Introduction
Over the last decade, therapy for glioblastoma (GBM)
has been centered on maximal safe surgical resection with adjuvant
radiotherapy and chemotherapy. However, despite this breakthrough
in combination therapy, the overall 5-year survival rate still
remains less than 10% due to frequent tumor relapse (1,2).
Radiotherapy is a typical and aggressive approach for the treatment
of many tumors, yet the inherent and acquired resistance of cancer
cells is a major hurdle of radiotherapy (3). Growing evidence has demonstrated that
radiation induces alterations in microRNA (miRNA) and gene profiles
of many tumors (4). However, the
underlying molecular mechanism of radioresistance is still not
completely understood.
Extracellular vesicles (EVs, exosomes and
microvesicles) are membrane-bound vessels that are actively
secreted by mammalian cells. They contain a specific cargo of
protein and nucleic acids (DNA, non-coding RNAs and mRNAs), which
reflect their cell types of origin and may be used as biomarkers
for the diagnosis of human diseases (5,6). EVs
have been recognized as important mediators of intercellular
communication and they may play an important role in transporting
radiation-induced RNAs and subsequently affect the proliferation
and radioresistance of tumor cells (7).
Circular RNAs (circRNAs) have recently emerged as a
comprehensive class of noncoding RNAs that may function as miRNA
sponges and regulate gene expression in mammals. circRNAs are
highly enriched in the brain and very stable within cells and body
fluids (8,9). Li et al reported that circRNAs
are enriched and stable in liver cancer exosomes (10). Xu et al also found
differential expression of circRNAs in EVs isolated from the serum
of patients with endometrial cancer when compared with normal
controls (11). A recent study
demonstrated that exosomal circRNAs from arsenite-transformed cells
were involved in the malignant transformation of human hepatic
cells (12).
To the best of our knowledge, no studies to date
have yet profiled the expression of circRNAs in EVs derived from
radioresistant glioma. The function of circRNAs in EVs from
radioresistant glioma is still unknown. Therefore, the aim of the
present study was to investigate the differential circRNA
expression in EVs isolated from radioresistant glioma cells and
normal glioma cells in detail, and to identify circRNAs in EVs
isolated from radioresistant glioma cells that may act as miRNA
sponges that may be associated with glioma radioresistance using
RNA sequencing and bioinformatics method.
Materials and methods
Cell culture
Human U-251MG cells (cat. no. KG050), obtained from
Nanjing KeyGen Biotech Co., Ltd. (Nangjing, China; http://www.keygentec.com.cn/prd-search-U251.html),
were cultured in Dulbecco's modified Eagle's medium (DMEM;
Gibco-BRL; Therno Fisher Scientific, Inc., Waltham, MA, USA)
containing 10% fetal bovine serum (FBS; Tianjin HaoYang Biological
Manufacture Co., Ltd, Tianjing, China) and supplemented with
penicillin-streptomycin. Cells were incubated at 37°C in 5%
CO2 and free of mycoplasma contamination after
testing.
Establishment of radioresistant U251
cells (RR-U251)
U251 cells were serially treated with 5 Gy of
radiation using a 60Co source (RuiDi Biotechnology,
Nanjing, China) until 60 Gy of irradiation was reached, as
previously described (13). Cell
viability, cell migration and cell invasion experiments confirmed
that radiosensitivity was decreased in RR-U251 cells and indicated
the successful establishment of the RR-U251 cell line (13).
EV isolation
EVs were obtained from supernatants of U251 and
RR-U251 cells cultured in DMEM with 10% exosome-free FBS. Cell
culture media were sequentially centrifuged at 500 × g for 15 min,
16,500 × g for 30 min to remove floating cells and cellular debris
followed by passing through a 0.22-µm filter. The filtrates were
further ultracentrifuged at 100,000 × g (Beckman Coulter Avanti
J-30I; Beckman Coulter, Inc., Brea, CA, USA) for 2 h at 4°C and
submitted to a second ultracentrifugation after washing in
phosphate-buffered saline (PBS). The final EV pellets were further
used immediately or resuspended in 100 µl PBS and stored at −80°C.
EVs isolated from normal U251 and RR-U251 cells were abbreviated as
Nor-EVs and RR-EVs, respectively.
Nanoparticle tracking analysis
Size determination of EVs was analyzed using
ZetaView (Particle Matrix GmbH, Microtrac, Meerbusch, Germany).
Transmission electron microscopy
EVs (10 µg) re-suspended in PBS were loaded onto
Formvar/carbon coated grids (Ted Pella Inc., Redding, CA, USA)
fixed in 2.5% glutaraldehyde, contrasted in 2% uranyl acetate and
visualized with FEI Tecnai G2 Spirit Bio TWIN (FEI, Hillsboro,
Oregon, USA).
RNA sequencing (RNA-seq)
For RNA-seq, total RNA was extracted from U251,
RR-U251 cells or EVs using the miRNeasy Micro kit (Qiagen GmbH,
Hilden, Germany). The ribosomal RNA was removed from total RNA.
After that, strand-specific RNA-seq library preparation was carried
out using the VAHTS Total RNA-seq (H/M/R) Library Prep Kit for
Illumina® (Vazyme Biotech Co., Ltd., Nanjing, China). In
brief, RNA samples were fragmented into small pieces, and then used
for first- and second-strand cDNA synthesis with random hexamer
primers. In second-strand cDNA synthesis, dUTP mix (without dTTP)
was used. After cDNA synthesis, end-repair was conducted, then a
single ‘A’ base was added, and finally adaptors were ligated onto
the cDNA products. Ligated cDNA products were purified and treated
with uracil DNA glycosylase to remove the second-strand cDNA. For
purified first-strand cDNA, PCR amplification was conducted
followed by library quality analysis using Bioanalyzer 4200
(Agilent Technologies Inc., Santa Clara, CA, USA). Then the
strand-specific RNA-seq libraries were sequenced on the HiSeq X10
system (Illumina, Inc., San Diego, CA, USA).
Differential expression analysis
HTSeq software version 0.6.1p2 (http://htseq.readthedocs.io/) was used to count reads
mapped to genes. Differential expression level of circRNAs or
miRNAs between the two groups was assessed by DEGseq algorithm in
the statistical program R version 3.3.2 (http://www.r-project.org/). We identified
differentially expressed circRNAs or miRNAs using a fold change
>1.5. Those miRNAs with a zero read count were filtered.
Real-time quantitative polymerase
chain reaction (RT-qPCR)
Total RNA was extracted and reverse transcribed
using random primers by RevertAid First Strand cDNA Synthesis kit
(Thermo Fisher Scientific, Inc., Waltham, MA, USA) according to the
manufacturer's protocol. The relative circRNA expression levels
were analyzed by FastStar Universal SYBR-Green Master (Roche
Diagnostics, Indianapolis, IN, USA) on ABI 7300 (Thermo Fisher
Scientific, Inc.). Divergent primer was designed for selected
circRNA (circATP8B4: left primer CCACAGATGACTATTTTCGCCA and right
primer GGTCTCAACAGGTGGCCT). β-actin served as the internal
reference to normalize the expression of circRNAs. The
2−ΔΔCq method (14) was
used to calculate the expression level. Analysis was performed on
three independent EV samples.
Prediction of circRNA-miRNA-mRNA
network
The putative circRNA/miRNA interaction was predicted
using miRanda software (August 2010 release, http://www.microrna.org/microrna/home.do) (15) and RNAhybrid software (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/)
(16). miRanda was set according to
the following parameters: gapopenpenalty was −9.0, gapextendpenalty
was −4.0, score threshold was 140.0, energy threshold was 1.0
kcal/mol and scaling parameter was 4.0. The following RNAhybrid
setting parameters were used: miRNA and circRNA binding energy
<-26 kcal/mol; perfect miRNA seed complementarity with circRNA
sequence, P-value <0.05. The candidate miRNA target gene was
analyzed using miRanda and TargetScan software (http://www.targetscan.org/). In order to reduce the
false positive rate, only the circRNA/miRNA interaction or miRNA
target gene predicted by both two independent tools were taken into
account.
EV labeling and coculture
EVs were labeled with the green fluorescent dye
PKH67 (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) according to
the manufacturer's protocol. Normal U251 cells were incubated with
the PKH67-labeled RR-Evs in 37°C for 24 h. Afterwards, the
cocultured cells were washed twice and fixed with 4%
paraformaldehyde for 30 min at room temperature. After
counterstained with DAPI dihydrochloride (Beyotime Institute of
Biotechnology, Haimen, China), fluorescence imaging was captured
using confocal laser scanning microscopy (Carl Zeiss LSM710; Carl
Zeiss, Oberkochen, Germany).
Cell viability assay
Cell viability was performed using MTT assay
(Sigma-Aldrich; Merck KGaA). Following radiotherapy of 5 Gy, normal
U251 and the cocultured cells were seeded onto 96-well plates
(5.0×103 cells/well; n=6 for each condition). After 48
h, 20 µl MTT was added to each well and incubated for 4 h prior to
the solution of 150 µl DMSO. Then the optical density (OD) in each
individual well was recorded at 570 nm using a Multiskan Ascent
microplate reader (Thermo Fisher Scientific, Shanghai, China). Cell
viability was calculated as follows: Cell viability (%) =
(ODtreatment/ODcontrol) × 100%.
Statistical analysis
Statistical analyses were performed using GraphPad
Prism 5 (GraphPad Software, Inc., La Jolla, CA, USA) and R software
version 3.3.2. Student's t-test was used to determine significant
differences between EVs from U251 and RR-U251 cells. A two-sided
P-value <0.05 was considered to indicate a statistically
significant result.
Results
Characterization of EVs released by
U251 and RR-U251 cells
EVs released by the U251 and RR-U251 cells were
isolated from the cell culture media (Materials and methods). They
were characterized by the size and morphology using transmission
electron microscopy and nanoparticle tracking analysis. The
analyses confirmed the typical EV shape and size (Fig. 1A and B).
Identification of differential circRNA
profiles and prediction of their characteristics
RNA-seq was performed to profile circRNA expression
in a pair of Nor-EV and RR-EV samples. A total of 1,235 circRNAs
were detected. Differentially expressed circRNAs were identified
using fold change filtering (fold change >1.5). There were 63
circRNAs significantly upregulated in RR-EVs and 48 circRNAs were
significantly downregulated in RR-EVs. The top 20 differentially
expressed circRNAs are presented in Table I. We detected the functions of
differentially expressed circRNAs in EV target genes using Gene
Oncology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG)
pathway analysis. It showed that the target genes that were related
to upregulated circRNAs participated in various biological
processes, such as protein binding, regulation of mast cell
apoptotic process and negative regulation of mRNA 3′-end processing
(Fig. 2A). The upregulated circRNAs
could also have an impact on several vital pathways, such as cell
cycle pathway (Fig. 2B).
 | Table I.Top 20 differentially expressed
circRNAs in EVs isolated from RR-U251 and U251 cells. |
Table I.
Top 20 differentially expressed
circRNAs in EVs isolated from RR-U251 and U251 cells.
| circRNA | Chromosome | Start | End | Strand | Exon count | Gene name | Regulation | Fold change |
|---|
| circATP8B4 | chr15 | 50038767 | 50107008 | − | 5 | ATP8B4 | Up | 2.972 |
| circCCDC134 | chr22 | 41808874 | 41810291 | + | 3 | CCDC134 | Up | 2.972 |
| circATP6V1H | chr8 | 53769617 | 53772167 | − | 2 | ATP6V1H | Up | 2.841 |
| circMARCH6 | chr5 | 10415487 | 10417404 | + | 2 | MARCH6 | Up | 2.754 |
| circLRRK1 | chr15 | 100983527 | 101015402 | + | 9 | LRRK1 | Up | 2.754 |
| circANAPC1 | chr2 | 111808946 | 111825855 | − | 8 | ANAPC1 | Up | 2.637 |
| circNSD2 | chr4 | 1918140 | 1918623 | + | 1 | NSD2 | Up | 2.637 |
| circUBAP2 | chr9 | 33948373 | 33956146 | − | 3 | UBAP2 | Up | 2.475 |
| circSEPT9 | chr17 | 77402058 | 77402703 | + | 1 | SEPT9 | Up | 2.475 |
| circTBC1D1 | chr4 | 38089931 | 38103157 | + | 3 | TBC1D1 | Up | 2.475 |
| circRNF216 | chr7 | 5641153 | 5652510 | − | 2 | RNF216 | down | 2.972 |
| circMTCL1 | chr18 | 8718423 | 8720496 | + | 2 | MTCL1 | down | 2.754 |
| circNR2C1 | chr12 | 95028386 | 95057878 | − | 7 | NR2C1 | down | 2.754 |
| circATAD2 | chr8 | 123333877 | 123336532 | − | 3 | ATAD2 | down | 2.754 |
| circCSPP1 | chr8 | 67105904 | 67116122 | + | 4 | CSPP1 | down | 2.667 |
| circUBQLN1 | chr9 | 83677726 | 83678599 | − | 2 | UBQLN1 | down | 2.667 |
| circ ZDHHC21 | chr9 | 14639895 | 14680162 | − | 6 | ZDHHC21 | down | 2.625 |
| circDLG1 | chr3 | 197090911 | 197119530 | − | 4 | DLG1 | down | 2.625 |
| circOSBPL10 | chr3 | 31876432 | 31879830 | − | 2 | OSBPL10 | down | 2.584 |
| circSNX2 | chr5 | 122801868 | 122803613 | + | 3 | SNX2 | down | 2.584 |
Identification of differential miRNA
profiles and prediction of their characteristics
We analyzed miRNA expression in both U251 and
RR-U251 cells based on RNA-seq. In total, 2,577 miRNAs were
detected. Among these, 1,165 were present in RR-U251 cells; 1,170
were detected in U251 cells. We detected 171 upregulated and 184
downregulated miRNAs in the RR-U251 cells compared to the U251
cells. The top 20 differentially expressed miRNAs are presented in
Table II. Heatmap was constructed
to demonstrate the differential miRNA expression between U251 and
RR-U251 cells (Fig. 2C). GO and
KEGG pathway analysis was conducted. We found that the negative
target mRNAs related to downregulated miRNAs were associated with
various biological processes, such as protein binding, regulation
of mast cell apoptotic process and positive regulation of cellular
response to X-ray (Fig. 2D). The
downregulated miRNAs could also have an impact on several vital
pathways, such as the p53 signaling pathway.
 | Table II.Top 20 differentially expressed
miRNAs in U251 and RR-U251 cells. |
Table II.
Top 20 differentially expressed
miRNAs in U251 and RR-U251 cells.
| miRNA | log2 fold
change | U251 | RR-U251 | Regulation |
|---|
|
hsa-miR-6787-3p | 3.887 | 0.937 | 13.867 | up |
| hsa-miR-665 | 3.646 | 0.937 | 11.734 | up |
|
hsa-miR-6851-3p | 3.186 | 0.937 | 8.534 | up |
|
hsa-miR-200b-5p | 2.771 | 0.937 | 6.400 | up |
|
hsa-miR-4709-3p | 2.771 | 0.937 | 6.400 | up |
|
hsa-miR-4717-3p | 2.771 | 0.937 | 6.400 | up |
| hsa-miR-3131 | 2.508 | 0.937 | 5.333 | up |
| hsa-miR-378g | 2.508 | 0.937 | 5.333 | up |
|
hsa-miR-7110-3p | 2.186 | 0.937 | 4.267 | up |
|
hsa-miR-3180-3p | 2.061 | 2.812 | 2.812 | up |
|
hsa-miR-6739-3p | −2.984 | 8.437 | 1.067 | down |
| hsa-miR-548s | −2.984 | 8.437 | 1.067 | down |
|
hsa-miR-6843-3p | −2.814 | 7.500 | 1.067 | down |
|
hsa-miR-6854-5p | −2.814 | 7.500 | 1.067 | down |
|
hsa-miR-5009-5p | −2.621 | 6.562 | 1.067 | down |
| hsa-miR-766-5p | −2.592 | 12.213 | 2.025 | down |
| hsa-miR-548u | −2.474 | 10.412 | 1.874 | down |
|
hsa-miR-548a-3p | −2.474 | 5.206 | 0.937 | down |
| hsa-miR-3175 | −2.403 | 15.185 | 2.871 | down |
|
hsa-miR-3130-5p | −2.403 | 7.593 | 1.436 | down |
Validation for the expression of
circRNAs
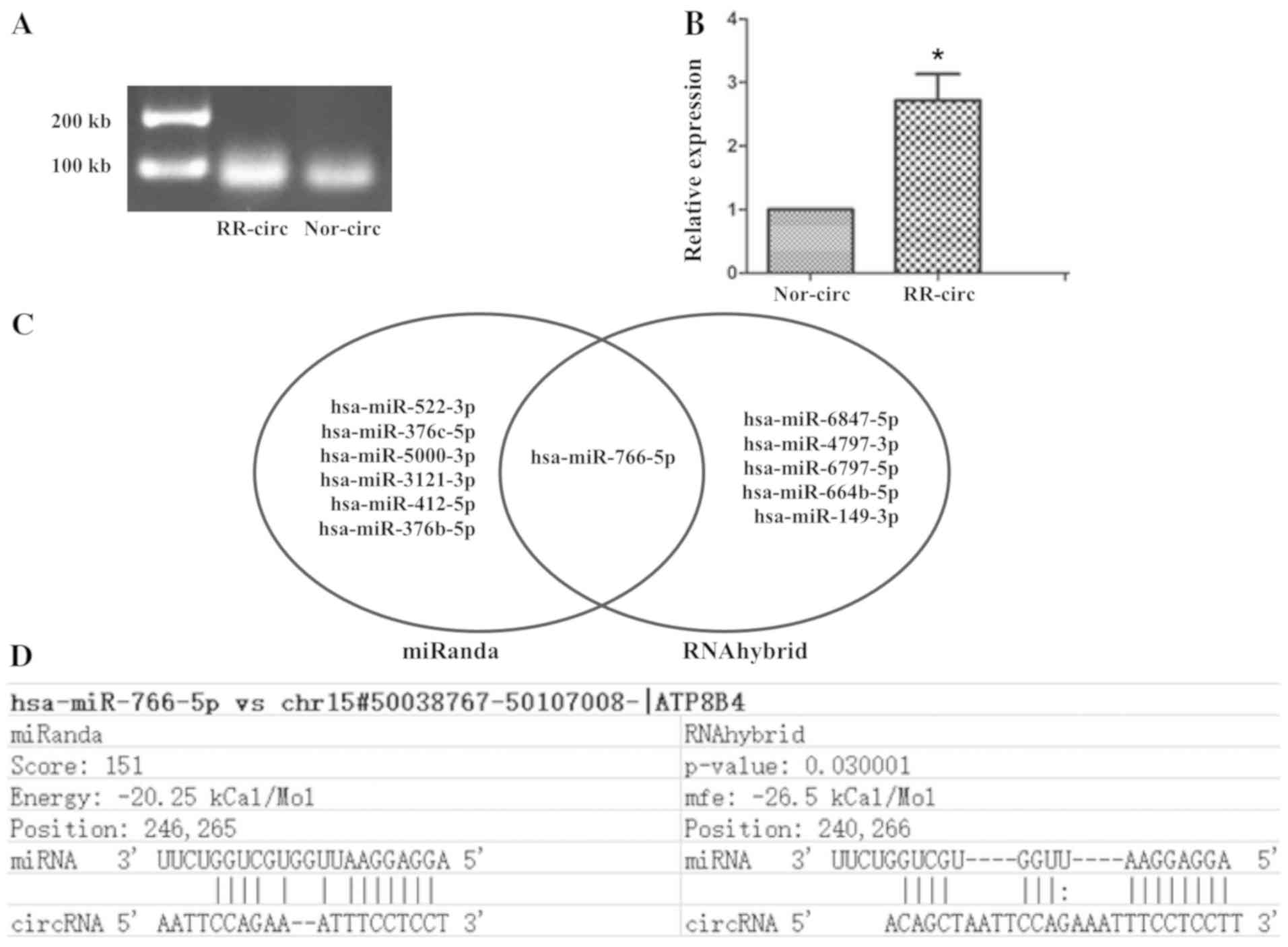
To validate the RNA-seq results, RT-qPCR analysis
was performed on one candidate circRNA in EVs with the top fold
change. The candidate circATP8B4 is made up of five exons. The
analysis was performed on three independent samples for RR-EVs and
Nor-EVs, respectively. It showed that the average expression level
of circATP8B4 was significantly higher in RR-EVs than those from
Nor-EVs (P<0.05; Fig. 3A and
B).
Prediction of circRNA-miRNA-mRNA
network
Literature has reported that circRNAs serve as miRNA
sponges in the microenvironment and regulate gene expression.
Therefore, we investigated those miRNAs with a fold change >1.5
in cells that could adsorb on circATP8B4. The sequence of
circATP8B4 FASTA format was saved, and then we used miRanda and
RNAhybrid softwares to predict the miRNAs interacting with
circATP8B4. Only the circATP8B4-miRNA interactions predicted by
both softwares were taken into account. The results showed that
circATP8B4 may regulate miR-766-5p function as a microRNA sponge
(Fig. 3C and D). Fourteen negative
target mRNAs of miR-766-5p were predicted using miRanda and
TargetScan software (Fig. 4A).
There are two potential binding sites of miR-766-5p present in
circATP8B4. GO analysis showed that the target genes were
associated with many biological pathways, cellular components and
molecular functions (Fig. 4B). They
also could participate in many pathways such as EGFR tyrosine
kinase inhibitor resistance according to KEGG analysis (Fig. 4C).
EV coculture and cell viability
assay
In order to verify ceRNA mechanism of circATP8B4
preliminarily, normal U251 cells were incubated with the
PKH67-labeled RR-EVs. After 24 h, RR-EVs were detected in
cocultured cells using confocal laser scanning microscopy (Fig. 5A). The normal U251 and cocultured
cells were treated with 5-Gy radiotherapy. MTT assay demonstrated
that cell viability of the cocultured cells was significantly
higher than that of the normal U251 cells (Fig. 5B).
Discussion
Circular RNAs (circRNAs) exist widely in various
organisms and are aberrantly expressed in epithelial tumors, such
as bladder (17), breast cancer
(18), basal cell carcinoma
(19) and colorectal cancer
(20) and in stromal tumors, such
as glioma (21). Studies have
reported that circRNAs are enriched in extracellular vesicles (EVs)
isolated from liver cancer or serum of endometrial cancer patients
(10,11,20).
Xu et al found that the number of upregulated circRNAs was
more than the number of downregulated circRNAs in EVs from patients
with endometrial cancer (11). In
the present study, we profiled expression levels of circRNAs
isolated from EVs in U251 and RR-U251 cells using RNA-seq, and a
total of 111 differential circRNAs were identified. We detected 63
upregulated and 48 downregulated circRNAs in RR-EVs. Our data were
similar to previous findings.
circRNAs are organization- and disease-specific, and
can thus be potential biomarkers for disease diagnosis (9,10,22–24).
Zhu et al found that circBRAF was significantly
downregulated in glioma patients with high pathological grade. The
level of circBRAF was an independent biomarker for predicting
survival in glioma patients (25).
In the present study, to validate the RNA-seq findings, we
performed RT-qPCR analysis for the most abundant candidate circRNA.
The expression level of circATP8B4 was significantly higher in
RR-EVs (EVs from radioresistant U251 cells) than this level in
Nor-EVs (EVs from U251 cells). Thus, circATP8B4 from EVs could be a
potential biomarker for glioma radioresistance.
Recent studies have found that circRNAs can regulate
miRNA function as miRNA sponges and play a significant role in
transcriptional control (26–29).
Therefore, we predicted the downregulated miRNAs in U251 cells that
could possibly interact with circATP8B4 using miRanda and RNAhybrid
softwares, and candidate miR-766-5p was selected. GO and KEGG
pathway analysis showed that the target genes that were related to
downregulated miRNAs participated in various biological processes,
such as cell apoptosis. Growing evidence indicates that miR-766
acts as a tumor promoter or suppressor in multiple cancer types
(30–32). Afgar et al found that miR-766
can reactivate the expression of tumor-suppressor genes in
colorectal cancer cell lines through DNMT3B gene inhibition
(33). This indicates that
circATP8B4 in RR-EVs might function as an miR-766 sponge to promote
the proliferation and radioresistance of normal U251 cells.
Acquired radioresistance has been considered as one
of the most important reasons for the treatment failure for GBM
patients. Studies have shown that the tumor microenvironment is
involved in the formation of glioma radioresistance (3,34). In
the tumor microenvironment, EVs are associated with cell-to-cell
signaling transmission and may influence processes in glioma
radioresistance. EVs derived from GBM cells were reported to
promote temozolomide (TMZ) chemotherapy resistance of recipient
cells via the transfer of cell-transforming proteins, mRNAs and
miRNAs (35). However, the role of
circRNAs in EVs associated with glioma radioresistant is still
unclear. The present study showed that RR-EVs could be transferred
to normal glioma U251 cells, and MTT assay showed those EVs are
involved in the radioresistance of normal glioma cells. This
implies that biomolecules such as ncRNAs or mRNAs in RR-EVs may
play an important role in glioma radioresistance. Our preliminary
experiments provide the possibility that circATP8B4 from RR-EVs may
promote glioma radioresistance.
It should be noted that in the present study, we
only identified one candidate upregulated circRNA in RR-EVs with
the top fold change. Future research into the ceRNA mechanism of
circATP8B4/miR-766/mRNA is still needed. The exact role of
circATP8B4 in RR-EVs should be confirmed by overexpression and
silencing experiments in vivo and in vitro. In
addition, future clinical studies should be performed to verify the
clinical significance of circATP8B4 in glioma radioresistance.
In conclusion, we identified 63 upregulated and 48
downregulated circRNAs in RR-EVs compared with those in Nor-EVs.
Gene Ontology and KEGG analyses were conducted to predict the
characteristics of the circRNAs in EVs. We performed RT-qPCR
analysis for candidate circATP8B4. CircATP8B4 from RR-EVs may be
transferred to normal glioma U251 cells and circATP8B4 may act as
an miR-766 sponge to promote cell radioresistance. Elucidation of
the function and mechanism warrants further research.
Acknowledgements
Not applicable.
Funding
The present study was funded by the General Project
of Nanjing Medical Science and Technology Development (no.
YKK16145) and the Jiangsu Provincial Medical Innovation Team (no.
CXTDA2017050).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
LG and HL designed and conceived the study. MZ and
JX performed the experiments; SZ, YL and HX assisted with the cell
culture and EV isolation; JX and YL prepared the figures; SZ was
responsible for the statistical analysis of the results. LG and MZ
wrote the manuscript. LG, HL and HX reviewed and edited the
manuscript and provided the funds. All authors read and approved
the manuscript and agree to be accountable for all aspects of the
research in ensuring that the accuracy or integrity of any part of
the work are appropriately investigated and resolved.
Ethics approval and consent
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
RR-U251
|
radioresistant U251 cells
|
|
circRNA
|
circular RNA
|
|
EVs
|
extracellular vesicles
|
|
Nor-EVs
|
EVs isolated from U251 cells
|
|
RR-EVs
|
EVs isolated from RR-U251 cells
|
|
RNA-seq
|
RNA sequencing
|
|
RT-qPCR
|
real-time quantitative PCR
|
|
miRNA
|
microRNA
|
|
GBM
|
glioblastoma
|
|
PBS
|
phosphate-buffered saline
|
|
GO
|
Gene Oncology
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
References
|
1
|
Shah AH, Graham R, Bregy A, Thambuswamy M
and Komotar RJ: Recognizing and correcting failures in glioblastoma
treatment. Cancer Invest. 32:299–302. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Quan JJ, Song JN and Qu JQ: PARP3
interacts with FoxM1 to confer glioblastoma cell radioresistance.
Tumour Biol. 36:8617–8624. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Han X, Xue X, Zhou H and Zhang G: A
molecular view of the radioresistance of gliomas. Oncotarget.
8:100931–100941. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Li W, Guo F, Wang P, Hong S and Zhang C:
miR-221/222 confers radioresistance in glioblastoma cells through
activating Akt independent of PTEN status. Curr Mol Med.
14:185–195. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Pant S, Hilton H and Burczynski ME: The
multifaceted exosome: Biogenesis, role in normal and aberrant
cellular function, and frontiers for pharmacological and biomarker
opportunities. Biochem Pharmacol. 83:1484–1494. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yang SJ, Wang DD, Li J, Xu HZ, Shen HY,
Chen X, Zhou SY, Zhong SL, Zhao JH and Tang JH: Predictive role of
GSTP1-containing exosomes in chemotherapy-resistant breast cancer.
Gene. 623:5–14. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tang Y, Cui Y, Li Z, Jiao Z, Zhang Y, He
Y, Chen G, Zhou Q, Wang W, Zhou X, et al: Radiation-induced
miR-208a increases the proliferation and radioresistance by
targeting p21 in human lung cancer cells. J Exp Clin Cancer Res.
12:35–37. 2016.
|
|
8
|
Barbagallo D, Caponnetto A, Cirnigliaro M,
Brex D, Barbagallo C, D'Angeli F, Morrone A, Caltabiano R,
Barbagallo GM, Ragusa M, et al: CircSMARCA5 inhibits migration of
glioblastoma multiforme cells by regulating a molecular axis
involving splicing factors SRSF1/SRSF3/PTB. Int J Mol Sci.
19:E4802018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li G, Yang H, Han K, Zhu D, Lun P and Zhao
Y: A novel circular RNA, hsa_circ_0046701, promotes carcinogenesis
by increasing the expression of miR-142-3p target ITGB8 in glioma.
Biochem Biophys Res Commun. 498:254–261. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J,
Chen D, Gu J, He X and Huang S: Circular RNA is enriched and stable
in exosomes: A promising biomarker for cancer diagnosis. Cell Res.
25:981–984. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Xu H, Gong Z, Shen Y, Fang Y and Zhong S:
Circular RNA expression in extracellular vesicles isolated from
serum of patients with endometrial cancer. Epigenomics. 10:187–197.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Dai X, Chen C, Yang Q, Xue J, Chen X, Sun
B, Luo F, Liu X, Xiao T, Xu H, et al: Exosomal circRNA_100284 from
arsenite-transformed cells, via microRNA-217 regulation of EZH2, is
involved in the malignant transformation of human hepatic cells by
accelerating the cell cycle and promoting cell proliferation. Cell
Death Dis. 9:4542018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Du HQ, Wang Y, Jiang Y, Wang CH, Zhou T,
Liu HY and Xiao H: Silencing of the TPM1 gene induces
radioresistance of glioma U251 cells. Oncol Rep. 33:2807–2814.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2-ΔΔCT method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Enright AJ, John B, Gaul U, Tuschl T,
Sander C and Marks DS: MicroRNA targets in Drosophila. Genome Biol.
5:R12003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Krüger J and Rehmsmeier M: RNAhybrid:
microRNA target prediction easy, fast and flexible. Nucleic Acids
Res. 34:W451–W454. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhong Z, Lv M and Chen J: Screening
differential circular RNA expression profiles reveals the
regulatory role of circTCF25-miR-103a-3p/miR-107-CDK6 pathway in
bladder carcinoma. Sci Rep. 6:309192016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Nair AA, Niu N, Tang X, Thompson KJ, Wang
L, Kocher JP, Subramanian S and Kalari KR: Circular RNAs and their
associations with breast cancer subtypes. Oncotarget.
7:80967–80979. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sand M, Bechara FG, Sand D, Gambichler T,
Hahn SA, Bromba M, Stockfleth E and Hessam S: Circular RNA
expression in basal cell carcinoma. Epigenomics. 8:619–632. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xie H, Ren X, Xin S, Lan X, Lu G, Lin Y,
Yang S, Zeng Z, Liao W, Ding YQ, et al: Emerging roles of
circRNA_001569 targeting miR-145 in the proliferation and invasion
of colorectal cancer. Oncotarget. 7:26680–26691. 2016.PubMed/NCBI
|
|
21
|
Yuan Y, Jiaoming L, Xiang W, Yanhui L, Shu
J, Maling G and Qing M: Analyzing the interactions of mRNAs,
miRNAs, lncRNAs and circRNAs to predict competing endogenous RNA
networks in glioblastoma. J Neurooncol. 137:493–502. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Dou Y, Cha DJ, Franklin JL, Higginbotham
JN, Jeppesen DK, Weaver AM, Prasad N, Levy S, Coffey RJ, Patton JG,
et al: Circular RNAs are down-regulated in KRAS mutant colon cancer
cells and can be transferred to exosomes. Sci Rep. 6:379822016.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhuo F, Lin H, Chen Z, Huang Z and Hu J:
The expression profile and clinical significance of circRNA0003906
in colorectal cancer. Onco Targets Ther. 25:5187–5193. 2017.
View Article : Google Scholar
|
|
24
|
Zhang Y, Liang W, Zhang P, Chen J, Qian H,
Zhang X and Xu W: Circular RNAs: Emerging cancer biomarkers and
targets. J Exp Clin Cancer Res. 36:1522017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhu J, Ye J, Zhang L, Xia L, Hu H, Jiang
H, Wan Z, Sheng F, Ma Y, Li W, et al: Differential expression of
circular RNAs in glioblastoma multiforme and its correlation with
prognosis. Transl Oncol. 10:271–279. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Rong D, Sun H, Li Z, Liu S, Dong C, Fu K,
Tang W and Cao H: An emerging function of circRNA-miRNAs-mRNA axis
in human diseases. Oncotarget. 8:73271–73281. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zheng Q, Bao C, Guo W, Li S, Chen J, Chen
B, Luo Y, Lyu D, Li Y, Shi G, et al: Circular RNA profiling reveals
an abundant circHIPK3 that regulates cell growth by sponging
multiple miRNAs. Nat Commun. 7:112152016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Nan A, Chen L, Zhang N, Liu Z, Yang T,
Wang Z, Yang C and Jiang Y: A novel regulatory network among
LncRpa, CircRar1, MiR-671 and apoptotic genes promotes lead-induced
neuronal cell apoptosis. Arch Toxicol. 91:1671–1684. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li YC, Li CF, Chen LB, Li DD, Yang L, Jin
JP and Zhang B: MicroRNA-766 targeting regulation of SOX6
expression promoted cell proliferation of human colorectal cancer.
Onco Targets Ther. 8:2981–2988. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li X, Shi Y, Yin Z, Xue X and Zhou B: An
eight-miRNA signature as a potential biomarker for predicting
survival in lung adenocarcinoma. J Transl Med. 12:1592014.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Liang H, Li X, Wang L, Yu S, Xu Z, Gu Y,
Pan Z, Li T, Hu M, Cui H, et al: MicroRNAs contribute to
promyelocyte apoptosis in As2O3-treated APL cells. Cell Physiol
Biochem. 32:1818–1829. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Afgar A, Fard-Esfahani P, Mehrtash A,
Azadmanesh K, Khodarahmi F, Ghadir M and Teimoori-Toolabi L:
MiR-339 and especially miR-766 reactivate the expression of tumor
suppressor genes in colorectal cancer cell lines through DNA
methyltransferase 3B gene inhibition. Cancer Biol Ther.
17:1126–1138. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Combs SE, Schmid TE, Vaupel P and Multhoff
G: Stress response leading to resistance in glioblastoma - The need
for innovative radiotherapy (iRT) concepts. Cancers. 8:E152016.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Mondal A, Kumari Singh D, Panda S and
Shiras A: Extracellular vesicles as modulators of tumor
microenvironment and disease progression in glioma. Front Oncol.
7:1442017. View Article : Google Scholar : PubMed/NCBI
|