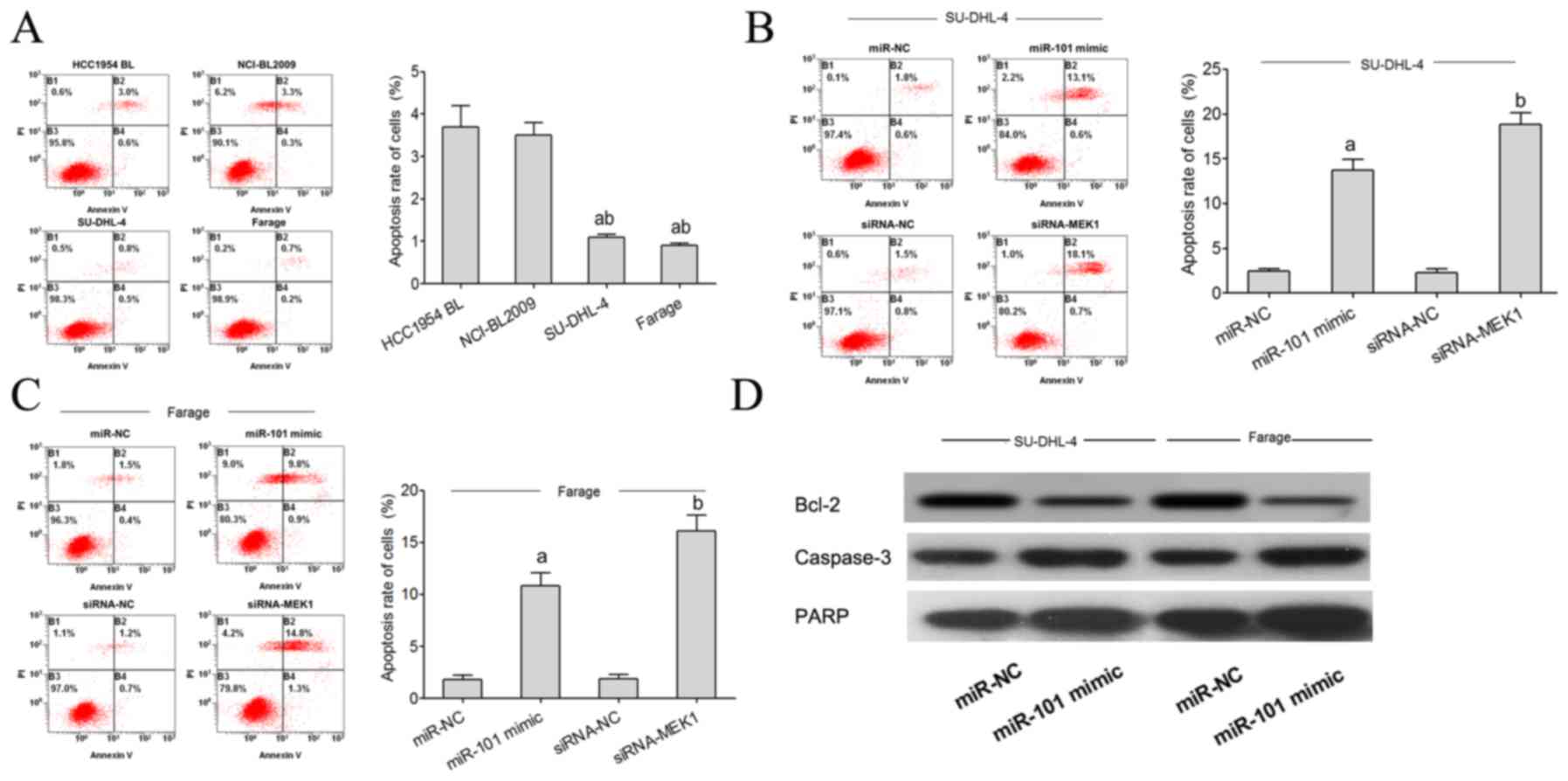
Flow cytometric analysis showed that the apoptosis
rate of SU-DHL-4 and Farage cells was significantly lower than that
noted in the HCC1954 BL and NCI-BL2009 cells (Fig. 5A). The flow cytometry test showed
that the apoptosis rate of SU-DHL-4 and Farage cells in the miR-101
mimic transfected group was significantly higher than that noted in
the miR-NC group. Compared with the siRNA-NC group, the apoptosis
rate of the SU-DHL-4 and Farage cells in the miR-101 minic and
siRNA-MEK1 group was significantly higher (Fig. 5B and C). Using western blot analysis
we found that miR-101 targets Bcl-2, PARP and caspase-3 expression
to regulate the apoptosis of lymphoma cells. When miR-101
expression was increased, expression of the anti-apoptotic factor
Bcl-2 protein was significantly decreased, and PARP and caspase-3
expression was increased (Fig.
5D).
Discussion
Lymphomas include Hodgkin lymphoma (HL) and
non-Hodgkin lymphoma (NHL), of which NHL is the most common
accounting for >90% of lymphomas (3). Based on the difference in cell origin,
NHL can be divided into B, T, and NK/T cell types. The B cell type
of NHL is the main type of all NHLs, and the incidence is >80%
(19). Diffuse large B cell
lymphoma (DLBCL) is the most common type of B-NHL, accounting for
approximately 50% of B-NHLs, and accounting for approximately 40%
of all NHL cases (4,5). DLBCL, the most common NHL in adults,
is an invasive lymphoma with a high degree of invasiveness, has a
poor response to treatment and a corresponding poor prognosis.
Therefore, an in-depth study of the pathogenesis of lymphoma is of
great significance for screening early diagnostic markers,
improving therapeutic effect, and improving the prognosis of
patients.
The MAPK signaling pathway is an important signal
transduction system that widely exists in eukaryotic cells, and can
be mediated and activated by cell factor receptors, the
intracellular receptor tyrosine kinase, G protein coupled
receptors, and cytokine receptors in a variety of extracellular
stimuli, such as cytokines, growth factors, neurotransmitters, and
G protein coupled receptor ligand interactions. The MAPK signaling
pathway consequently activates a variety of transcription factors
in the nucleus, regulates various protein kinase reactions in
cells, regulates the transcription and expression of target genes,
and ultimately affects cell survival, proliferation, migration,
apoptosis, and other physiologic and biological processes (20,21).
The MAPK signaling pathway family primarily consists of the
following 4 pathways: ERK; c-Jun N-terminal kinase (JNK); p38
mitogen-activated protein kinase (p38 MAPK); and large mitogen
activated protein kinase pathway (BMK1). Furthermore, the ERK/MAPK
signaling pathway mediated by the MAPK signal transduction pathway
is the classic MAPK signaling pathway, which plays a major role in
the MAPK signaling transduction pathway. The ERK/MAPK signaling
pathway is widely expressed in many tissues and cells, and can
regulate cell proliferation, the cell cycle, apoptosis, migration
and invasion, and many other biological processes (22,23).
Excessive activation of the ERK/MAPK signaling pathway can cause
abnormal proliferation, apoptosis and differentiation of cells, and
thus is associated with tumor occurrence, progression and
metastasis, including gallbladder carcinoma (22), esophageal cancer (24), and breast cancer (24–27).
MEK1 is a double-specific protein kinase, which can
act upstream of ERK protein and phosphorylate the Tyr/Thr residues
of ERK protein, thereby activating the ERK/MAPK signaling pathway
(28). Many studies have shown that
the abnormal increase in MEK1 expression or functional activity is
related to the occurrence, progression, drug resistance and poor
prognosis of many types of tumors, such as pancreatic, ovarian and
lung cancers (6–8). Moreover, it has been shown that the
expression or enhancement of the function of MEK1 may also play an
important role in lymphoma (9). The
abnormal decrease in expression of miR-101 is associated with the
incidence of lymphoma, suggesting a possible tumor-suppressor
effect. Bioinformatic analysis showed that there was a target
complementary binding site between miR-101 and MEK1. The aim of the
present study was to determine whether or not miR-101 plays a role
in regulating MEK1 expression, ERK/MAPK pathway activity, and the
proliferation and apoptosis in DLBCL cell lines.
The results of the double luciferase gene report
showed that the miR-101 mimic can significantly decrease the
relative luciferase activity of wild-type pMIR-MEK1-WT transfected
into 293T cells, but the mutant pMIR-MEK1-MUT transfected 293T cell
relative luciferase activity was not affected, indicating that
there was targeted regulation of the interaction between miR-101
and MEK1 mRNA. The results of the present study showed that
compared with the control group (hyperplastic lymph node tissues),
expression of miR-101 in DLBCL tumor tissues was significantly
decreased, and the more advanced the clinical stage of the
patients, the higher the IPI score and the lower the expression of
miR-101, suggesting that the abnormal reduction in miR-101
expression was related to DLBCL features. Furthermore, the survival
curve also showed that the survival rate of the patients with low
miR-101 expression was significantly less than patients with high
miR-101 expression. Compared with the lymph node tissues of the
control group, the expression of MEK and p-MEK1 was significantly
increased in the tumor tissues of patients with DLBCL. Moreover,
the survival and prognosis of patients with low expression of MEK1
mRNA were significantly better than patients with a high level of
MEK1 mRNA expression, which was in contrast to the relationship
between miR-101 expression and patient survival and indirectly
indicated the differential significance of miR-101 and MEK1 for
DLBCL.
In a previous study of the relationship between
miR-101 and lymphoma, Sasaki et al (29) showed that the decreased expression
of miR-101 in the peripheral blood of lymphoma patients may play an
important role in upregulating the expression of its target gene,
EZH2, and promoting the pathogenesis of lymphoma. Moreover,
Ferreira et al (30)
reported that compared with reactive proliferative lymph nodes, the
expression of miR-101 in tumor tissues of patients with NHL was
significantly decreased, suggesting that miR-101 may be a
tumor-suppressor gene in lymphoma. Furthermore, Merkel et al
(31) showed that compared with
normal human CD3T lymphocytes and lymph node tissues of healthy
persons, the expression of miR-101 in anaplastic large cell
lymphoma in tumor tissues was reduced by >2- to 20-fold. Leich
et al (17) reasoned that
the abnormally reduced expression of miR-101 in tumor tissues of T
(11,15) transposition-negative follicular
lymphoma may be related to the enhancement of tumor cell
proliferation and malignancy. Ng et al (18) showed that compared with normal lymph
node tissues, the expression of miR-101 in tumor tissues and NK
tumor cell lines of NK/T cell lymphoma patients was significantly
decreased. Although there are many reports concerning the
relationship between miR-101 and the pathogenesis of lymphoma, most
were T cell-derived or NK cell-derived lymphomas; however, research
concerning the relationship between miR-101 and DLBCL is limited.
The results of the present study showed that the expression of
miR-101 in DLBCL patients was significantly lower than that noted
in the lymphoid tissue, and was correlated with disease stage, IPI
score, survival, and prognosis. In addition to lymphoma, the
relationship between miR-101 and the incidence of lymphoblastic
leukemia has also been reported. Qian et al (32) showed that the expression of miR-101
in the peripheral blood of patients with lymphoblastic leukemia was
decreased significantly compared with healthy individuals. In
addition, they also found that the expression of miR-101 in the
peripheral blood cells of patients with acute T lymphoblastic
leukemia was significantly less than miR-101 expression in the
healthy thymus and bone marrow cells. Furthermore, Liu et al
(33) showed that the expression of
miR-101 in bone marrow tissues of patients with acute lymphoblastic
leukemia was abnormally reduced. These studies indicated that the
abnormal decrease in miR-101 expression was a contributing factor
in the pathogenesis of lymphoma and lymphocytic leukemia; our study
findings mutually corroborated and were supported by these
studies.
The Bcl-2 gene of B lymphocytoma is an
anti-apoptotic factor (34) which
plays an important role in the regulation of the
mitochondrial-dependent apoptotic pathway. Bcl-2 protein is
localized in the endoplasmic reticulum, mitochondrial membrane, and
nuclear membrane, and plays a role in anti-apoptosis through a
variety of mechanisms. These include inhibition of pro-apoptosis
mitochondrial cytochrome c, release to the cytoplasm, blockage of
the destruction of cell components by oxygen-free radicals,
affecting the transmembrane transport of calcium ions, promoting
the formation of heterologous two dimers with the pro-apoptotic
proteins of the Bcl-2 family, and promoting the localization and
distribution of pro-apoptotic factors (35–38).
The results of the present study showed that transfection of
miR-101 mimic or siRNA-MEK1 significantly reduced the expression of
MEK1, p-MEK1 and p-ERK1/2 in SU-DHL-4 and Farage cells. In
addition, the expression of the anti-apoptotic factor, Bcl-2, was
significantly reduced, lymphoma cell proliferation was
significantly decreased, and apoptosis was significantly increased.
Qian et al (32) showed that
in the process of the pathogenesis of lymphocytic leukemia, miR-101
acts as a tumor suppressor. Moreover, overexpression of miR-101 was
found to decrease the proliferation and invasion of Jurkat
lymphocytic leukemia cells, significantly increase apoptosis, and
increase the sensitivity of the chemotherapy drug, doxorubicin, by
targeting inhibition of the Notch1 gene. Ferreira et al
(30) found that histone
deacetylase inhibitor treatment inhibited the proliferation of Raji
cell lymphoma and induced cell cycle arrest, which was accompanied
by significantly increased expression of miR-101, suggesting that
miR-101 may have a tumor-suppressor role for the reduction of
malignant lymphoma cell biological characteristics. Moreover,
Merkel et al (31) showed
that overexpression of miR-101 inhibited the expression of mTOR,
attenuated the proliferation of SR-786 and Karpas-299 cells,
induced cell cycle G1 phase arrest, and significantly promoted cell
apoptosis. Ng et al (18)
showed that there is a target regulatory relationship between
miR-101 and STMN1 in the NK-YS cell line of NK/T cell lymphomas.
Furthermore, miR-101 plays a role in downregulating STMN1 gene
expression and inhibiting the proliferation of NK-YS cells. The
results of this study showed that miR-101 inhibited the
proliferation of lymphoma cells and promoted the inhibition of
apoptosis of lymphoma cells, similar to the above results. In the
study of the relationship between MEK1 and the biological behavior
of lymphoma cells, Kumar et al (39) showed that inhibition of MEK1
downregulated the expression of PD-L1 in B cell lymphoma cells,
giving credence to the effect of antitumor immunity, which could be
a potential target in the treatment of lymphoma. Further detection
by Nguyen et al demonstrated that (9) the downregulation of MEK1 by PD184352
or shRNA could significantly enhance the effect of the
chemotherapeutic drug, sorafenib, on inducing apoptosis of DLBCL
cells, demonstrating that the enhancement function of MEK1 is
related to the malignancy of DLBCL cells, while the antagonistic
function of MEK1 was found to reduce the malignant characteristics
of DLBCL cells. Kumar et al and Nguyen et al
(9,39) showed that the enhancement of EMK1
expression or functional activity is a tumor-promoting factor in
lymphoma, while inhibiting the expression and function of MEK1
reduces the malignant effect of lymphoma, which is in agreement
with our results. At present, the role of miR-155 in regulating
MEK1 expression and DLBCL cell proliferation and apoptosis has been
rarely reported. The present study confirmed the relationship
between miR-101 and MEK1 and their associated function with DLBCL.
The present study also confirmed that miR-101 plays an important
role in targeting inhibition of MEK1 expression, affecting the
activity of the ERK/MAPK pathway, and regulating the proliferation
and apoptosis of lymphoma cells.
In conclusion, downregulation of miR-101 is related
to the pathogenesis and prognosis of DLBCL. miR-101 overexpression
can inhibit the proliferative ability of lymphoma cells and promote
cell apoptosis through targeting inhibition of MEK1 expression.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Fujian
Provincial Natural Science Foundation 2018J01203.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
RZ and YH conceived and designed the study. YH, RZ,
YZ, LL and XM performed the experiments. YH, YZ and RZ wrote the
paper. YH, RZ and LL reviewed and edited the manuscript. All
authors read and approved the manuscript and agree to be
accountable for all aspects of the research in ensuring that the
accuracy or integrity of any part of the work are appropriately
investigated and resolved.
Ethics approval and consent to
participate
All of the patients signed informed consent, which
was approved by the Ethics Committee of Zhangzhou Affiliated
Hospital of Fujian Medical University (protocol no. 2010032).
Patient consent for publication
Not applicable.
Competing interests
The authors state that they have no competing
interests.
References
|
1
|
Luminari S: Bridging the gap between
epidemiology and clinical research in lymphoma. Leuk Lymphoma.
54:1855–1856. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Smedby KE and Hjalgrim H: Epidemiology and
etiology of mantle cell lymphoma and other non-Hodgkin lymphoma
subtypes. Semin Cancer Biol. 21:293–298. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Skrabek P, Turner D and Seftel M:
Epidemiology of non-Hodgkin lymphoma. Transfus Apher Sci.
49:133–138. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hamlin PA, Satram-Hoang S, Reyes C, Hoang
KQ, Guduru SR and Skettino S: Treatment patterns and comparative
effectiveness in elderly diffuse large B-cell lymphoma patients: A
surveillance, epidemiology, and end results-medicare analysis.
Oncologist. 19:1249–1257. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Castillo JJ, Winer ES and Olszewski AJ:
Sites of extranodal involvement are prognostic in patients with
diffuse large B-cell lymphoma in the rituximab era: An analysis of
the surveillance, epidemiology and end results database. Am J
Hematol. 89:310–314. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gysin S, Paquette J and McMahon M:
Analysis of mRNA profiles after MEK1/2 inhibition in human
pancreatic cancer cell lines reveals pathways involved in drug
sensitivity. Mol Cancer Res. 10:1607–1619. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Pénzváltó Z, Lánczky A, Lénárt J,
Meggyesházi N, Krenács T, Szoboszlai N, Denkert C, Pete I and
Győrffy B: MEK1 is associated with carboplatin resistance and is a
prognostic biomarker in epithelial ovarian cancer. BMC Cancer.
14:8372014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Song JY, Kim CS, Lee JH, Jang SJ, Lee SW,
Hwang JJ, Lim C, Lee G, Seo J, Cho SY and Choi J: Dual inhibition
of MEK1/2 and EGFR synergistically induces caspase-3-dependent
apoptosis in EGFR inhibitor-resistant lung cancer cells via BIM
upregulation. Invest New Drugs. 31:1458–1465. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Nguyen TK, Jordan N, Friedberg J, Fisher
RI, Dent P and Grant S: Inhibition of MEK/ERK1/2 sensitizes
lymphoma cells to sorafenib-induced apoptosis. Leuk Res.
34:379–386. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zheng W, Liu Z, Zhang W and Hu X: miR-31
functions as an oncogene in cervical cancer. Arch Gynecol Obstet.
292:1083–1089. 2005. View Article : Google Scholar
|
|
11
|
Wang LQ, Zhang Y, Yan H, Liu KJ and Zhang
S: MicroRNA-373 functions as an oncogene and targets YOD1 gene in
cervical cancer. Biochem Biophys Res Commun. 459:515–520. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mou Z, Xu X, Dong M and Xu J:
MicroRNA-148b acts as a tumor suppressor in cervical cancer by
inducing G1/S-phase cell cycle arrest and apoptosis in a
caspase-3-dependent manner. Med Sci Monit. 22:2809–2815. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Fan D, Wang Y, Qi P, Chen Y, Xu P, Yang X,
Jin X and Tian X: MicroRNA-183 functions as the tumor suppressor
via inhibiting cellular invasion and metastasis by targeting MMP-9
in cervical cancer. Gynecol Oncol. 141:166–174. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chen LG, Xia YJ and Cui Y: Upregulation of
miR-101 enhances the cytotoxic effect of anticancer drugs through
inhibition of colon cancer cell proliferation. Oncol Rep.
38:100–108. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lin C, Huang F, Li QZ and Zhang YJ:
miR-101 suppresses tumor proliferation and migration, and induces
apoptosis by targeting EZH2 in esophageal cancer cells. Int J Clin
Exp Pathol. 7:6543–6550. 2014.PubMed/NCBI
|
|
16
|
Liu L, Guo J, Yu L, Cai J, Gui T, Tang H,
Song L, Wang J, Han F, Yang C, et al: miR-101 regulates expression
of EZH2 and contributes to progression of and cisplatin resistance
in epithelial ovarian cancer. Tumour Biol. 35:12619–12626. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Leich E, Zamo A, Horn H, Haralambieva E,
Puppe B, Gascoyne RD, Chan WC, Braziel RM, Rimsza LM, Weisenburger
DD, et al: MicroRNA profiles of t(14;18)-negative follicular
lymphoma support a late germinal center B-cell phenotype. Blood.
118:5550–5558. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ng SB, Yan J, Huang G, Selvarajan V, Tay
JL, Lin B, Bi C, Tan J, Kwong YL, Shimizu N, et al: Dysregulated
microRNAs affect pathways and targets of biologic relevance in
nasal-type natural killer/T-cell lymphoma. Blood. 118:4919–4929.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chihara D, Nastoupil LJ, Williams JN, Lee
P, Koff JL and Flowers CR: New insights into the epidemiology of
non-Hodgkin lymphoma and implications for therapy. Expert Rev
Anticancer Ther. 15:531–544. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Pancione M, Giordano G, Parcesepe P,
Cerulo L, Coppola L, Curatolo AD, Conciatori F, Milella M and
Porras A: Emerging insight into MAPK inhibitors and immunotherapy
in colorectal cancer. Curr Med Chem. 24:1383–1402. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang X, Liu K, Zhang T, Wang Z, Qin X,
Jing X, Wu H, Ji X, He Y and Zhao R: Cortactin promotes colorectal
cancer cell proliferation by activating the EGFR-MAPK pathway.
Oncotarget. 8:1541–1554. 2017.PubMed/NCBI
|
|
22
|
Buchegger K, Silva R, López J, Ili C,
Araya JC, Leal P, Brebi P, Riquelme I and Roa JC: The ERK/MAPK
pathway is overexpressed and activated in gallbladder cancer.
Pathol Res Pract. 213:476–482. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liao T, Wen D, Ma B, Hu JQ, Qu N, Shi RL,
Liu L, Guan Q, Li DS and Ji QH: Yes-associated protein 1 promotes
papillary thyroid cancer cell proliferation by activating the
ERK/MAPK signaling pathway. Oncotarget. 8:11719–11728. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zheng ST, Huo Q, Tuerxun A, Ma WJ, Lv GD,
Huang CG, Liu Q, Wang X, Lin RY, Sheyhidin I and Lu XM: The
expression and activation of ERK/MAPK pathway in human esophageal
cancer cell line EC9706. Mol Biol Rep. 38:865–872. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chen P, Xu W, Luo Y, Zhang Y, He Y, Yang S
and Yuan Z: MicroRNA 543 suppresses breast cancer cell
proliferation, blocks cell cycle and induces cell apoptosis via
direct targeting of ERK/MAPK. Onco Targets Ther. 10:1423–1431.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Koyama T, Ogawara K, Kasamatsu A, Okamoto
A, Kasama H, Minakawa Y, Shimada K, Yokoe H, Shiiba M, Tanzawa H,
et al: ANGPTL3 is a novel biomarker as it activates ERK/MAPK
pathway in oral cancer. Cancer Med. 4:759–769. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lu Z, Ding L, Hong H, Hoggard J, Lu Q and
Chen YH: Claudin-7 inhibits human lung cancer cell migration and
invasion through ERK/MAPK signaling pathway. Exp Cell Res.
317:1935–1946. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wong KK: Recent developments in
anti-cancer agents targeting the Ras/Raf/MEK/ERK pathway. Recent
Pat Anticancer Drug Discov. 4:28–35. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sasaki D, Imaizumi Y, Hasegawa H, Osaka A,
Tsukasaki K, Choi YL, Mano H, Marquez VE, Hayashi T, Yanagihara K,
et al: Overexpression of Enhancer of zeste homolog 2 with
trimethylation of lysine 27 on histone H3 in adult T-cell
leukemia/lymphoma as a target for epigenetic therapy.
Haematologica. 96:712–719. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ferreira AC, Robaina MC, Rezende LM,
Severino P and Klumb CE: Histone deacetylase inhibitor prevents
cell growth in Burkitt's lymphoma by regulating PI3K/Akt pathways
and leads to upregulation of miR-143, miR-145, and miR-101. Ann
Hematol. 93:983–993. 2014.PubMed/NCBI
|
|
31
|
Merkel O, Hamacher F, Laimer D, Sifft E,
Trajanoski Z, Scheideler M, Egger G, Hassler MR, Thallinger C,
Schmatz A, et al: Identification of differential and functionally
active miRNAs in both anaplastic lymphoma kinase (ALK)+
and ALK− anaplastic large-cell lymphoma. Proc Natl Acad
Sci USA. 107:16228–16233. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Qian L, Zhang W, Lei B, He A, Ye L, Li X
and Dong X: MicroRNA-101 regulates T-cell acute lymphoblastic
leukemia progression and chemotherapeutic sensitivity by targeting
Notch1. Oncol Rep. 36:2511–2516. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Liu X, Zou L, Zhu L, Zhang H, Du C, Li Z,
Gao C, Zhao X, Bao S and Zheng H: miRNA mediated up-regulation of
cochaperone p23 acts as an anti-apoptotic factor in childhood acute
lymphoblastic leukemia. Leuk Res. 36:1098–1104. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Gahl RF, Dwivedi P and Tjandra N: Bcl-2
proteins bid and bax form a network to permeabilize the
mitochondria at the onset of apoptosis. Cell Death Dis.
7:e24242016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Jagani H, Kasinathan N, Meka SR and
Josyula VR: Antiapoptotic Bcl-2 protein as a potential target for
cancer therapy: A mini review. Artif Cells Nanomed Biotechnol.
44:1212–1221. 2016.PubMed/NCBI
|
|
36
|
Um HD: Bcl-2 family proteins as regulators
of cancer cell invasion and metastasis: a review focusing on
mitochondrial respiration and reactive oxygen species. Oncotarget.
7:5193–5203. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Sivakumar D and Sivaraman T: A review on
structures and functions of Bcl-2 family proteins from homo
sapiens. Protein Pept Lett. 23:932–941. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yang D, Chen MB, Wang LQ, Yang L, Liu CY
and Lu PH: Bcl-2 expression predicts sensitivity to chemotherapy in
breast cancer: A systematic review and meta-analysis. J Exp Clin
Cancer Res. 32:1052013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kumar SR, Kim DY, Henry CJ, Bryan JN,
Robinson KL and Eaton AM: Programmed death ligand 1 is expressed in
canine B cell lymphoma and downregulated by MEK inhibitors. Vet
Comp Oncol. 15:1527–1536. 2017. View Article : Google Scholar : PubMed/NCBI
|