Introduction
Nasopharyngeal carcinoma (NPC) is a prevalent head
and neck tumor which has a mortality rate of 15–50 per 100,000
individuals in Southeast Asia, especially in the southern area of
China (1,2). Globally, there are 80,000 new cases of
NPC diagnosed each year and approximately 50,000 patients succumb
to this neoplasm annually (3). A
variety of factors, including viral infection, genetics and
environment, contribute to the occurrence of NPC (4). NPC can be divided into three
histologic subtypes based on the World Health Organization (WHO)
guidelines: Type I, keratinizing squamous cell carcinoma; type II,
non-keratinizing differentiated carcinoma; and type III,
nonkeratinizing undifferentiated carcinoma (5). It is proposed that NPC tumorigenesis
and progression is a multistep process which involves multiple
genetic and epigenetic changes (6,7).
Although the outcome of NPC treatment has been improved by advanced
radiotherapy and chemotherapy, the 5-year survival rate of NPC
patients is still at 50–70% (8) and
NPC therapy is mainly limited by tumor recurrence and distant
metastasis (9,10). Therefore, it is of great importance
to identify novel biomarkers and new targets for the therapy of
NPC.
Long non-coding RNAs (lncRNAs) are a family of
non-coding RNAs (ncRNAs) that have less than 200 bases in length
and possess limited ability for protein-coding (11–14).
Numerous studies have confirmed that dysregulation of lncRNA
expression is associated with the development of a number of tumors
(15–17). It is well established that lncRNAs
play important roles in the regulation of a battery of cellular and
biological processes, including development, differentiation,
de-differentiation, proliferation, autophagy and apoptosis
(18). Cancer susceptibility
candidate 2 (CASC2), a newly identified lncRNA which is located on
chromosome 10q2, was found to play a tumor-suppressive role in some
types of tumors such as hepatocellular carcinoma, bladder, breast
cancer, osteosarcoma, gastric cancer, lung adenocarcinoma and
endometrial cancer (19–25). MicroRNAs (miRNAs), a set of RNAs
which are 19–22 nt in length with no coding ability, function as
key regulators in tumorigenesis and progression (26). Recently, it has been proposed that
one of the functions of lncRNAs is to sponge specific miRNAs and
thus to affect the mRNA expression of target genes, the effect of
which is believed to play a role in the tumorigenic process
(27,28).
Bioinformatic software predicted that there were
binding sites of miR-18a-5p in the 3′ untranslated region (UTR) of
CASC2. However, the expression and functional role of CASC2 and
miR-18a-5p in the pathogenesis of NPC remain unclear. The goal of
the present study was to identify the pattern of CASC2 and
miR-18a-5p expression in NPC, and to evaluate the role of CASC2 and
miR-18a-5p interaction in the regulation of NPC cell proliferation
and apoptosis. In the present study, we found a reduction in CASC2
expression and an increase in miR-18a-5p expression in NPC tissues
and cells. We further revealed the critical roles of CASC2 and
miR-18a-5p in the modulation of the apoptosis and proliferation of
NPC cells. We identified that CASC2 inhibited the tumorigenesis and
malignant potential of NPC via targeting the miR-18a-5p/C-terminal
binding protein interacting protein (CtIP)/RBBP8 axis.
Materials and methods
Ethics statement
The present study was conducted according to the
principles expressed in the Declaration of Helsinki, and the use of
clinical sample tissues was approved by the Ethics Committee of the
First Affiliated Hospital of Xinxiang Medical University (Weihui,
China). Written informed consent was obtained from all the enrolled
patients.
Tissue samples and cells
Twenty-five NPC patients (age, 25–72 years; 14
female and 11 male patients) who underwent surgery at the First
Affiliated Hospital of Xinxiang Medical University (from November
3, 2014 to August 23, 2015) were included in the study. NPC tissues
and normal nasopharyngeal tissues were obtained from the patients.
All patients had received no therapy prior to biopsy. All
histologic diagnoses were conducted by the two independent
pathologist. Informed consent was received from all the
patients.
Normal nasopharyngeal epithelial cells (N69) and
three NPC cell lines (SUNE1, SUNE2 and 6–10B) were purchased from
the American Type Culture Collection (ATCC; Manassas, VA, USA).
293T cells were purchased from the Shanghai Institutes for
Biological Sciences Cell Resource Centre (Shanghai, China). The
cell lines were incubated in RPMI-1640 medium (Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal
bovine serum (FBS; Gibco, Thermo Fisher Scientific, Inc.) in a
humidified atmosphere at 37°C with 5% CO2.
Transfection of plasmids
PCR was performed to amplify the CASC2 full length
sequence and then the pcDNA3.1 vector (Thermo Fisher Scientific,
Inc.) was used to establish the pcDNA-CASC2 overexpression plasmid
(CASC2). The siRNAs [si-CASC2 and its negative control (si-NC)],
microRNA inhibitors [anti-miR-18a-5p and its negative control
(anti-miR-NC)] and microRNA mimics [miR-18a-5p and its negative
control (miR-NC)] were obtained from Shanghai GenePharma Co., Ltd.
(Shanghai, China). The transfection of oligonucleotides and
plasmids was performed using Lipofectamine™ 2,000 reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions.
Evaluation of cell proliferation
Proliferation was evaluated using Cell Counting
Kit-8 (CCK-8; Beyotime Institute of Biotechnology, Haimen, China)
assay kit. After the treatment, medium was removed and fresh medium
was added followed by the addition of 10 µl of CCK-8 solution.
Then, the cells were incubated at 37°C for 2 h. Finally, the
absorbance at 450 nm was assessed using a Bio-Rad iMark microplate
absorbance reader (Bio-Rad Laboratories Inc., Hercules, CA, USA).
CCK-8 assay was performed in triplicate experiments.
Apoptosis
TUNEL assay kits (Roche Diagnostics, Basel,
Switzerland) were used to detect apoptotic cell death in SUNE1 and
6-10B cells. In brief, cells were trypsinized and resuspended at
the concentration of 1×106 cells/ml 48 h after
transfection. The apoptotic rate of NPC cells was analyzed using a
flow cytometer (FACSCalibur; BD Biosciences, Franklin Lakes, NJ,
USA) and quantified by CellQuest software (BD Biosciences).
Relative apoptosis was expressed as a percentage of apoptotic
cells.
Luciferase assay
PCR was performed to amplify the partial sequences
of CASC2 which contained the putative binding sites of miR-18a-5p.
The sequences were then cloned into the pmirGLO Dual-Luciferase
miRNA Target Expression Vector (Promega Corp., Fitchburg, WI, USA).
GeneArt™ Site-Directed Mutagenesis system (Thermo Fisher
Scientific, Inc.) was used to induce site-directed mutagenesis of
miR-18a-5p complementary bases in the sequences of CASC2. Then,
293T cells were transfected with the constructed wild-type (WT) and
mutant (MUT) reporter vectors, respectively, in the presence of
anti-miR-18a-5p or anti-miR-NC and miR-18a-5p mimics or miR-NC. The
activity of luciferase was measured using the Dual-Luciferase Assay
system (Promega) and then normalized to Renilla luciferase
activity based on the manufacturer's instructions. The assay was
performed in triplicate experiments.
RNA extraction, reverse transcription
and RT-qPCR
Total RNA was extracted from cells and tissues using
TRIzol reagent (Life Technologies; Thermo Fisher Scientific, Inc.)
following the manufacturer's instructions. After that, the quality
of RNA was assessed using the NanoDrop 1,000 (NanoDrop
Technologies; Thermo Fisher Scientific, Inc.). For the detection of
mRNA and lncRNA expression, real-time PCR (RT-qPCR) reaction was
conducted using SYBR-Green (Takara Biotechnology, Co., Lt., Dalian,
China). The amplification was carried out on an ABI 7,500 Real-Time
PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.).
GAPDH and β-actin were regarded as the housekeeping genes for the
normalization of target genes. For the measurement of miRNA,
synthesis of cDNA was carried out using the qScript microRNA cDNA
Synthesis kit (Quantabio, Beverly, MA, USA) following the
manufacturer's protocols. MicroRNA RT-qPCR was performed using a
miScript SYBR-Green PCR kit (Qiagen, Hilden, Germany) as per the
manufacturer's instructions. U6 was regarded as the housekeeping
gene for the normalization of target miRNAs. 2−ΔΔCq
method was used to evaluate the relative fold change of gene
expression (29).
RNA immunoprecipitation assay
RNA immunoprecipitation (RIP) assay was performed to
detect the interaction between lncRNA and miRNA, using an Imprint
RNA immunoprecipitation kit (Sigma-Aldrich; Merck KGaA, Darmstadt,
Germany). In brief, anti-IgG (negative control) and anti-Argomaute2
(anti-Ago2) were added to the cell lysate and incubated overnight
at 4°C. After that, Protein A magnetic beads were added to the
mixture to form a complex of immunoprecipitation. After that, extra
protein and DNA were removed to purify the complex. Finally, the
enrichment of CASC2 and miR-18a-5p in the immunoprecipitated
complex was evaluated by RT-qPCR assay.
Lentivirus production and
infection
The CASC2-shRNA lentivirus vector (LV-shCASC2) was
constructed by Shanghai GenePharma Biotech Co., Ltd. LV-shCASC2 or
empty vector (LV-shNC) lentiviruses were used to transfect SUNE1
cells. Then, the transfected cells were purified with 5 µg/ml
puromycin to obtain cells with stable CASC2 knockdown.
Xenograft mouse model
The animal experiment was carried out following the
Guidelines for the Care and Use of Laboratory Animals of the
National Institutes of Health. The study was approved by the Ethics
Committee of the First Affiliated Hospital of Xinxiang Medical
University. Twelve male BALB/c nude mice (18–22 g, 6–8 weeks old)
were purchased from the Animal Center of of Xinxiang Medical
University. The mice were housed in a specific pathogen free (SPF)
animal laboratory under temperature (23±2°C) and humidity (55±5%)
condition with a standard light cycle (12 h light/dark) and free
access to food and water. To establish the xenograft model,
8×106 SUNE1 cells with or without stable knockdown of
CASC2 were subcutaneously inoculated into the mice. The
experimental period was 6 weeks. Tumor volume was measured using a
caliper and calculated according to the following formula: volume =
0.5 × length × width × width. After the animal experiment, mice
were euthanized using isoflurane and tumor tissues were excised for
the evaluation of weight and then stored at −80°C for further
analysis.
Statistical analysis
All data are expressed as mean ± SD. Differences
between groups were analyzed with one-way analysis of variance
(ANOVA) followed by Tukey's test or Student's t- test. P-values
<0.05 were considered to be statistically significant, and all
analyses were two-sided. All statistical analyses were performed
using GraphPad Prism 6.1 software (GraphPad Software Inc., San
Diego, CA, USA).
Results
CASC2 expression is downregulated and
miR-18a-5p expression is upregulated in NPC tissues and cell
lines
The mRNA expression patterns of CASC2 and miR-18a-5p
in NPC tissues and cells were firstly assessed. We showed that the
expression of CASC2 was significantly reduced in NPC tissues
compared with that in normal nasopharyngeal tissues (Fig. 1A). Similarly, CASC2 expression was
significantly decreased in NPC cell lines compared with that in
epithelial N69 cells (Fig. 1B). In
contrast with the pattern of CASC2 expression, the level of
miR-18a-5p in NPC tissues was significantly higher than that in
normal nasopharyngeal tissues (Fig.
1C). Additionally, miR-18a-5p expression was significantly
increased in NPC cell lines compared with that in epithelial N69
cells (Fig. 1D). The findings
indicated that dysregulation of CASC2 and miR-18a-5p expression may
be involved in the tumorigenesis of NPC.
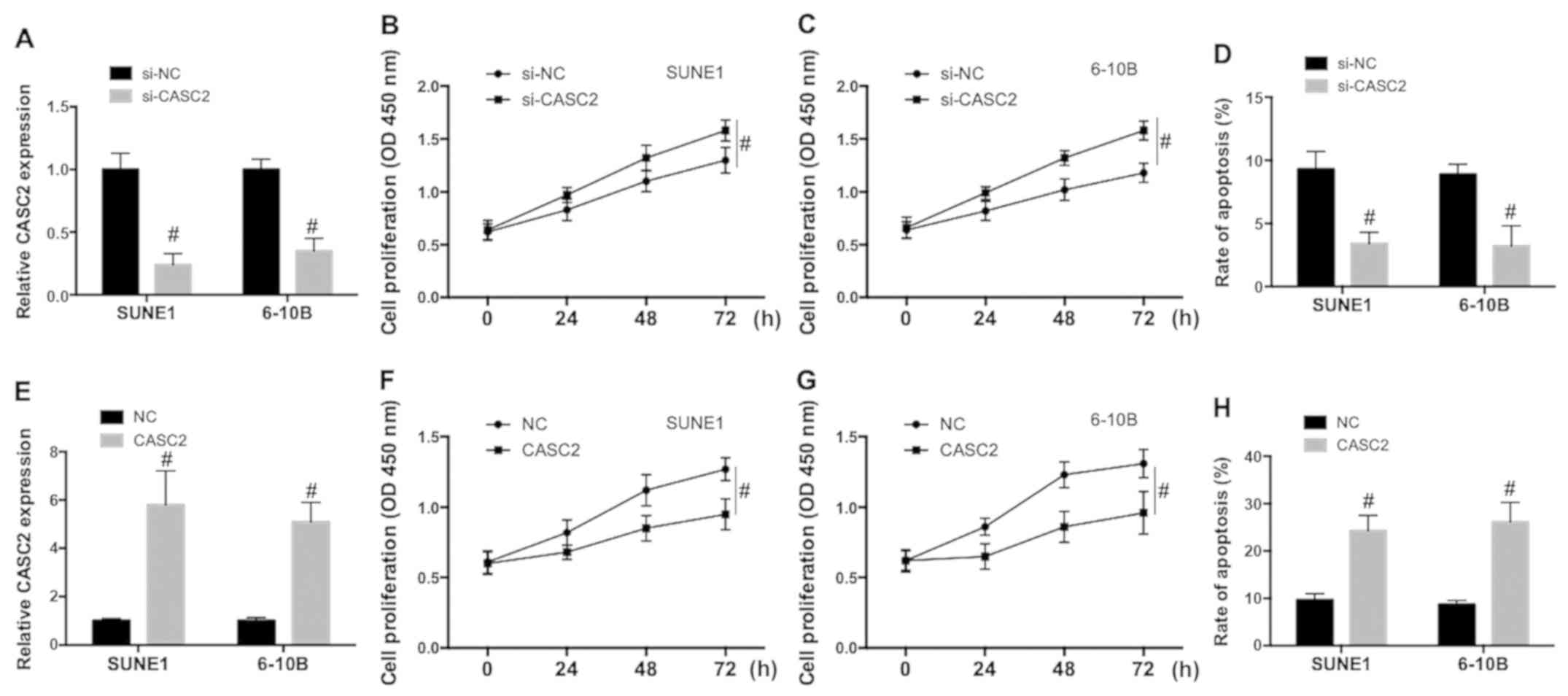
CASC2 downregulation promotes
proliferation and inhibits apoptosis in NPC cells
In order to explore the role of CASC2 in the
regulation of NPC malignant potential, SUNE1 and 6-10B cells were
transfected with small interference RNA of CASC2 (si-CASC2) or a
plasmid carrying the CASC2 sequence to downregulate or upregulate
CASC2 expression. The efficiency of the knockdown and
overexpression was examined. As shown in Fig. 2A and E, the CASC2 expression level
was significantly decreased by si-CASC2 and increased by the
plasmid carrying the CASC2 sequence in SUNE1 and 6-10B cells.
Knockdown of CASC2 significantly increased cell proliferation in
SUNE1 (Fig. 2B) and 6-10B (Fig. 2C) cells and inhibited spontaneous
apoptosis in NPC cells (Fig. 2D).
Upregulation of CASC2 significantly decreased cell proliferation in
SUNE1 (Fig. 2F) and 6-10B cells
(Fig. 2G) cells and increased
apoptosis in NPC cells (Fig. 2H).
The data indicated that CASC2 downregulation promoted proliferation
and inhibited apoptotic cell death, while CASC2 upregulation
decreased proliferation and promoted apoptosis in NPC cells.
CASC2 directly inhibits miR-18a-5p
expression
In order to examine the possible mechanism
underlying the antitumor role of CASC2 in NPC, bioinformatic
analysis was employed to explore CASC2-associated miRNAs. We showed
that CASC2 contained putative binding sites for miR-18a-5p
(Fig. 3A), indicating that
miR-18a-5p may interact with CASC2. This assumption was validated
using Dual-Luciferase reporter assay. 293T cells were
co-transfected with the constructed wild-type (WT) and mutant (MUT)
CASC2 luciferase vectors, anti-miR-NC or anti-miR-18a-5p and miR-NC
or miR-18a-5p mimics. The results also showed that miR-18a-5p
mimics significantly reduced the luciferase activities of WT-CASC2,
while miR-18a-5p inhibitors increased WT-CASC2 luciferase
activities (Fig. 3B). In contrast,
miR-18a-5p mimics and miR-18a-5p inhibitors did not significantly
affect MUT-CASC2 luciferase activities (Fig. 3B). Considering the important role of
Ago2 in the formation of RNA-induced silencing complex and the
maturation of miRNAs, we performed RIP assay to measure whether
there was endogenous interaction between CASC2 and miR-18a-5p. As
evidenced in Fig. 3C, CASC2 and
miR-18a-5p expression were significantly increased in the Ago2
antibody complex compared with that in the IgG antibody complex in
SUNE1 cells. In order to further evaluate the role of CASC2 in the
regulation of miR-18a-5p, SUNE1 cells were transfected with
CASC2-overexpression plasmid or si-CASC2 (Fig. 3D). Upregulation of CASC2
significantly decreased miR-18a-5p expression in SUNE1 cells, while
CASC2 knockdown significantly promoted miR-18a-5p expression
(Fig. 3E). These data demonstrated
that CASC2 may act as a sponge for miR-18a-5p in the NPC cells.
To further explore the role of miR-18a-5p in
CASC2-induced regulation of proliferation and apoptosis, SUNE1 and
6-10B cells were co-transfected with CASC2-overexpression plasmid
and miR-18a-5p mimics. We revealed that CASC2
overexpression-induced reduction of proliferation was significantly
suppressed by miR-18a-5p mimics (Fig.
4A and B). In addition, CASC2 overexpression-induced increase
of apoptosis was significantly blocked by miR-18a-5p mimics
(Fig. 4C and D). The data suggest
that CASC2 acts as a tumor suppressor by acting as ceRNA of
miR-18a-5p in NPC cells.
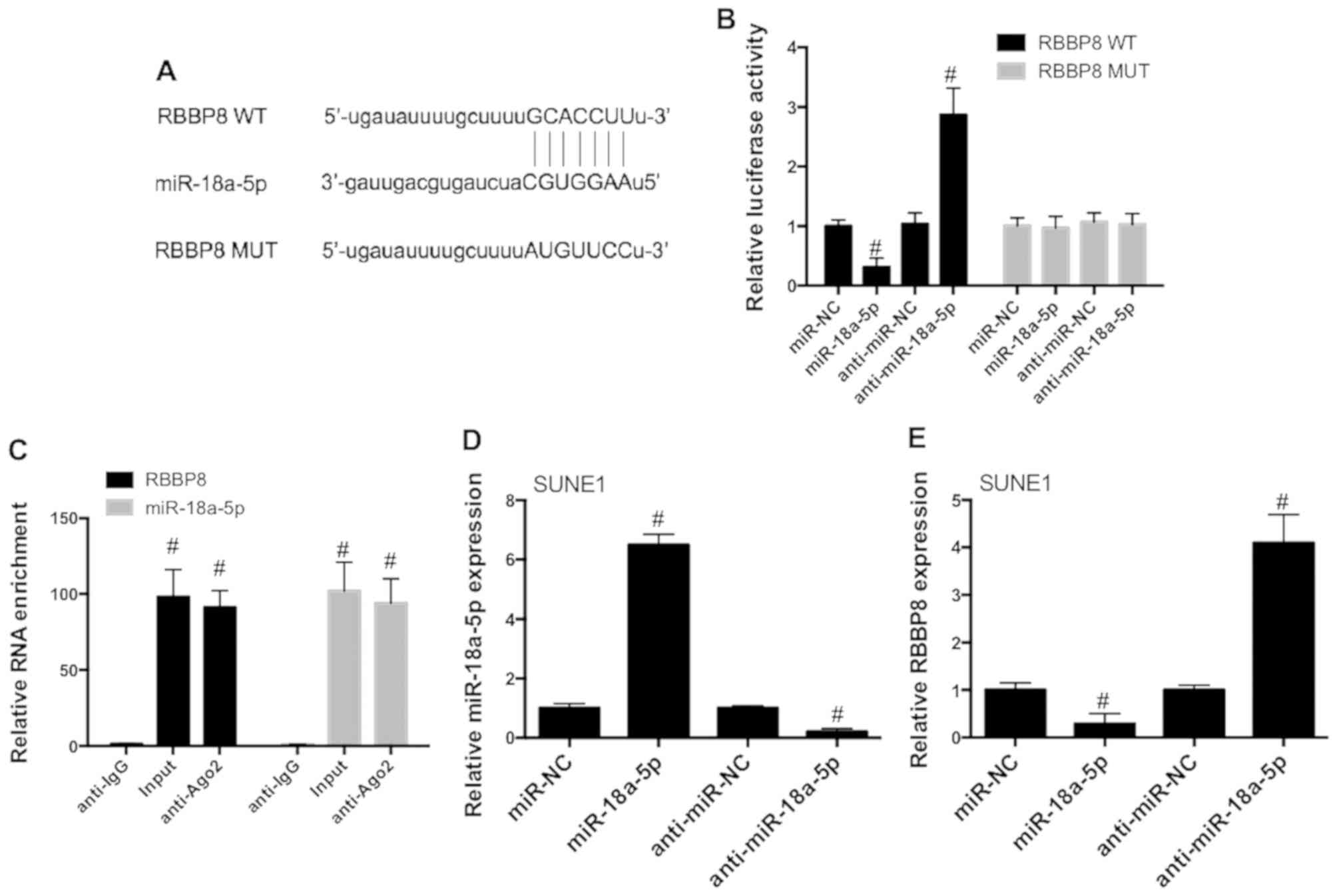
miR-18a-5p promotes cell proliferation
and inhibits apoptosis by the regulation of RBBP8
To elucidate the underlying mechanism of
miR-18a-5p-induced regulation of NPC progression, we aimed to
identify possible targets of miR-18a-5p using bioinformatic
analysis. The results revealed that RBBP8 possesses complementary
sites of miR-18a-5p (Fig. 5A),
indicating that there is a possible interaction between miR-18a-5p
and RBBP8. 293T cells were co-transfected with constructed WT and
MUT RBBP8 luciferase vectors with or without anti-miR-NC or
anti-miR-18a-5p and miR-NC or miR-18a-5p mimics. The results
revealed that miR-18a-5p mimics significantly decreased the
luciferase activities of RBBP8 (WT), while miR-18a-5p inhibitors
exhibited an opposite effect (Fig.
5B). However, neither miR-18a-5p mimics nor its inhibitors
affected MUT RBBP8 reporter luciferase activities (Fig. 5B). As shown in Fig. 5C, RBBP8 and miR-18a-5p mRNA level
were significantly increased in the Ago2 antibody complex in SUNE1
cells. To assess the role of miR-18a-5p in the regulation of RBBP8,
SUNE1 cells were transfected with miR-18a-5p mimics or inhibitors
(Fig. 5D). miR-18a-5p mimics
significantly decreased RBBP8 expression in SUNE1 cells, while
miR-18a-5p inhibitors promoted RBBP8 expression (Fig. 5E). These data demonstrated that
RBBP8 may be a target of miR-18a-5p in NPC cells.
Next, SUNE1 and 6-10B cells were transfected with
miR-18a-5p inhibitors together with LV-shNC or LV-shRBBP8.
miR-18a-5p inhibitors decreased cell proliferation, and this effect
was inhibited by RBBP8 knockdown (Fig.
6A and B). In addition, inhibition of miR-18a-5p significantly
increased apoptosis and RBBP8 knockdown suppressed this effect
(Fig. 6C and D). The results
indicated that RBBP8 functions as a target of miR-18a-5p in NPC
cells.
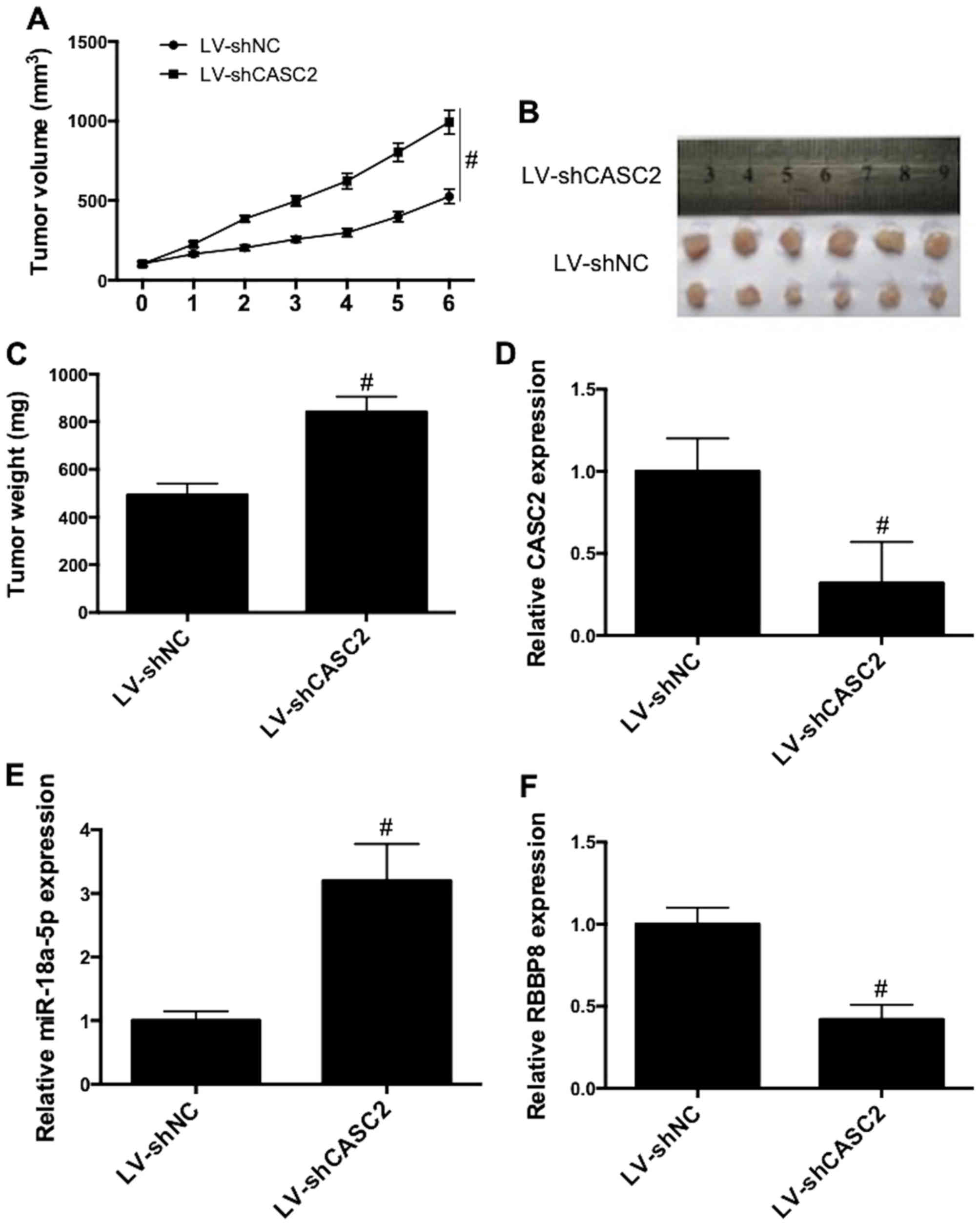
CASC2 knockdown promotes tumor growth,
promotes miR-18a-5p expression and inhibits RBBP8 expression in
vivo
To further examined the role of CASC2 in
vivo, tumor growth in xenograft mice was investigated. CASC2
knockdown significantly promoted the growth of tumors, as
illustrated by the increase in tumor volume (Fig. 7A) and tumor weight (Fig. 7B and C). Additionally, CASC2
knockdown (Fig. 7D) increased
miR-18a-5p expression (Fig. 7E) and
decreased RBBP8 expression (Fig.
7F) in tumor tissues. These results revealed that CASC2
knockdown promoted tumor growth, promoted miR-18a-5p expression and
inhibited RBBP8 expression in vivo.
Discussion
Cancer susceptibility candidate 2 (CASC2), a newly
identified lncRNA, is located on chromosome 10q26 (30). It was initially found that CASC2
expression is reduced in endometrial carcinoma and downregulation
of CASC2 provides a growth advantage in endometrial cancer cells
(30,31). Subsequently, CASC2 was found to play
a tumor suppressive role in several human types of cancers,
including hepatocellular carcinoma, bladder, breast cancer,
osteosarcoma, gastric cancer, lung adenocarcinoma and endometrial
cancer (19–25). We revealed, in the present study, a
significant decrease in CASC2 expression in nasopharyngeal
carcinoma (NPC) tissues and cells (Fig.
1). Moreover, downregulation of CASC2 promoted proliferation
and inhibited apoptotic cell death in SUNE1 and 6-10B cells, while
CASC2 upregulation inhibited cell proliferation and promoted
apoptosis (Fig. 2). The
tumor-suppressive role in vivo was also confirmed in
xenograft mice (Fig. 7). The data
suggested that CASC2 may be a key regulator in the tumorigenesis of
NPC.
The regulatory mechanisms of lncRNAs include
signals, decoys, guides and scaffolds (32). As a decoy, lncRNAs function via
binding microRNAs or proteins to modulate the functions of key
molecules (33). Numerous
investigations have suggested that CASC2 may function to sponge
miRNAs, which leads to the inhibition of miRNA-induced regulation
of target mRNAs and thus the regulation of many cancers (20,22–24,34).
For example, CASC2 downregulation was found to promote the growth
and invasion of osteosarcoma through regulation of miR-181a
(22). In hepatocellular carcinoma,
CASC2 regulates miR-24-3p, leading to a decrease in cell viability
and an increase in apoptosis (23).
CASC2 was found to modulate docetaxel-induced sensitivity in
prostate cancer cells via regulation of miR-183/Sprouty2 signaling
(34).
MicroRNAs are a class of short (18–24 nt), single
stranded and non-coding RNAs. These RNAs could directly bind with
the target mRNAs, leading to transcriptional regulation (35). A battery of studies have shown that
dysfunction of miR-18a-5p is associated with cancer pathogenesis.
Recently, it was reported that GAS5 could regulate miR-18a-5p and
thus modulate proliferation, migration and invasion in glioma cells
(36). In breast cancer, miR-18a-5p
could target SREBP1 and modulate epithelial-mesenchymal transition
(37). miR-18a-5p was also reported
to function as an oncogene and prognostic biomarker in the
development of RCC (38). In lung
cancer, miR-18a-5p plays an oncogenic role via direct regulation of
IRF2 (39). In the present study,
we revealed that miR-18a-5p is a target of CASC2, and CASC2
suppresses the expression of miR-18a-5p. miR-18a-5p expression was
significantly increased in NPC tissues and cells (Fig. 1). miR-18a-5p mimics inhibited CASC2
overexpression-induced reduction in proliferation and increase in
apoptosis (Fig. 3). The data
suggest that miR-18a-5p promoted NPC tumorigenesis.
The possible underlying mechanisms of the oncogenic
role of miR-18a-5p in NPC were investigated. Bioinformatic analysis
suggested the possible interaction between miR-18a-5p and RBBP8
(Fig. 5). It was previously
described that CtIP/RBBP8 is a transcriptional corepressor
(40), which was shown to bind
several other transcription factors such as TRB3, LMO4 and Ikaros
that are associated with cancer (41,42).
Frameshift mutations induced by CtIP/RBBP8 microsatellite have been
found in colorectal (43) and
endometrial cancer (44). From a
mechanistic point, CtIP/RBBP8 regulates the cell cycle by
interacting with RB1 (45). In
breast cancer, a reduction in CtIP/RBBP8 expression is associated
with an increase in disease-free survival under the condition of
co-treatment of hormone, radiotherapy and chemotherapy (46). In the present study, the results
suggested that miR-18a-5p targeted RBBP8 and suppressed its
expression (Fig. 5). RBBP8
knockdown blocked the decrease in cell proliferation and increase
in apoptosis induced by miR-18a-5p inhibitors (Fig. 6).
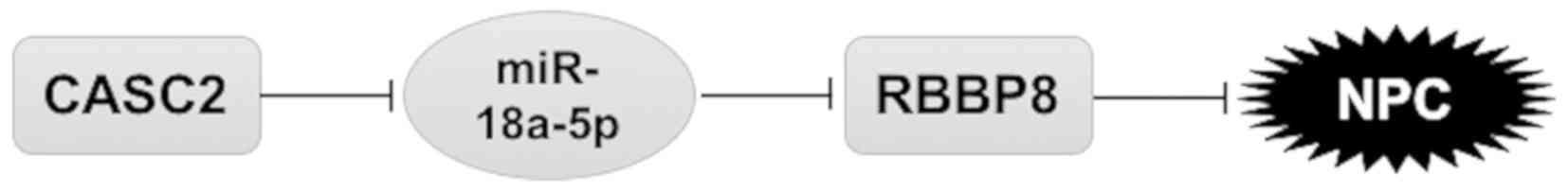
In summary, the present study identified that
downregulation of CASC2 is critical for the cell proliferation and
tumor growth of NPC. CASC2 regulates the malignant potential of NPC
through modulation of RBBP8 via sponging miR-18a-5p (Fig. 8). Our findings highlight the
CASC2/miR-18a-5p/RBBP8 axis in NPC pathogenesis and provide new
biomarkers and potential targets for the treatment of NPC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
Data will be avaible on request.
Authors' contributions
WJM and QQJ conceived and designed the study. QQJ,
DJY, GZZ, QL and HMM performed the experiments and analyzed the
data. WJM, QQJ and DJY prepared the figures. WJM and QQJ wrote the
manuscript. All authors read and approved the manuscript and agree
to be accountable for all aspects of the research in ensuring that
the accuracy or integrity of any part of the work are appropriately
investigated and resolved.
Ethics approval and consent to
participate
The present study was conducted according to the
principles expressed in the Declaration of Helsinki, and the use of
clinical sample tissues was approved by the Ethics Committee of the
First Affiliated Hospital of Xinxiang Medical University (Weihui,
China). The animal experiment was carried out following the
Guidelines for the Care and Use of Laboratory Animals of the
National Institutes of Health. The study was approved by the Ethics
Committee of the First Affiliated Hospital of Xinxiang Medical
University. Written informed consent was obtained from all the
enrolled patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chen W and Hu GH: Biomarkers for enhancing
the radiosensitivity of nasopharyngeal carcinoma. Cancer Biol Med.
12:23–32. 2015.PubMed/NCBI
|
|
2
|
Vokes EE, Liebowitz DN and Weichselbaum
RR: Nasopharyngeal carcinoma. Lancet. 350:1087–1091. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Petersson F: Nasopharyngeal carcinoma: A
review. Semin Diagn Pathol. 32:54–73. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhou X, Xiao X, Huang T, Du C, Wang S, Mo
Y, Ma N, Murata M, Li B, Wen W, et al: Epigenetic inactivation of
follistatin-like 1 mediates tumor immune evasion in nasopharyngeal
carcinoma. Oncotarget. 7:16433–16444. 2016.PubMed/NCBI
|
|
5
|
Aldape K, Zadeh G, Mansouri S,
Reifenberger G and von Deimling A: Glioblastoma: Pathology,
molecular mechanisms and markers. Acta Neuropathol. 129:829–848.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shi W, Bastianutto C, Li A, Perez-Ordonez
B, Ng R, Chow KY, Zhang W, Jurisica I, Lo KW, Bayley A, et al:
Multiple dysregulated pathways in nasopharyngeal carcinoma revealed
by gene expression profiling. Int J Cancer. 119:2467–2475. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Altun M, Fandi A, Dupuis O, Cvitkovic E,
Krajina Z and Eschwege F: Undifferentiated nasopharyngeal cancer
(UCNT): Current diagnostic and therapeutic aspects. Int J Radiat
Oncol Biol Phys. 32:859–877. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tan WL, Tan EH, Lim DW, Ng QS, Tan DS,
Jain A and Ang MK: Advances in systemic treatment for
nasopharyngeal carcinoma. Chin Clin Oncol. 5:212016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Parkin DM and Muir CS: Cancer incidence in
five continents. Comparability and quality of data. IARC Sci Publ.
120:45–173. 1992.
|
|
10
|
Lee AW, Sze WM, Au JS, Leung SF, Leung TW,
Chua DT, Zee BC, Law SC, Teo PM, Tung SY, et al: Treatment results
for nasopharyngeal carcinoma in the modern era: The Hong Kong
experience. Int J Radiat Oncol Biol Phys. 61:1107–1116. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cai X, Liu Y, Yang W, Xia Y, Yang C, Yang
S and Liu X: Long noncoding RNA MALAT1 as a potential therapeutic
target in osteosarcoma. J Orthop Res. 34:932–941. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gao KT and Lian D: Long non-coding RNA
MALAT1 is an independent prognostic factor of osteosarcoma. Eur Rev
Med Pharmacol Sci. 20:3561–3565. 2016.PubMed/NCBI
|
|
13
|
Huo Y, Li Q, Wang X, Jiao X, Zheng J, Li Z
and Pan X: MALAT1 predicts poor survival in osteosarcoma patients
and promotes cell metastasis through associating with EZH2.
Oncotarget. 8:46993–47006. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li Q, Pan X, Wang X, Jiao X, Zheng J, Li Z
and Huo Y: Long noncoding RNA MALAT1 promotes cell proliferation
through suppressing miR-205 and promoting SMAD4 expression in
osteosarcoma. Oncotarget. 8:106648–106660. 2017.PubMed/NCBI
|
|
15
|
Fan L, Huang C, Li J, Gao T, Lin Z and Yao
T: Long non-coding RNA urothelial cancer associated 1 regulates
radioresistance via the hexokinase 2/glycolytic pathway in cervical
cancer. Int J Mol Med. 42:2247–2259. 2018.PubMed/NCBI
|
|
16
|
Wu X, Tudoran OM, Calin GA and Ivan M: The
many faces of long noncoding RNAs in cancer. Antioxid Redox Signal.
29:922–935. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang H, Cai Y, Zheng L, Zhang Z, Lin X
and Jiang N: Long noncoding RNA NEAT1 regulate papillary thyroid
cancer progression by modulating miR-129-5p/KLK7 expression. J Cell
Physiol. 233:6638–6648. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Martens-Uzunova ES, Bottcher R, Croce CM,
Jenster G, Visakorpi T and Calin GA: Long noncoding RNA in
prostate, bladder, and kidney cancer. Eur Urol. 65:1140–1151. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang Y, Zhu M, Sun Y, Li W, Wang Y and Yu
W: Up-regulation of lncRNA CASC2 suppresses cell proliferation and
metastasis of breast cancer via inactivating of the TGF-β signaling
pathway. Oncol Res. Mar 9–2018.(Epub ahead of print). doi:
10.3727/096504018X15199531937158. View Article : Google Scholar
|
|
20
|
Zhao L and Zhang Y: Long noncoding RNA
CASC2 regulates hepatocellular carcinoma cell oncogenesis through
miR-362-5p/Nf-κB axis. J Cell Physiol. 233:6661–6670. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhou J, Huang H, Tong S and Huo R:
Overexpression of long non-coding RNA cancer susceptibility 2
inhibits cell invasion and angiogenesis in gastric cancer. Mol Med
Rep. 16:5235–5240. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ba Z, Gu L, Hao S, Wang X, Cheng Z and Nie
G: Downregulation of lncRNA CASC2 facilitates osteosarcoma growth
and invasion through miR-181a. Cell Prolif. 51:e124092018.
View Article : Google Scholar
|
|
23
|
Fan JC, Zeng F, Le YG and Xin L: lncRNA
CASC2 inhibited the viability and induced the apoptosis of
hepatocellular carcinoma cells through regulating miR-24-3p. J Cell
Biochem. 119:6391–6397. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li H, Shi H, Gao M, Ma N and Sun R: Long
non-coding RNA CASC2 improved acute lung injury by regulating
miR-144-3p/AQP1 axis to reduce lung epithelial cell apoptosis. Cell
Biosci. 8:152018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang D, Gao ZM, Han LG, Xu F, Liu K and
Shen Y: Long noncoding RNA CASC2 inhibits metastasis and epithelial
to mesenchymal transition of lung adenocarcinoma via suppressing
SOX4. Eur Rev Med Pharmacol Sci. 21:4584–4590. 2017.PubMed/NCBI
|
|
26
|
Calin GA and Croce CM: Chromosomal
rearrangements and microRNAs: A new cancer link with clinical
implications. J Clin Invest. 117:2059–2066. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liz J and Esteller M: lncRNAs and
microRNAs with a role in cancer development. Biochim Biophys Acta
1859. 169–176. 2016.
|
|
28
|
Yoon JH, Abdelmohsen K and Gorospe M:
Functional interactions among microRNAs and long noncoding RNAs.
Semin Cell Dev Biol. 34:9–14. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Baldinu P, Cossu A, Manca A, Satta MP,
Sini MC, Rozzo C, Dessole S, Cherchi P, Gianfrancesco F, Pintus A,
et al: Identification of a novel candidate gene, CASC2, in a region
of common allelic loss at chromosome 10q26 in human endometrial
cancer. Hum Mutat. 23:318–326. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Baldinu P, Cossu A, Manca A, Satta MP,
Sini MC, Palomba G, Dessole S, Cherchi P, Mara L, Tanda F and
Palmieri G: CASC2a gene is down-regulated in endometrial cancer.
Anticancer Res. 27:235–243. 2007.PubMed/NCBI
|
|
32
|
Wang KC and Chang HY: Molecular mechanisms
of long noncoding RNAs. Mol Cell. 43:904–914. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Qu L, Ding J, Chen C, Wu ZJ, Liu B, Gao Y,
Chen W, Liu F, Sun W, Li XF, et al: Exosome-transmitted lncARSR
promotes sunitinib resistance in renal cancer by acting as a
competing endogenous RNA. Cancer Cell. 29:653–668. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Gao W, Lin S, Cheng C, Zhu A, Hu Y, Shi Z,
Zhang X and Hong Z: Long non-coding RNA CASC2 regulates Sprouty2
via functioning as a competing endogenous RNA for miR-183 to
modulate the sensitivity of prostate cancer cells to docetaxel.
Arch Biochem Biophys. 2018 Jan 23;(pii): S0003-9861(17)30840-8.
doi: 10.1016/j.abb.2018.01.013. PubMed/NCBI
|
|
35
|
Iorio MV and Croce CM: MicroRNA
dysregulation in cancer: Diagnostics, monitoring and therapeutics.
A comprehensive review. EMBO Mol Med. 9:8522017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Liu Q, Yu W, Zhu S, Cheng K, Xu H, Lv Y,
Long X, Ma L, Huang J, Sun S and Wang K: Long noncoding RNA GAS5
regulates the proliferation, migration, and invasion of glioma
cells by negatively regulating miR-18a-5p. J Cell Physiol.
234:775–768. 2018.
|
|
37
|
Zhang N, Zhang H, Liu Y, Su P, Zhang J,
Wang X, Sun M, Chen B, Zhao W, Wang L, et al: SREBP1, targeted by
miR-18a-5p, modulates epithelial-mesenchymal transition in breast
cancer via forming a co-repressor complex with snail and HDAC1/2.
Cell Death Differ. Jul 9–2018.(Epub ahead of print). doi:
10.1038/s41418-018-0158-8.
|
|
38
|
Zhou L, Li Z, Pan X, Lai Y, Quan J, Zhao
L, Xu J, Xu W, Guan X, Li H, et al: Identification of miR-18a-5p as
an oncogene and prognostic biomarker in RCC. Am J Transl Res.
10:1874–1886. 2018.PubMed/NCBI
|
|
39
|
Liang C, Zhang X, Wang HM, Liu XM, Zhang
XJ, Zheng B, Qian GR and Ma ZL: MicroRNA-18a-5p functions as an
oncogene by directly targeting IRF2 in lung cancer. Cell Death Dis.
8:e27642017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Schaeper U, Subramanian T, Lim L, Boyd JM
and Chinnadurai G: Interaction between a cellular protein that
binds to the C-terminal region of adenovirus E1A (CtBP) and a novel
cellular protein is disrupted by E1A through a conserved PLDLS
motif. J Biol Chem. 273:8549–8552. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Koipally J and Georgopoulos K: Ikaros-CtIP
interactions do not require C-terminal binding protein and
participate in a deacetylase-independent mode of repression. J Biol
Chem. 277:23143–23149. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Sum EY, Peng B, Yu X, Chen J, Byrne J,
Lindeman GJ and Visvader JE: The LIM domain protein LMO4 interacts
with the cofactor CtIP and the tumor suppressor BRCA1 and inhibits
BRCA1 activity. J Biol Chem. 277:7849–7856. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Ikenoue T, Togo G, Nagai K, Ijichi H, Kato
J, Yamaji Y, Okamoto M, Kato N, Kawabe T, Tanaka A, et al:
Frameshift mutations at mononucleotide repeats in RAD50
recombinational DNA repair gene in colorectal cancers with
microsatellite instability. Jpn J Cancer Res. 92:587–591. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Bilbao C, Ramirez R, Rodriguez G, Falcón
O, León L, Díaz-Chico N, Perucho M and Díaz-Chico JC: Double strand
break repair components are frequent targets of microsatellite
instability in endometrial cancer. Eur J Cancer. 46:2821–2827.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Fusco C, Reymond A and Zervos AS:
Molecular cloning and characterization of a novel
retinoblastoma-binding protein. Genomics. 51:351–358. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Yamaguchi N, Osaki M, Onuma K, Yumioka T,
Iwamoto H, Sejima T, Kugoh H, Takenaka A and Okada F:
Identification of microRNAs involved in resistance to sunitinib in
renal cell carcinoma cells. Anticancer Res. 37:2985–2992.
2017.PubMed/NCBI
|