Introduction
Protein disulfide isomerase (PDI) is a member of the
thioredoxin superfamily that catalyzes the oxidation, reduction and
isomerization of protein disulfide bonds via two redox active sites
consisting of -Cys-Gly-His-Cys- (CXXC motif), which are located in
two thioredoxin fold domains. The functions of PDI are important
for the correct folding of newly synthesized polypeptides in the
endoplasmic reticulum (ER) (1). At
present, 21 genes have been classified as encoding PDI and
associated proteins, and these PDI family proteins have
thioredoxin-like folds or thioredoxin-like active sequences, with a
CXXC motif in their amino acid sequences (2). PDI P5 (P5, also known as PDIA6)
contains two redox active -Cys-Gly-His-Cys- sequences, which are
indispensable for the isomerase activity of this enzyme. In
addition, it has chaperone activity, although the activities of P5
are weaker than those of PDI (3).
Currently, the detailed physiological roles of P5 in cells are
unknown. It was previously reported that P5 is associated with
major histocompatibility complex class I polypeptide-related
sequence A (MICA) on the surface of cancer cells and is required
for the secretion of soluble MICA from cancer cells, which promotes
immune evasion by these cells (4).
Furthermore, P5 is localized not only to the ER but also to the
mitochondria, and MTS-P5 cells, in which P5 is stably expressed in
the mitochondria of Saos-2 cells, are resistant to
H2O2- or rotenone-induced cell death
(5,6). These findings suggested that there are
still unidentified roles for P5 in cells. Previous studies
regarding PDIs revealed important roles for these enzymes in cancer
cells and suggested that they may be promising targets for cancer
therapy (7–10); however, the functional roles of P5
and the significance of targeting this enzyme in cancer cells are
currently unclear.
In order to regulate the activity of P5 in cancer
cells, our previous study screened for specific inhibitors of P5
using a chemical compound library and revealed that anacardic acid
is able to inhibit the reductase activity of P5, but does not
inhibit the activity of other PDI family proteins, such as PDI,
ERp57 or thioredoxin (11).
Furthermore, anacardic acid is able to decrease the secretion of
soluble MICA from cancer cells (11). These results suggested that
targeting P5 on cancer cells may lead to the identification of
novel types of cancer chemotherapy; however, further investigation
into the detailed functional roles of P5 in cancer cells is
required.
The present study detected the expression levels of
P5 on the surface of several normal and cancer cell lines. The
results revealed that P5 expression was increased on the surface of
cancer cells compared with normal cell lines. Furthermore, the
effects of P5 knockdown on cancer cells were determined with
regards to Bip promoter activation, cell growth and migration.
Screening for P5-specific binding proteins was conducted in cancer
cells compared with normal cells, and vimentin was identified as
one such protein. Finally, the effects of P5 knockdown on cancer
cells were determined with regards to the expression levels of
vimentin and epithelial-mesenchymal transition (EMT) markers. Bip
promoter activation and cell morphology were assessed
simultaneously at the single-cell level using a real-time
monitoring method, which utilizes bioluminescence and fluorescence.
The present study also discussed the potency of targeting P5 in
cancer cells as a novel type of cancer treatment.
Materials and methods
Materials
The affinity-purified rabbit anti-P5 antibody used
in this study was kindly provided by Dr T. Komiya (Nagahama
Institute of Bio-Science and Technology, Nagahama, Japan) (6). The rabbit anti-PDI polyclonal antibody
was prepared as previously described (12). Protein G PLUS-agarose was purchased
from Santa Cruz Biotechnology, Inc. (Dallas, TX, USA). Recombinant
human vimentin protein was purchased from PeproTech, Inc. (Rocky
Hill, NJ, USA). Anacardic acid, ribostamycin, bovine serum albumin
(BSA) and purified PDI were purchased from Sigma-Aldrich (Merck
KGaA, Darmstadt, Germany). Temozolomide (TMZ) was purchased from
LKT Laboratories, Inc. (St. Paul, MN, USA). Pre-stained protein
markers for SDS-PAGE and thapsigargin (Tg) were purchased from
Nacalai Tesque, Inc. (Kyoto, Japan). The other reagents were mostly
obtained from Nacalai Tesque, Inc. All reagents used were of
research grade.
Cells and cell culture
The normal and cancer cell lines used in the present
study are presented in Table I.
A172, SNB19, T98G, BT20, MDA-MB-231, T47D, H322, H460, H526,
Panc-1, SU86.86, Caki-1, LNCaP, PA-1, HeLa and WI38 cell lines were
obtained from the American Type Culture Collection (Manassas, VA,
USA). BxPC3, HCT116, SK-OV-3 and OE19 cell lines were obtained from
the European Collection of Authenticated Cell Cultures (Salisbury,
UK). HepG2, HuCCT-1, KMRC-1, DLD-1, LoVo, SW837, PC-3, U937, HL-60,
K562 and THP-1 cell lines were obtained from Health Science
Research Resources Bank (Osaka, Japan). SW48 and SW480 cells were
purchased from DS Pharma Biomedical Co., Ltd. (Osaka, Japan).
Astrocytes (ACBRI371), pancreatic epithelial (PE) cells (ACBRI515),
and hepatocytes (ACBRI3716) were purchased from Cell Systems
(Kirkland, WA, USA) via DS Pharma Biomedical Co., Ltd. U251 and
293T cell lines were obtained from the National Cancer Institute,
Frederick Cancer Research Facility, Division of Cancer Treatment
Tumor Repository Program (Frederick, MD, USA) and RIKEN BioResource
Center (Tsukuba, Japan), respectively. The ML-RCC cell line was
kindly provided by Dr R. Puri (Center for Biological Evaluation,
Food and Drug Administration, Silver Spring, MD, USA); the cell
line was established as previously described (13). The cells were cultured in RPMI-1640
(Nacalai Tesque, Inc.) (U251, A172, SNB19, BT20, MDA-MB-231, T47D,
H322, H460, H526, BxPC3, SU86.86, HuCCT-1, ML-RCC, DLD-1, SW837,
LNCap, OE19, U937, HL-60, K562 and THP-1), McCoy's 5A (Thermo
Fisher Scientific, Inc., Waltham, MA, USA) (Caki-1, HCT-116 and
SK-OV-3), Dulbecco's modified Eagle's medium (DMEM) (Nacalai
Tesque, Inc.) (Panc-1, KMRC-1, HeLa and 293T), MEM (Nacalai Tesque,
Inc.) (T98G, HepG2, PC-3, PA-1 and WI38), DMEM/F12 (Nacalai Tesque,
Inc.) (LoVo, SW48 and SW480) or Complete medium kit (cat no.
4Z0-500-R; Cell Systems) (astrocytes, PE cells and hepatocytes)
containing 10% fetal bovine serum (cat no. 172012-500ML;
Sigma-Aldrich; Merck KGaA), 100 µg/ml penicillin and 100 µg/ml
streptomycin at 37°C in an atmosphere containing 5% CO2
and 95% ambient air. All of the cell lines were used for flow
cytometry and western blotting to assess the expression levels of
P5. U251/Luc, SNB19, PE and astrocytes were used for the cell
growth assay post-transfection with siRNA. U251, PE, BT20, H322,
DLD-1, PA-1 and U937 cells were used for immunoprecipitation.
U251/Luc cells were used for all other experiments described in
this study.
 | Table I.Normal and cancer cell lines used to
analyze the expression levels of P5. |
Table I.
Normal and cancer cell lines used to
analyze the expression levels of P5.
| Organ | Cell lines |
|---|
| Cancer |
|
Brain | U251, A172, SNB19,
T98G |
|
Breast | BT20, MDA-MB-231,
T47D |
|
Lung | H322, H460,
H526 |
|
Pancreas | BxPC3, Panc-1,
SU86.86 |
|
Liver | HepG2, HuCCT-1 |
|
Kidney | Caki-1, KMRC-1,
ML-RCC |
|
Colon | DLD-1, HCT116,
LoVo, SW48, SW480 |
|
Rectum | SW837 |
|
Prostate | LNCaP, PC-3 |
|
Ovary | PA-1, SK-OV-3 |
| Uterine
cervix | HeLa |
|
Esophagus | OE19 |
|
Blood | U937, HL-60, K562,
THP-1 |
| Normal |
|
Brain | Astrocyte |
|
Lung | WI38 |
|
Pancreas | Pancreatic
epithelial cells |
|
Liver | Hepatocyte
cells |
|
Kidney | 293T |
Expression and purification of human
P5
Recombinant human P5 protein was purified using a
Ni2+-chelating Resin Column (GE Healthcare Bio-Sciences,
Pittsburgh, PA, USA), as previously described (11,12).
Specificity of the anti-P5 antibody to
P5 protein
PDI, P5 and BSA proteins (1.6 µg) were separated by
12.5% SDS-PAGE, and western blotting was performed using rabbit
anti-PDI (1:6,000) or anti-P5 (1:6,000) antibodies as primary
antibodies, and horseradish peroxidase-conjugated donkey
anti-rabbit immunoglobulin G (IgG) (1:2,000; cat. no. NA934-1ML; GE
Healthcare Bio-Sciences) as a secondary antibody. The staining of
gel and membranes following SDS-PAGE and western blotting was
performed using Coomassie Brilliant Blue (Nacalai Tesque, Inc.) and
the Peroxidase Stain DAB kit (Nacalai Tesque, Inc.),
respectively.
Western blotting
Western blotting was conducted as previously
described (14). Briefly, total
protein extracts were prepared from cells lysed with reporter lysis
buffer (Promega Corporation, Madison, WI, USA). Subsequently, total
protein concentration was quantified by measuring the absorbance at
280 nm using a NanoDrop 1000 spectrophotometer (NanoDrop; Thermo
Fisher Scientific, Inc., Wilmington, DE, USA). Proteins (20
µg/lane) were separated by 12.5% SDS-PAGE and transferred to
nitrocellulose membranes using the iBlot system (Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocol. The
membranes were blocked with 5% skim milk (w/v) in PBS for 90 min at
room temperature, after which, the membranes were probed with
anti-P5 (1:6,000), anti-Bip (1:500; cat. no. MAB4846; R&D
Systems, Inc. Minneapolis, MN, USA), anti-vimentin (1:800; cat. no.
ab20346; Abcam, Cambridge, UK) or anti-β-actin (1:4,000; cat no.
A5316-.2ML; Sigma-Aldrich; Merck KGaA) as primary antibodies
overnight at 4°C, and with a horseradish peroxidase-conjugated
donkey anti-rabbit IgG (1:2,000; cat. no. NA934-1ML; GE Healthcare
Bio-Sciences) or sheep anti-mouse IgG (1:2,000; cat. no. NA931-1ML;
GE Healthcare Bio-Sciences) secondary antibody for 3 h at room
temperature. The blots were then analyzed with Chemi-Lumi One Super
reagent (Nacalai Tesque, Inc.) using a LAS-3000 LuminoImage
analyzer (Fujifilm, Tokyo, Japan). Densitometric analysis of the
bands obtained from western blotting was performed using Multi
Gauge software V3.0 (Fujifilm).
Flow cytometry
Flow cytometry was performed as previously described
(15). Briefly, the
affinity-purified rabbit anti-P5 antibody was labeled with
6-(fluorescein-5-carboxamido) hexanoic acid succinimidyl ester
(FAM-X) using a FAM-X labeling kit (KPL, Inc., Gaithersburg, MD,
USA), according to the manufacturer's protocol. Subsequently,
1.0×105 normal or cancer cells were incubated with the
labeled antibody for 2 h at room temperature. After incubation, the
cells were washed twice with PBS and flow cytometry was performed
using FACSCalibur (BD Biosciences, San Jose, CA, USA). The data
were analyzed using CellQuest software version 6.0 (BD
Biosciences).
Small interfering (si)RNA
transfection
Stealth RNA interference (RNAi) duplexes, negative
control medium GC (cat. no. 12935300; Thermo Fisher Scientific,
Inc.) and P5 siRNA (5′-CCAUAUCCUUGAUACUGGAGCUGCA-3′ and
5′-UGCAGCUCCAGUAUCAAGGAUAUGG-3′) (Invitrogen; Thermo Fisher
Scientific, Inc.) were used for P5 knockdown experiments, as
previously described (16).
Briefly, U251/Luc, SNB19, PE cells and astrocytes were grown to
40–50% confluence on a 6-well plate or 35 mm glass-bottomed dish;
the cells were and transfected with 0.125 or 0.25 µM siRNA in
incomplete medium without FBS, penicillin and streptomycin. siRNA
transfection was performed using Lipofectamine RNAiMAX (Invitrogen;
Thermo Fisher Scientific, Inc.), according to the manufacturer's
protocol (6,16). After transfection, cells were kept
at 37°C for 3 h, and medium was replaced with complete medium. A
total of 24 h post-transfection, bioluminescence imaging was
performed, and 48 or 72 h post-transfection, cell samples were
prepared for western blotting or reverse transcription-polymerase
chain reaction (RT-PCR). A total of 72 h post-transfection, cells
were seeded in 96-well plates for cell growth and migration
assays.
Cell growth, viability and migration
assays
Cell growth was assessed using the WST-8 assay, as
previously described (15).
Briefly, cells were seeded in 96-well plates at a concentration of
3,000 cells/well, and the number of living cells was measured using
Cell Count Reagent SF (Nacalai Tesque, Inc.). Absorbance was
measured at a wavelength of 450 nm using a microplate reader (GE
Healthcare Bio-Sciences). Cell viability was also assessed using
the WST-8-assay, as previously described (15). Briefly, U251/Luc cells were seeded
in 96-well plates as aforementioned, and were treated with various
concentrations (0, 25, 50, 100 and 200 µM) of TMZ in the presence
or absence of 25 µM anacardic acid, 1 mM ribostamycin, or a
combination of these compounds. Cell viability was calculated after
48 h; the viability of untreated control cells was set to 100%.
Cell migration was assessed using an Oris™ Cell migration assay
(Platypus Technologies, LLC, Madison, WI, USA), according to the
manufacturer's protocol.
Immunoprecipitation
Immunoprecipitation was performed using an anti-P5
antibody, as previously described (14). Briefly, cancer and normal cells were
washed with ice-cold PBS and were lysed with
radioimmunoprecipitation assay buffer (Nacalai Tesque, Inc.) on ice
for 15 min. Cell lysates were collected after centrifugation at 300
× g for 5 min at 4°C, and the total protein concentration of the
supernatant was determined spectrophotometrically using a NanoDrop
1000 spectrophotometer (NanoDrop; Thermo Fisher Scientific, Inc.).
The supernatant from the cell lysate containing 100 µg of protein
was incubated with 50 µl Protein G PLUS-agarose solution at 4°C for
1 h, and following centrifugation at 19,000 × g for 3 min, the
supernatant was transferred to a new tube. The supernatant was
incubated at 4°C for 1 h following addition of an anti-P5 antibody
(1:100), and was further incubated at 4°C overnight following the
addition of 50 µl protein G PLUS-agarose solution. The agarose was
collected by centrifugation at 19,000 × g for 3 min, washed at
least three times with cold PBS, and boiled in SDS-PAGE sample
buffer at 98°C for 5 min. The samples were then separated by 12.5%
SDS-PAGE, and the bound proteins were visualized by silver staining
using Sil-Best Stain One (Nacalai Tesque, Inc.), according to the
manufacturer's protocol. To examine the binding of vimentin with P5
in cells by immunoprecipitation, total cell lysates from U251, PE,
BT20, H322, DLD-1, PA-1 and U937 cells were prepared, and
immunoprecipitation was performed using the anti-P5 antibody as
aforementioned. The samples were separated by 12.5% SDS-PAGE, and
then western blotting was performed using anti-P5 and anti-vimentin
antibodies.
Mass spectrometry and protein
identification
The bands of interest were excised from the gel
using a box cutter after silver staining, and in-gel digestion was
performed using the In-gel tryptic digestion kit (Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocol. Gel
trypsin digestion and protein identification by liquid
chromatography-tandem mass spectrometry (LC/MS/MS) were performed
as previously described (14) at
the Medical Research Support Center, Kyoto University (Kyoto,
Japan). Acquired datasets were analyzed using ProteinPilot Software
v. 4.5 Beta (SCIEX, Tokyo, Japan) with the Paragon algorithm and a
combined database of UniProtKB/Swiss-Prot data and known
contaminants (SCIEX).
Biomolecular interactions
Surface plasmon resonance experiments were performed
using the Biacore T100 system (GE Healthcare Bio-Sciences), as
previously described (11).
Briefly, recombinant human vimentin protein was immobilized on the
surface of a CM5 sensor chip with N-hydroxysuccinimide and
N-ethyl-N'-(dimethylaminopropyl) carbodiimide
activation chemistry, according to the manufacturer's protocol. As
an analyte, purified recombinant P5 protein was injected over the
flow cell, and HBS-EP buffer (0.01 M HEPES, 0.15 M NaCl, 0.005%
Tween-20, 3 mM EDTA, pH 7.4) was used as a running buffer to
inhibit nonspecific binding. An approximate equilibrium
dissociation constant (Kd) value was obtained as previously
described using Biacore T100 evaluation software ver. 2.0.2 (GE
Healthcare Bio-Sciences) (17).
RT-PCR
RT-PCR was performed as previously described
(18). Briefly, following isolation
of total RNA using a NucleoSpin RNA kit (MACHEREY-NAGEL GmbH &
Co. KG, Düren, Germany), RT was conducted using a ReverTra Ace qPCR
RT kit (Toyobo Life Science, Osaka, Japan), according to the
manufacturer's protocol. Subsequently, each 1-µl aliquot of cDNA
was amplified in a final 50-µl PCR mixture containing Titanium taq
DNA Polymerase (1:100; Takara Bio, Inc., Otsu, Japan) and 0.2 mM
dNTP mixture solution from the Long Range PCR kit (Qiagen GmbH,
Hilden, Germany). The PCR thermocycling conditions were as follows:
Initial denaturation at 94°C for 1 min, followed by 30–35 cycles at
94°C for 30 sec, 60–61°C for 30 sec and 72°C for 30 sec, and a
final extension step at 72 °C for 30 sec. For upregulation of the
CHOP gene, which is a positive control for ER stress, U251/Luc
cells were treated with 0.5 µM Tg for 6 h at 37°C, and cells were
collected for the isolation of total RNA as aforementioned.
Specific primers for RT-PCR were as follows: CCAAT-enhancer-binding
protein homologous protein (CHOP), forward
5′-ATCAAAAATCTTCACCACTCTTGAC-3′, reverse
5′-ACTTTCCTTTCATTCTCCTGTTCTT-3′; vimentin, forward
5′-GGTACAAATCCAAGTTTGCTGACC-3′, reverse
5′-CTCAATGTCAAGGGCCATCTTAAC-3′; Snail, forward
5′-CAAGGCCATGTCCGGACCCACACTGGCG-3′, reverse
5′-CTTCCTGCTGGAGCTGGGGAAGGCTGTC-3′; Slug, forward
5′-GGCCAAACATAAGCAGCTGCACTGCG-3′, reverse
5′-CAGATGAGCCCTCAGATTTGACCTGTC-3′; Twist, forward
5′-GCAGACGCAGCGGGTCATGGCCAACG-3′, reverse
5′-CATCCTCCAGACCGAGAAGGCGTAGC-3′; N-cadherin, forward
5′-GACCCATCCACGCCGAGCCCCAGTATCCG-3′, reverse
5′-CACCCTGAAGTTCAGTCATCACCTCCACC-3′; E-cadherin, forward
5′-GCTAGTCTGAGCTCCCTGAACTCCTCAG-3′, reverse
5′-GGGGCCCGCCTCTCTCGAGTCCCCTAGTCG-3′; and GAPDH, forward
5′-GTCTTCACCACCATGGAGAAGGCT-3′ and reverse
5′-CATGCCAGTGAGCTTCCCGTTCA-3′. GAPDH was used as an internal
control. PCR products were run on a 1% agarose gel, which was
stained with GelRed (Biotium, Fremont, CA USA) for UV analysis.
Bioluminescence imaging
U251 cells stably transfected with pBipPro-Luc
(U251/Luc), in which the Bip promoter region was cloned into the
pGL4.14 vector (Promega Corporation), were prepared. Successful
transfection was confirmed using the LV200 imaging system (Olympus
Corporation, Tokyo, Japan), through the observation of
bioluminescence, as previously described (19). Luminescence images at the
single-cell level were obtained using the LV200 imaging system, as
previously described (19).
Briefly, 24 h post-transfection of U251/Luc cells with negative
control or P5 siRNA, images were captured using a ×40 objective
lens at 10-min intervals, and Bip promoter activity was observed
following the addition of D-luciferin. An expression vector of
vimentin fused with PSmOrange (20)
(vim/Orange) was provided by Addgene, Inc. (Cambridge, MA, USA).
For the observation of vim/Orange, BP535-555HQ (Olympus
Corporation) and 570-625RFP (Olympus Corporation) were used as
excitation and emission filters, respectively. U251/Luc cells
stably transfected with vim/Orange (U251/Luc/Orange) were also
prepared in selective medium containing 200 µg/ml hygromycin B
(Nacalai Tesque, Inc.) and 1,000 µg/ml G-418 (Wako Pure Chemical
Industries, Ltd., Osaka, Japan). Successful transfection was
confirmed using the LV200 system, through the observation of both
bioluminescence and fluorescence using the filters. After the
establishment of stable U251/Luc and U251/Luc/Orange cells, they
were transfected with the negative control and P5 siRNAs, as
aforementioned. All data analysis was performed using AQUACOSMOS
ver. 2.6 software (Hamamatsu Photonics, Hamamatsu, Japan).
Statistical analysis
Experiments were repeated at least two times.
Statistical significance was determined using Student's t-test for
pairwise comparisons, whereas multiple comparisons were evaluated
by one-way analysis of variance followed by Dunnett's test using
JMP Pro version 14 (SAS Institute, Inc., Cary, NC, USA). P<0.05
was considered to indicate a statistically significant
difference.
Results
Expression levels of P5 on the surface
of normal and cancer cells
The present study examined the expression levels of
P5 on the surface of normal and cancer cells by
fluorescence-activated cell sorting (FACS) analysis using an
affinity-purified anti-P5 antibody labeled with FAM-X, which was
confirmed to be specific to P5 protein and did not cross-react with
PDI or bovine serum albumin (Fig.
1A). The expression levels of P5 were detected on the cell
lines shown in Table I using FACS
analysis; the results revealed that P5 expression was markedly
increased on the surface of several cancer cell lines, including
U251, T98G, H322, DLD-1, HCT116, SW837, PA-1, U937, K562 and THP-1
cells compared with normal cell lines, and it was expressed at the
highest levels on the surface of K562 and THP-1 cells, which are
chronic myelogenous and acute monocytic leukemia cell lines
(21,22), respectively (Fig. 1B). A constant increase in the
surface expression of P5 was observed in glioblastoma (U251, A172,
SNB19, and T98G), breast (BT20, MDA-MB-231, T47D), colon (DLD-1,
HCT116, LOVO, SW480, SW837), and ovarian and uterine cervical
(PA-1, SK-OV-3, HeLa) cancer, and leukemia (U937, HL-60, K562,
THP-1) cell lines (Fig. 1B). In
addition, the expression levels of P5 were compared between normal
and cancer cell lines; its expression was significantly increased
on the surface of cancer cell lines, compared with normal cells;
leukemia cell lines were not included in this analysis (Fig. 1C). The present study also
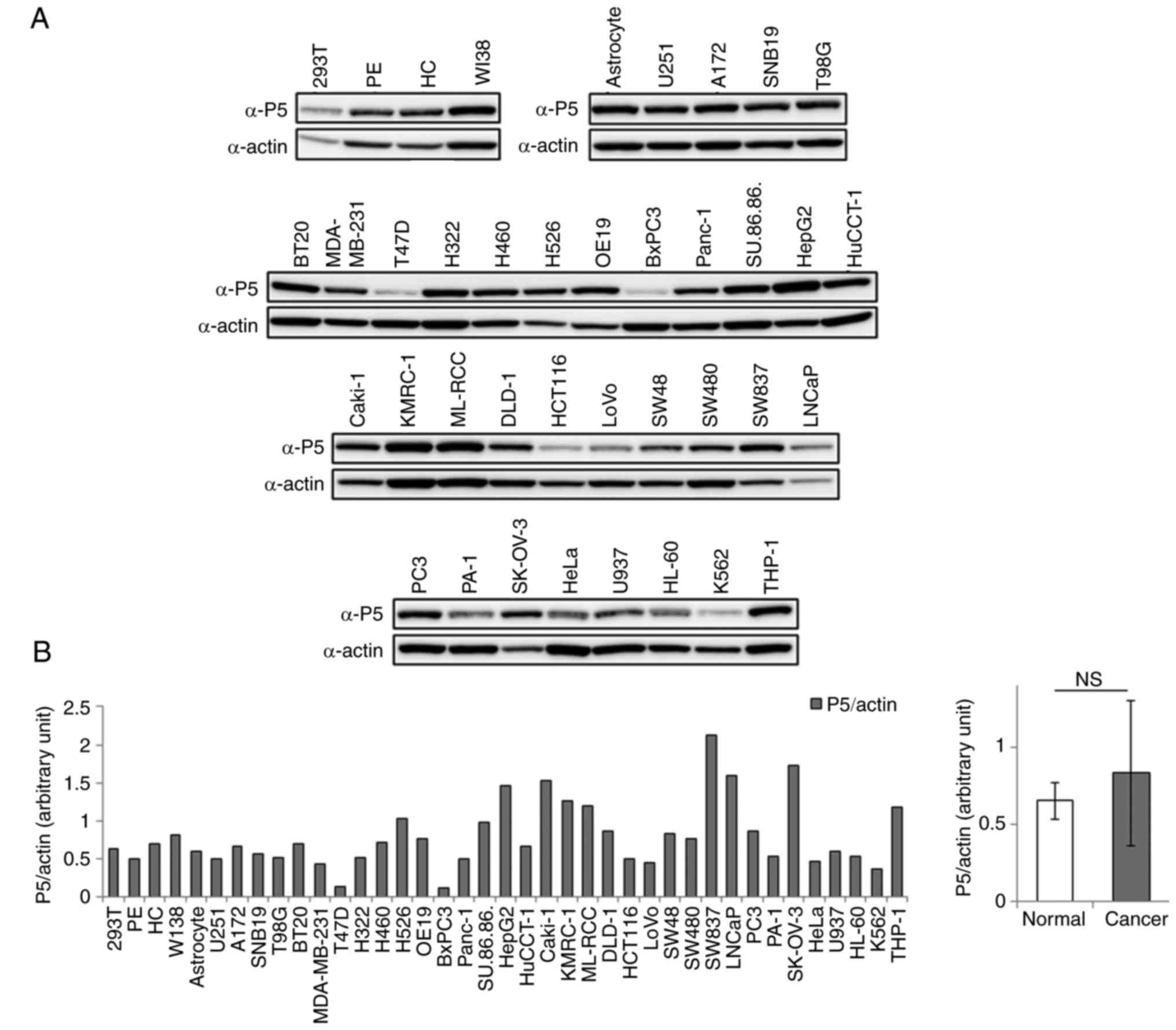
investigated the expression of P5 in these cell lines by western
blotting using an anti-P5 antibody. P5 was expressed in all cell
lines examined; however, its expression levels differed among the
cell lines (Fig. 2A and B). When
the expression levels of P5 in total cell lysates were compared
between normal and cancer cell lines, it was revealed that its
expression was not significantly different between normal and
cancer cells; leukemia cell lines were not included in this
analysis (Fig. 2B). These results
suggested that P5 expression may be increased on the surface of
several cancer cells compared with normal cells; however, the
enzyme was expressed at a constant level within both normal and
cancer cells.
 | Figure 1.Expression levels of P5 on the
surface of normal and cancer cells. (A) Specificity of the
affinity-purified rabbit anti-P5 antibody. All proteins were
separated by SDS-PAGE. Western blotting was performed using a
rabbit anti-PDI raised against purified bovine PDI or
affinity-purified anti-P5 antibody. Lane 1, prestained marker
proteins; lane 2, purified PDI protein; lane 3, purified
recombinant P5 protein from Escherichia coli; lane 4, bovine
serum albumin. (B) Expression levels of P5 on the surface of normal
and cancer cells, as determined by fluorescence-activated cell
sorting analysis using an affinity-purified anti-P5 antibody
labeled with FAM-X. The control group was not incubated with
anti-P5, and was set at 1.0 for all cell lines. The assay was
repeated two times to confirm the results. Data were not subjected
to statistical analysis. (C) Fold fluorescence intensity of anti-P5
staining on the surface of normal cell lines compared with cancer
cell lines, with the exception of the leukemia cell lines (U937,
HL-60, K562 and THP-1). Data are presented as the means ± standard
deviation. **P<0.01. FAM-X, 6-(fluorescein-5-carboxamido)
hexanoic acid succinimidyl ester; HC, hepatocytes; PDI, protein
disulfide isomerase; PE, pancreatic epithelial cells. |
Effects of P5 knockdown in cancer
cells on activation of the Bip promoter during cancer cell
growth
Alongside a previous report regarding the role of P5
in cancer cells (4), the present
study indicated that P5 may have a significant role in cancer
cells. Therefore, the present study further investigated the
functional roles of P5 in cancer cells. The effects of P5 knockdown
in cancer cells on activation of the Bip promoter during cancer
cell growth were analyzed, as our previous study indicated that the
Bip promoter is periodically activated, according to real-time
monitoring using bioluminescence at the single-cell level in U251
glioblastoma cells (19). Following
siRNA-induced knockdown of P5, western blotting confirmed that its
expression was reduced in cancer cells (Fig. 3A). Real-time bioluminescence
monitoring at the single-cell level demonstrated that Bip promoter
activation was observed in U251/Luc cells even post-transfection
with negative control siRNA during cancer cell growth (Fig. 3B and C), as shown in our previous
study (19); however, it was not
activated post-transfection with P5 siRNA, although Bip promoter
activity was maintained and luminescence was still observed in the
cancer cells (Fig. 3B and C). Since
the Bip promoter is periodically activated, particularly in
dividing cells during cell growth (19), these results suggested that knocking
down P5 in cancer cells may affect Bip promoter activity and cancer
cell division during cell growth. The present study investigated
the effects of P5 knockdown in U251/Luc cells on the expression of
Bip and CHOP, which are markers of ER stress responses; there was
no significant difference in their expression between the groups
transfected with the negative control or P5 siRNAs (Fig. 3D). Therefore, knocking down P5 in
cancer cells may not induce an ER stress response.
 | Figure 3.Effects of siRNA-induced P5 knockdown
in cancer cells on activation of the Bip promoter. (A) Expression
levels of P5 following siRNA transfection. U251/Luc cells were
transfected with or without negative control or P5 siRNAs, and at
48 and 72 h post-transfection, the expression levels of P5 were
detected by western blotting. β-actin was used as the loading
control. (B) Luminescence images of U251/Luc cells transfected with
negative control or P5 siRNAs obtained by the LV200 system at 0,
24, 48 and 72 h. Squares in the luminescence images indicate ROIs,
in which luminescence intensity was measured for time-lapse
analysis at the single-cell level. Scale bars, 100 µm. (C) Time
course analysis of Bip promoter activation on single-cell level
imaging. Time course analysis of Bip promoter activation in
U251/Luc cells post-transfection with negative control or P5 siRNAs
was performed using the LV200 system. (D) Effects of siRNA-induced
P5 knockdown on the expression of Bip and CHOP. Upper panels,
U251/Luc cells were transfected with or without negative control or
P5 siRNAs, and at 48 and 72 h post-transfection, the protein
expression levels of Bip protein were detected by western blotting.
β-actin was used as the loading control. Lower panels, U251/Luc
cells were transfected with or without negative control or P5
siRNAs, and at 48 and 72 h post-transfection, total RNA was
extracted from these cells and reverse transcription-polymerase
chain reaction was conducted. Treatment of U251/Luc cells with 0.5
µM Tg for 6 h was used as a positive control for upregulation of
CHOP expression. α, anti; CHOP, CCAAT-enhancer-binding protein
homologous protein; RLU, relative luminescence unit; ROI, region of
interest; siRNA, small interfering RNA; Tg, thapsigargin. |
Effects of P5 knockdown in cancer
cells on cell growth and migration
Since Bip promoter activation during cancer cell
growth was not observed post-transfection with P5 siRNA, the
present study examined the effects of siRNA-induced P5 knockdown in
U251/Luc cancer cells on cell growth and migration. P5 knockdown in
cancer cells significantly inhibited cell growth, as assessed using
the WST-8 assay, compared with in cells transfected with or without
negative control siRNA (Fig. 4A);
this phenomenon was also observed when using another glioblastoma
cell line, SNB19 (data not shown). However, transfection of normal
PE or astrocyte cells with P5 siRNA, in which the knockdown of P5
expression was confirmed by western blotting, did not inhibit cell
growth (Fig. 4A). The present study
also investigated the effects of P5 knockdown in U251/Luc cancer
cells on cell migration; P5 knockdown reduced cancer cell migration
compared with in cells transfected with or without negative control
siRNA (Fig. 4B). These results
suggested that P5 may serve an important role in cancer cell growth
and migration.
 | Figure 4.Effects of siRNA-induced P5 knockdown
on cancer and normal cell growth, and cancer cell migration. (A)
Effects of P5 knockdown on cancer (upper graph) and normal (middle
and lower graphs) cell growth. U251/Luc cells, and normal PE cells
and astrocytes were transfected with or without negative control or
P5 siRNAs; 72 h post-transfection, cell growth was monitored using
the WST-8 assay. Data are presented as the means ± SD from three
independent experiments, and each assay was performed in
triplicate. **P<0.01. Inset blots indicate the expression of P5
in PE cells or astrocytes 72 h post-transfection with siRNA. (B)
Effects of P5 knockdown on cancer cell migration. U251/Luc cells
were transfected with or without negative control or P5 siRNAs; 72
h post-transfection, the cells were seeded onto a 96-well migration
assay plate and cell migration was assessed by confocal microscopy
(left images) and a fluorescence microplate reader (right graphs),
after staining with cell tracker green at the indicated time
points. Data are presented as the means ± standard deviation of
technical 8 replicate wells on a 96-well plate, and the assay was
repeated two times to confirm the results. Data were not subjected
to statistical analysis. Scale bars, 500 µm. α, anti; n.s., not
significant; OD, optical density; siRNA, small interfering RNA. |
Identification of a P5-binding protein
in cancer cells
Since the present findings suggested that P5 serves
important roles in cancer cells, further functional analysis of
this enzyme was conducted by screening for specific proteins that
bind to P5 in cancer cells compared with normal cells.
Immunoprecipitation was performed using an anti-P5 antibody, and
several proteins were revealed to specifically bind to P5 in cancer
cells compared with normal cells (Fig.
5A). After performing the immunoprecipitation experiments
several times, one of the protein bands visualized by SDS-PAGE and
silver staining was identified as a specific protein that bound to
P5 in cancer cells (Fig. 5A). To
identify this binding protein, the band was excised from the gel,
in-gel digested and analyzed by LC-MS/MS. The band was identified
as human vimentin protein (data not shown). Subsequently,
immunoprecipitation with an anti-P5 antibody using a total cell
lysate from U251 cells, and western blotting using an anti-vimentin
antibody, confirmed that vimentin could bind to P5. In addition,
vimentin was revealed to bind to P5 predominantly in U251
glioblastoma cells (Fig. 5B).
Subsequently, the present study performed a biomolecular
interaction analysis with purified recombinant P5 and vimentin
proteins using the Biacore T100 system; vimentin immobilized on the
sensor chip was revealed to interact with P5 protein and the Kd
value of this interaction was 1.13±0.26×10−5 M (Fig. 5C). These results suggested that
vimentin may bind to P5 specifically and is a P5-binding protein
predominantly in glioblastoma cells.
 | Figure 5.Isolation and identification of a
specific P5-binding protein in cancer cells. (A)
Immunoprecipitation of proteins associated with P5 in cancer and
normal cells. Whole cell lysates were prepared from normal (PE) and
cancer (U251) cells, and associated proteins were
immunoprecipitated using an α-P5 antibody. Following SDS-PAGE for
the separation of proteins bound to P5, silver staining was
performed to visualize these proteins. The arrow indicates a
P5-binding protein predominantly detected in cancer cells, which
was identified by liquid chromatography-tandem mass spectrometry.
(B) Immunoprecipitation of P5 with vimentin. Whole cell lysates
from U251 and PE cells were prepared, and associated proteins were
captured using protein G agarose resin. Vimentin was detected by
western blotting using an α-vimentin antibody. Total cell U251
indicates the positive control for the detection of vimentin and
P5, in which 30 µg protein from whole U251 cell lysates was loaded
onto SDS-PAGE without immunoprecipitation, and detected with
α-vimentin and α-P5 antibodies (upper panel). Whole cell lysates
from U251, BT20, H322, DLD-1, PA-1, U937 and PE cells were also
immunoprecipitated using an α-P5 antibody, and western blotting was
performed for the detection of vimentin (lower panel). Detection of
P5 in western blotting was performed as a control for
immunoprecipitation. PE, pancreatic epithelial. (C) Sensorgrams of
the binding of P5 protein to immobilized vimentin as determined by
biosensor analysis. Recombinant vimentin protein was immobilized on
the surface of sensor chips, and Purified human P5 protein was
injected at various concentrations. Thin and thick arrows indicate
the beginning and end of sample injection, respectively. α,
anti. |
Effects of knocking down P5 in cancer
cells on cell morphology and EMT marker proteins
Since vimentin was revealed to predominantly bind to
P5 in glioblastoma cells, the present study examined the effects of
siRNA-induced P5 knockdown on the expression of vimentin in
glioblastoma cells. P5 knockdown did not affect the expression of
vimentin, as assessed by RT-PCR (Fig.
6A). Conversely, P5 knockdown affected the morphology of
glioblastoma cells compared with cells transfected with negative
control siRNA (Fig. 6B). The
present study also conducted real-time monitoring, and
simultaneously observed luminescence and fluorescence at the
single-cell level using U251/Luc/Orange cells and the LV200 system;
activity of the Bip promoter was decreased in cancer cells
transfected with P5 siRNA and the expression pattern of vimentin
was different from that in cells transfected with negative control
siRNA (Fig. 6C). In addition, P5
knockdown inhibited the division of glioblastoma cells and induced
death of cells that could not divide, as observed by the LV200
system (data not shown). These results suggested that P5 serves
important roles in the division of glioblastoma cells and the
localization of vimentin in these cells. Since vimentin is well
known as a member of the intermediate filament family (23), and is widely used as a marker of
EMT, which occurs during the metastasis of cancer cells (24,25),
the effects of siRNA-induced P5 knockdown in glioblastoma cells on
the expression of EMT marker proteins, such as Snail, Slug, Twist,
N-cadherin and E-cadherin, were determined. P5 knockdown in
glioblastoma cells decreased the expression of Snail and increased
the expression of Slug; however, it had no effect on the expression
of Twist, N-cadherin and E-cadherin, as assessed by RT-PCR
(Fig. 6D).
 | Figure 6.Effects of P5 knockdown on the
expression of vimentin and EMT marker proteins in cancer cells. (A)
Effect of siRNA-induced P5 knockdown on the expression of vimentin
in cancer cells. U251/Luc cells were transfected with or without
negative control or P5 siRNAs, and at 48 or 72 h post-transfection,
total RNA was extracted from the cells and RT-PCR was performed.
(B) Effects of siRNA-induced P5 knockdown on cancer cell
morphology. U251/Luc cells were transfected with negative control
or P5 siRNAs, and 48 h post-transfection, differential interference
contrast images were obtained using an Olympus FV1000 confocal
laser scanning microscope with ×32 objective lens. Scale bars, 100
µm. (C) Images of real-time monitoring by simultaneous observation
of bioluminescence and fluorescence. U251/Luc/Orange cells were
transfected with negative control or P5 siRNAs, and at 48 h
post-transfection, real-time monitoring of activity of the Bip
promoter and the expression pattern of vimentin in cancer cells was
performed by simultaneous observation of bioluminescence and
fluorescence at the single-cell level using the LV200 system with a
high magnification lens (×100). Green and orange in the images
indicate Luc (bioluminescence) and Orange (fluorescence),
respectively. Scale bars, 50 µm. (D) Effects of siRNA-induced P5
knockdown on the expression of EMT markers. U251/Luc cells were
transfected with or without negative control or P5 siRNAs, and at
48 or 72 h post-transfection, total RNA was extracted from these
cells. Reverse transcription-polymerase chain reaction for Snail,
Slug, Twist, N-cadherin and E-cadherin was then performed. GAPDH
was used as an internal control. At least three independent
transfections were performed and confirmed in all assays. EMT,
epithelial-mesenchymal transition; siRNA, small interfering
RNA. |
Effects of anacardic acid and
ribostamycin on the cytotoxic activity of TMZ against glioblastoma
cells
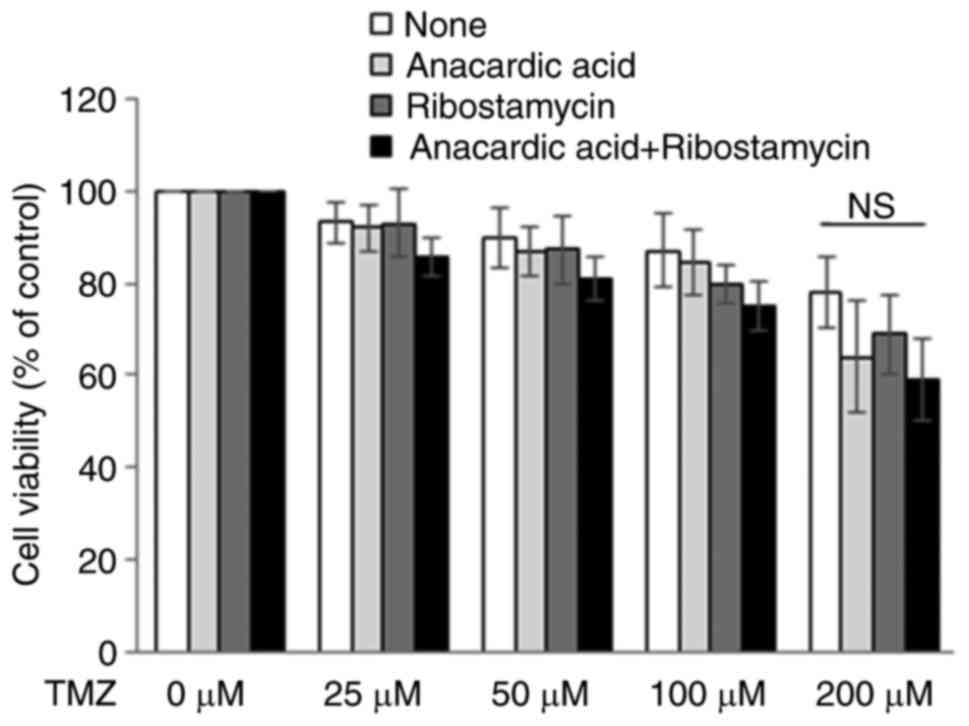
The present study investigated the effects of
anacardic acid on the cytotoxic activity of TMZ, in order to
evaluate the availability of P5 for molecular targeting. In
response to anacardic acid, the cytotoxic activity of TMZ against
glioblastoma cells was not significantly affected (Fig. 7). However, a slight increase in the
cytotoxic activity of TMZ was detected in the presence of both
anacardic acid and ribostamycin; however, this finding was still
not significant (Fig. 7).
Discussion
The present study examined the expression levels of
P5 on the surface of several normal and cancer cell lines, and
demonstrated that its expression was increased in numerous cancer
cell lines compared with normal cell lines. However, the expression
of P5 in cell lysates, as assessed by western blotting, did not
significantly differ between normal and cancer cells. Taken
together with a previous report by Kaiser et al, in which P5
was suggested to serve significant roles in the secretion of
soluble MICA from the surface of cancer cells (4), these results indicated that P5 on the
surface of cancer cells may be a novel anticancer target.
Glioblastoma is the most common and aggressive
malignancy of the central nervous system, and its prognosis remains
dismal, despite recent developments in its treatment (26). Therefore, the identification and
functional analysis of novel candidate targets in glioblastoma
cells will be indispensable for the improvement of chemotherapy.
Since the expression levels of P5 on the surface of glioblastoma
cells were increased compared with normal cell lines; however,
marked differences in expression were not observed among these
glioblastoma cell lines. Therefore, it was suggested that the
expression levels of P5 would be constantly increased on the
surface of glioblastoma, thus the present study focused on the
functional analysis of P5 in glioblastoma cells. Among the
glioblastoma cell lines used in this study, U251 cells are commonly
used as experimental models of glioblastoma, and there are many
reports and publications using this cell line. In addition, we
previously reported that the transfection efficiency of plasmid DNA
into U251 was sufficiently successful among the cell lines analyzed
(27), suggesting that U251 cells
may be used for experiments that introduce target genes into cells
via plasmid DNA to establish a stable cell line, or for knockdown
of genes with siRNA. The present study successfully established
U251 cell lines stably transfected with a reporter vector, in which
the Bip promoter region was cloned into a pGL4.14 vector for
detection by reporter assay and bioluminescence imaging using the
LV200 system; our previous study also reported that this cell line
was available for the evaluation of chemical chaperones by
real-time bioluminescence imaging (19). The present study used U251 and
U251/Luc cells for functional analysis of P5, including
bioluminescence imaging at the single-cell level. Although P5 has
an ER retention signal sequence (Lys-Asp-Glu-Leu) at its
C-terminus, several reports have demonstrated that PDI family
proteins, including P5, localize not only to the ER but also to the
nucleolus (28), cell surface
(4,29,30),
and mitochondria (5,6). It was recently reported that Bip
(78-kDa glucose-regulated protein), which also has an ER retention
signal sequence at its C-terminus, mainly exists as a peripheral
protein on the plasma membrane of stressed cancer cells through its
interaction with other cell surface proteins, such as
glycosylphosphatidylinositol-anchored proteins, and that Bip
expression on the cell surface requires its substrate-binding
activity (31). Since several types
of cancer cells activate numerous signal transduction pathways
during the ER stress response to maintain ER homeostasis (32), and P5 also possesses chaperone
activity, it has been suggested that P5 may be translocated to the
surface of stressed cancer cells via a mechanism similar to that of
Bip, by its substrate-binding site.
It has recently been reported that Bip has a key
role as a prosurvival component in cancer cells, which can provide
protection from cell death (33).
In addition, it has been hypothesized that Bip is a novel target
for increasing chemosensitivity in malignant gliomas (34). On the basis of these reports, our
previous study performed real-time monitoring of Bip promoter
activity during U251 glioblastoma cell growth using a
bioluminescence imaging technique at the single-cell level, and
reported how such bioluminescence imaging techniques could be used
to analyze real-time promoter activity (19). In addition, it has been reported
that P5 forms a noncovalent complex with Bip and cooperates with
the chaperone protein toward client proteins (35). Taken together with these previous
reports and observations, the present study investigated the
effects of P5 knockdown in U251 glioblastoma cells on Bip promoter
activation by bioluminescence imaging at the single-cell level; the
results revealed that activation of the Bip promoter during cancer
cell growth was affected by transfection with P5 siRNA compared
with negative control siRNA. P5 knockdown in glioblastoma cells
also inhibited cell growth and migration, although it did not
affect the growth of normal PE cells and astrocytes. Therefore, it
has been suggested that P5 may serve an important role in the
growth of glioblastoma cells. Since Bip knockdown in glioblastoma
cells also induces the ER stress response, as determined by CHOP
upregulation following Bip knockdown (34), the present study examined the
effects of siRNA-induced P5 knockdown on the expression levels of
Bip and CHOP, and revealed that it had no effect on their
expression. Therefore, knocking down P5 in cancer cells may not
induce the ER stress response; this phenomenon may be considered
another advantage for targeting P5 in glioblastoma cells, which
operates via a mechanism different from that of Bip knockdown,
since the ER stress response in cancer can also increase cell
survival and protect against cell death (33).
To further elucidate the functional roles of P5 in
glioblastoma cells, the present study screened for specific
P5-binding proteins in glioblastoma cells compared with in normal
cells, and successfully identified vimentin as a P5-binding protein
in glioblastoma cells. Vimentin is a well-known member of the
intermediate filament family (23),
and it has been reported that vimentin induces cell shape
alterations during EMT (24).
Notably, P5 knockdown induced a morphological alteration in
glioblastoma cells and affected the expression of EMT markers Snail
and Slug; however, it did not affect the expression of Twist,
N-cadherin and E-cadherin. Since vimentin did not bind to P5 in
normal cells, and P5 knockdown did not affect normal cell growth,
these results suggested that P5 may participate in the
stabilization or regulation of vimentin via protein-protein
interactions, and in EMT homeostasis, in glioblastoma cells.
Further investigations are required to validate this hypothesis.
Since Snail has an oncogenic role in glioblastoma by promoting EMT
(25) and P5 knockdown in
glioblastoma cells reduced the expression of Snail, targeting P5 in
glioblastoma cells may also increase the efficacy of treatment for
this malignancy.
The present study demonstrated that simultaneous
real-time monitoring of bioluminescence and fluorescence at the
single-cell level using the LV200 system could provide useful
information about the effects of P5 knockdown in glioblastoma cells
on Bip promoter activity and the expression pattern of vimentin.
Bioluminescence imaging has several advantages, such as low
background and high quantification (36), which are necessary for the analysis
of promoter activity with a reporter gene; the avoidance of low
levels of damage to living cells from excitation illumination
(37); and longer available
observation periods (38).
Fluorescence imaging also has advantages for time-lapse imaging,
such as the observation of expression patterns and localization of
proteins in cells, in which it is possible to obtain clear images
compared with bioluminescence even with short observation periods.
Therefore, the method of simultaneous observation of fluorescence
and bioluminescence at the single-cell level described in the
present study may be considered a novel and attractive approach for
the functional analysis of promoter activity and protein
localization in various cells, including cancer cells.
The present study also investigated the effects of
anacardic acid, which was previously identified as an inhibitor of
the reductase activity of P5 (11),
on the cytotoxic activity of TMZ, which is used clinically for the
treatment of glioblastoma (39), in
order to evaluate the availability of P5 for molecular targeting.
In response to anacardic acid, the cytotoxic activity of TMZ
against glioblastoma cells was not affected; however, the cytotoxic
activity of TMZ was slightly increased in the presence of anacardic
acid and ribostamycin; ribostamycin was previously revealed to
inhibit the chaperone activity of PDI in cells (40). Although these findings were not
significantly different, these results suggested that inhibitors of
both the isomerase and chaperone activities of P5 may be suitable
drugs for the effective enhancement of chemotherapy against
glioblastoma, although further study of P5 in glioblastoma cells is
required to support this suggestion. Therefore, P5 may be
considered a novel, attractive and potent target for the treatment
of glioblastoma.
In conclusion, to the best of our knowledge, this is
the first study to report that one of the proteins that binds to P5
in U251 glioblastoma cells is vimentin, and the present findings
indicated that P5 may be an attractive target for novel molecular
targeted therapy of glioblastoma. However, the results of P5
functional analyses in glioblastoma cells were obtained from
limited cell lines, and further studies are required to confirm the
detailed functional roles of P5 in glioblastoma cells. Taken
together with previous research into the roles of vimentin and EMT
in glioblastoma cells, these observations may be provide useful
information for further studies into the mechanism underlying the
functional roles of P5 in cancer cells, which might assist in the
development of novel treatments for glioblastoma.
Acknowledgements
The authors would like to thank Dr Shinji Ito
(Medical Research Support Center, Kyoto University) for providing
assistance with mass spectrometry and protein identification; Ms.
Mitsuko Tachi, Ms. Nanako Okushima and Ms. Aki Matsumoto for
providing technical assistance with cell culturing (Department of
Pharmacoepidemiology, Kyoto University); and Dr Thoru Komiya for
providing the affinity-purified rabbit anti-P5 antibody. They would
also like to thank Mr. Ryutaro Akiyoshi and Ms. Yoko Hatta-Ohashi
(Olympus Corporation) for providing technical assistance and advice
on bioluminescence imaging using the LV200 system.
Funding
The present study was supported by Grants-in-Aid for
Young Scientists (B) (grant no. 26870287), Grants-in-Aid for
Scientific Research (B) (grant no. 26290052) and Grants-in-Aid for
Scientific Research (C) (grant no. 16K07170) from the Japan Society
for the Promotion of Science. The present study was also supported
in part by a collaboration research fund from Olympus
Corporation.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
TH and KK conceived the study, designed the
experimental methods, and supervised the study. AT and YM performed
the purification of recombinant proteins, FACS and western blot
analysis. AT established the stable cell lines, and performed the
cell migration assay and bioluminescence imaging using the LV200
system. TH and AT performed the rest of the experiments. TH, AT and
KK contributed to the interpretation of the data. TH wrote the
manuscript, and AT and KK reviewed and edited the manuscript. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
Koji Kawakami serves as a scientific advisor to
Olympus Corporation. None of the other authors have any potential
competing interests.
References
|
1
|
Laurindo FR, Pescatore LA and Fernandes
Dde C: Protein disulfide isomerase in redox cell signaling and
homeostasis. Free Radic Biol Med. 52:1954–1969. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Galligan JJ and Petersen DR: The human
protein disulfide isomerase gene family. Hum Genomics. 6:62012.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kikuchi M, Doi E, Tsujimoto I, Horibe T
and Tsujimoto Y: Functional analysis of P5, a protein disulfide
isomerase homologue. J Biochem. 132:451–455. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kaiser BK, Yim D, Chow IT, Gonzalez S, Dai
Z, Mann HH, Strong RK, Groh V and Spies T:
Disulphide-isomerase-enabled shedding of tumor-associated NKG2D
ligands. Nature. 447:482–486. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kimura T, Horibe T, Sakamoto C, Shitara Y,
Fujiwara F, Komiya T, Yamamoto A, Hayano T, Takahashi N and Kikuchi
M: Evidence for mitochondrial localization of P5, a member of the
protein disulphide isomerase family. J Biochem. 144:187–196. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shitara Y, Tonohora Y, Goto T, Yamada Y,
Miki T, Makino H, Miwa M and Komiya T: Mitochondrial P5, a member
of protein disulphide isomerase family, suppresses oxidative
stress-induced cell death. J Biochem. 152:73–85. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lovat PE, Corazzari M, Armstrong JL,
Martin S, Pagliarini V, Hill D, Brown AM, Piacentini M,
Birch-Machin MA and Redfern CP: Increasing melanoma cell death
using inhibitors of protein disulfide isomerases to abrogate
survival response to endoplasmic reticulum stress. Cancer Res.
68:5363–5369. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bonome T, Levine DA, Shih J, Randonovich
M, Pise-Masison CA, Bogomolniy F, Ozbun L, Brady J, Barrett JC,
Boyd J, et al: A gene signature predicting for survival in
suboptimally debulked patients with ovarian cancer. Cancer Res.
68:5478–5486. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Talantov D, Mazumder A, Yu JX, Briggs T,
Jiang Y, Backus J, Atkins D and Wang Y: Novel genes associated with
malignant melanoma but not benign melanocytic lesions. Clin Cancer
Res. 11:7234–7242. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xu S, Butkevich AN, Yamada R, Zhou Y,
Debnath B, Duncan R, Zandi E, Petasis NA and Neamati N: Discovery
of an orally active small-molecule irreversible inhibitor of
protein disulfide isomerase for ovarian cancer treatment. Proc Natl
Acad Sci USA. 109:16348–16353. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Horibe T, Torisawa A, Okuno Y and Kawakami
K: Discovery of protein disulfide isomerase P5 inhibitors that
reduce the secretion of MICA from cancer cells. ChemBioChem.
15:1599–1606. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kimura T, Nishida A, Ohara N, Yamagishi D,
Horibe T and Kikuchi M: Functional analysis of the CXXC motif using
phage antibodies that cross-react with protein disulphide-isomerase
family proteins. Biochem J. 382:169–176. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Obiri NI, Hillman CG, Haas GP, Sud S and
Puri RK: Expression of high affinity interleukin-4 receptors on
human renal cell carcinoma cells and inhibition of tumor cell
growth in vitro by interleukin-4. J Clin Invest. 91:88–93. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kohno M, Horibe T, Ohara K, Ito S and
Kawakami K: The membrane-lytic peptide K8L9 and melittin enter
cancer cells via receptor endocytosis following subcytotoxic
exposure. Chem Biol. 21:1522–1532. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kohno M, Horibe T, Haramoto M, Yano Y,
Ohara K, Nakajima O, Matsuzaki K and Kawakami K: A novel hybrid
peptide targeting EGFR-expressing cancers. Eur J Cancer.
47:773–783. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Honjo Y, Horibe T, Torisawa A, Ito H,
Nakanishi A, Mori H, Komiya T, Takahashi R and Kawakami K: Protein
disulfide isomerase P5-immunopositive inclusions in patients with
Alzheimer's disease. J Alzheimers Dis. 38:601–609. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
MacLeod TJ, Kwon M, Filipenko NR and
Waisman DM: Phospholipid-associated annexin A2-S100A10
heterotetramer and its subunits: Characterization of the
interaction with tissue plasminogen activator, plasminogen, and
plasmin. J Biol Chem. 278:25577–25584. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yang L, Horibe T, Kohno M, Haramoto M,
Ohara K and Kawakami K: Targeting interleukin-4 receptor α with
hybrid peptide for effective cancer therapy. Mol Cancer Ther.
11:235–243. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Horibe T, Okushima N, Torisawa A, Akiyoshi
R, Hatta-Ohashi Y, Suzuki H and Kawakami K: Evaluation of chemical
chaperones based on the monitoring of Bip promoter activity and
visualization of extracellular vesicles by real time
bioluminescence imaging. Luminescence. 33:249–255. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Subach OM, Patterson GH, Ting LM, Wang Y,
Condeelis JS and Verkhusha VV: A photoswitchable orange-to-far-red
fluorescent protein, PSmOrange. Nat Methods. 8:771–777. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lozzio CB and Lozzio BB: Human chronic
myelogenous leukemia cell-line with positive Philadelphia
chromosome. Blood. 45:321–334. 1975.PubMed/NCBI
|
|
22
|
Tsuchiya S, Yamabe M, Yamaguchi Y,
Kobayashi Y, Konno T and Tada K: Establishment and characterization
of a human acute monocytic leukemia cell line (THP-1). Int J
Cancer. 26:171–176. 1980. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Satelli A and Li S: Vimentin in cancer and
its potential as a molecular target for cancer therapy. Cell Mol
Life Sci. 68:3033–3046. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mendez MG, Kojima S and Goldman RD:
Vimentin induces changes in cell shape, motility, and adhesion
during the epithelial to mesenchymal transition. FASEB J.
24:1838–1851. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Myung JK, Choi SA, Kim SK, Wang KC and
Park SH: Snail plays an oncogenic role in glioblastoma by promoting
epithelial mesenchymal transition. Int J Exp Pathol. 7:1977–1987.
2014.
|
|
26
|
Jhanwar-Uniyal M, Labagnara M, Friedman M,
Kwasnicki A and Murali R: Glioblastoma: Molecular pathways, stem
cells and therapeutic targets. Cancers. 7:538–555. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Horibe T, Torisawa A, Akiyoshi R,
Hatta-Ohashi Y, Suzuki H and Kawakami K: Transfection efficiency of
normal and cancer cell lines and monitoring of promoter activity by
single-cell bioluminescence imaging. Luminescence. 29:96–100. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Turano C, Coppari S, Altieri F and Ferraro
A: Proteins of the PDI family: Unpredicted non-ER locations and
functions. J Cell Physiol. 193:154–163. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Barbouche R, Miquelis R, Jones IM and
Fenouillet E: Protein-disulfide isomerase-mediated reduction of two
disulfide bonds of HIV envelope glycoprotein 120 occurs post-CXCR4
binding and is required for fusion. J Biol Chem. 278:3131–3136.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ryser HJ, Levy EM, Mandel R and DiSciullo
GJ: Inhibition of human immunodeficiency virus infection by agents
that interfere with thiol-disulfide interchange upon virus-receptor
interaction. Proc Natl Acad Sci USA. 91:4559–4563. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tsai YL, Zhang Y, Tseng CC, Stanciauskas
R, Pinaud F and Lee AS: Characterization and mechanism of
stress-induced translocation of 78-kilodalton glucose-regulated
protein (GRP78) to the cell surface. J Biol Chem. 290:8049–8064.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Davenport EL, Morgan GJ and Davies FE:
Untangling the unfolded protein response. Cell Cycle. 7:865–869.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Schönthal AH: Pharmacological targeting of
endoplasmic reticulum stress signaling in cancer. Biochem
Pharmacol. 85:653–666. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Pyrko P, Schönthal AH, Hofman FM, Chen TC
and Lee AS: The unfolded protein response regulator GRP78/BiP as a
novel target for increasing chemosensitivity in malignant gliomas.
Cancer Res. 67:9809–9816. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Jessop CE, Watkins RH, Simmons JJ, Tasab M
and Bulleid NJ: Protein disulfide isomerase family members show
distinct substrate specificity: P5 is target to BiP client
proteins. J Cell Sci. 122:4287–4295. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Choy G, O'Connor S, Diehn FE, Costouros N,
Alexander HR, Choyke P and Libutti SK: Comparison of noninvasive
fluorescent and bioluminescent small animal optical imaging.
Biotechniques. 35:1022–1026. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Dixit R and Cyr R: Cell damage and
reactive oxygen species production induced by fluorescence
microscopy: Effect on mitosis and guidelines for non-invasive
fluorescence microscopy. Plant J. 36:280–290. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Strayer C, Oyama T, Schultz TF, Raman R,
Somers DE, Más P, Panda S, Kreps JA and Kay SA: Cloning of the
Arabidopsis clock gene TOC1, an autoregulatory response regulator
homolog. Science. 289:768–771. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Venur VA, Peereboom DM and Ahluwalia MS:
Current medical treatment of glioblastoma. Cancer Treat Res.
163:103–115. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ko MK and Kay EP: PDI-mediated ER
retention and proteasomal degradation of procollagen I in corneal
endothelial cells. Exp Cell Res. 295:25–35. 2004. View Article : Google Scholar : PubMed/NCBI
|