Introduction
Pancreatic cancer is an aggressive disease with an
extremely poor clinical outcome and a 5-year survival rate of
<5% (1,2). Approximately 37,170 new cases of
pancreatic cancer were diagnosed in 2007 and nearly 33,370
mortalities were caused by this disease in the US (3). More than 80% of pancreatic ductal
adenocarcinoma (PDAC) patients (accounting for 85% of pancreatic
cancer) are diagnosed at a stage that is already regional or
distant metastasized (4). The
5-year survival rate for PDAC, improves from 2 to 23% if the
disease is diagnosed at its localized stage compared to a distant
metastatic stage (5). Chemotherapy
is an alternative for unresectable PDAC; however, limited survival
benefit still exists. Thence, it is extremely significant to
unearth novel diagnostic biomarkers for PDAC, which can indicate
the use of curative surgical treatment (6).
Cysteine proteases, expressed widely in tissues, are
a group of intracellular proteins with protein degradation activity
which are associated with a wide variety of biological processes,
including inflammation, modulation of the immune response and
facilitating the progression of malignant tumors (7–9).
Previous clinical studies have revealed that the cystatin
superfamily proteins inhibit the proteolytic activity of cysteine
proteases specifically in attenuating the aggressiveness of various
malignant tumors (10–12).
Among the three distinct subfamilies belonging to
the cystatin superfamily, the family 1 cystatins, represented by
cystatin A (CSTA) and B (CSTB), lack disulfite bonds as well as
signal peptides and only function intracellularly. Family 2
cystatin subunits are secreted proteins, all composed of 115–120
amino acids with two interchain disulfide bonds. L- and
H-kininogens of family 3 cystatin subunits are complex glycosylated
cytoplasmic proteins with type-2-like cystatin domains and
bradykinin moiety (13–15).
There are seven members in the family 2 cystatins,
cystatin SN (CST1), SA (CST2), C (CST3), S (CST4), D (CST5), E/M
(CST6), and F (CST7). Family 2 cystatins are types of cysteine
protease inhibitors found in various human fluids and secretions
that appear to provide protection. To date, the expression level of
the family 2 cystatins has been reported to be associated with
tumor biology function, progression and prognosis in breast,
small-cell lung and colorectal cancer (16–19).
However, the key family 2 cystatins subunit in PDAC patients
remains unknown.
In the present study, we aimed to investigate the
clinical implications of the family 2 cystatins in early-stage PDAC
using bioinformatics and survival analysis. Finally, CST7 was
identified as a key cystatin subunit. Furthermore, a prognostic
model for patients with early-stage PDAC was constructed.
Materials and methods
Biological function of the cystatin
gene family
To investigate the potential molecular roles and
related pathways of the cystatin gene family, we first analyzed
gene-gene interactions using GeneMANIA (http://www.genemania.org/). Protein-protein
interactions of cystatin genes were ascertained by the Search Tool
for the Retrieval of Interacting Genes/Proteins (STRING, http://string-db.org/). Gene Ontology (GO) terms
annotation and gene function enrichment analysis of cystatin genes
were performed using the Database for Annotation, Visualization and
Integrated Discovery (DAVID, http://david.ncifcrf.gov/home.jsp) version 6.8.
Cystatin gene family expression in
tissues
Expression of CST mRNA between paraneoplastic
tissues and primary cancer tissues was performed by Metabolic gEne
RApid Visualizer (MERAV, http://merav.wi.mit.edu/). To investigate the cystatin
gene family expression level in various normal tissues, we
generated a heatmap using the Genotype-Tissue Expression (GTEx)
portal (https://www.gtexportal.org/).
Co-expression relationships between CST genes in tumor tissues were
assessed by the Pearson correlation coefficient. Pearson
correlation was performed using corrplot R package (available from
http://github.com/taiyun/corrplot).
Cystatin gene family in survival
analysis of PDAC patients
In the survival analysis, patients were divided into
low- and high-level groups according to median mRNA expression
level. We screened key cystatin isoforms that were associated with
the prognosis of early-stage PDAC in The Cancer Genome Atlas (TCGA;
http://portal.gdc.cancer.gov/). All the
expression data for cystatin and the relevant clinical parameters
were obtained from the University of California, Santa Cruz Xena
browser (UCSC Xena: http://xena.ucsc.edu/). The inclusion criteria of
patients enrolled in the present study were as follows: i)
available survival data; ii) patients who underwent
pancreaticoduodenectomy and the histology type was confirmed as
PDAC; iii) stage I or II PDAC according to the 7th American Joint
Committee on Cancer (AJCC); and iv) the tissues samples were
collected by resection or biopsy prior to receiving chemotherapy.
The number of AJCC stage III or IV patients was 8. To control bias,
PDAC patients with AJCC stage III or IV and those non-PDAC
histology types were excluded. Inclusion and exclusion criteria
were implemented in accordance with a previous study (20). Finally, CST7 was selected as a key
cystatin isoform in further analysis. Overall survival (OS) and
disease-free survival (DFS) was calculated using SPSS version 24.0.
Prognosis risk was established on CST7 expression level and trend
direction depended on a regression coefficient (β) as the positive
and negative correlation that was derived from a univariate Cox
proportional hazards regression model. Hazard ratios (HR) and 95%
confidence intervals (CIs) were also computing in the hazards
regression model. Combined survival analysis was performed to
identify the relationship between CST7 expression and
clinicopathological features in the prognosis of patients with
early-stage PDAC. Multivariate regression analysis of CST7
expression in PDAC patients was adjusted for prognosis-related
clinical factors from univariate Cox proportional hazards
regression model.
Prognostic nomogram conduction
Based on the examination and transformation of
variables evaluated in a univariate Cox proportional hazards
regression model, we formulated a prognostic nomogram model using
the rms (21) R package. The
prognostic performance of the nomogram was measured by concordance
index (C-index). The C-index was calculated to evaluate the
performance of each model in the survival data and was considered a
measure of predictive ability (22). Bootstraps with the resamples of PDAC
patients were applied to predict survival probability and to obtain
a more realistic bias-corrected estimate of the model coefficients
and the C-index.
Gene set enrichment analysis (GSEA)
for CST7 in PDAC patients
Setting CST gene expression levels as population
phenotypes in GSEA (http://software.broadinstitute.org/gsea/index.jsp), we
further analyzed gene expression omics predictions and assessed
related pathways and molecular mechanisms in PDAC patients. A
nominal P-value <0.05 and false discovery rate (FDR) <0.25 of
the enrichment gene sets in the analysis were considered
statistically significant.
Statistical analysis
All statistical analyses were performed using SPSS
version 24.0 (SPSS; IBM Corp., Armonk, NY, USA) and R3.4.1
(www.r-project.org). A two-sided P-value of
<0.05 was considered statistically significant. The survival
curves and heatmap were depicted by GraphPad Prism7.01 (GraphPad
Software, Inc., La Jolla, CA, USA). The Kaplan-Meier survival
curves were compared by the log-rank test.
Results
The pathways, interaction networks and
GO term analysis of the cystatin gene family
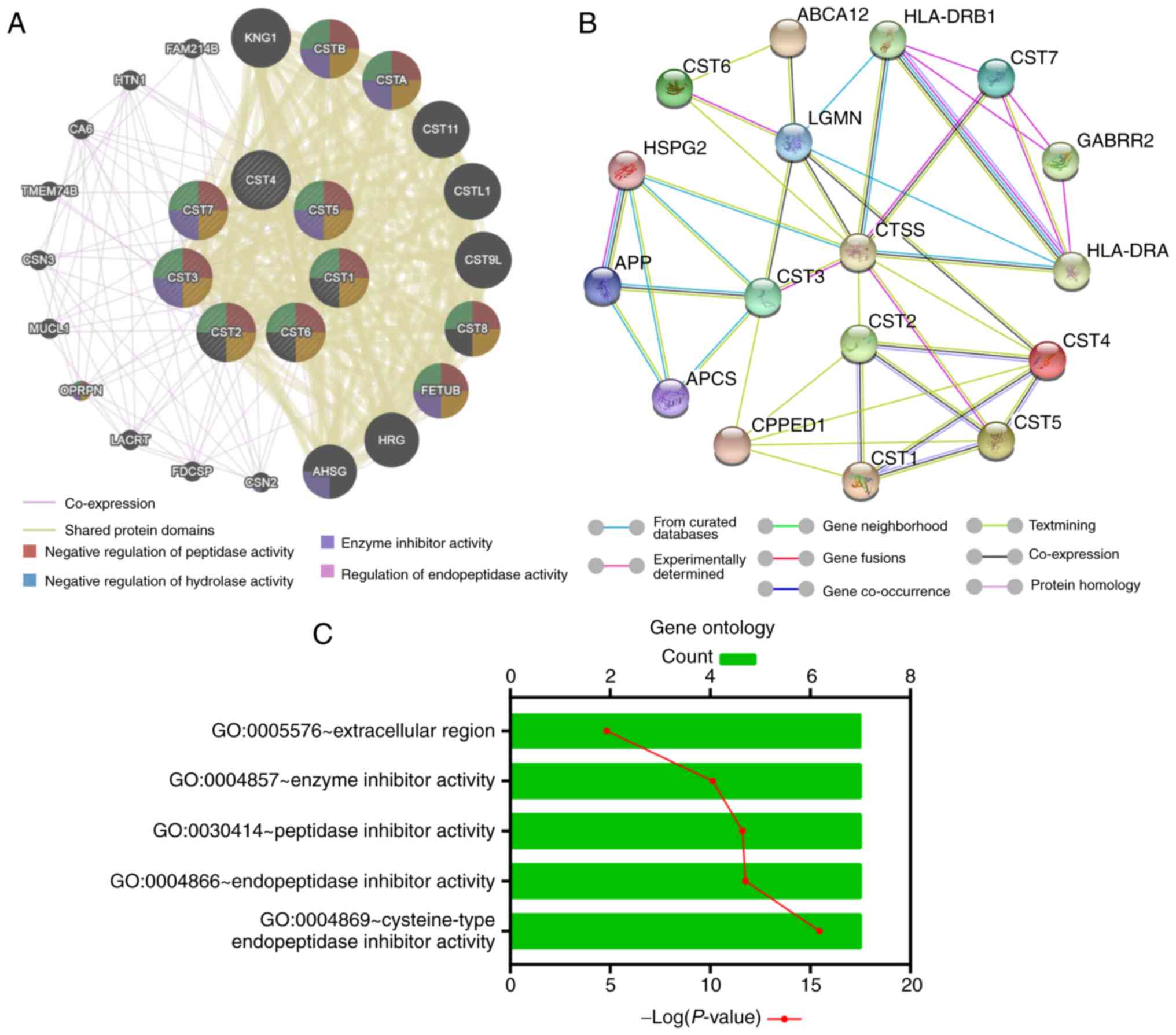
Using the gene-gene interaction analysis in
GeneMANIA, we identified that the cystatin gene family mainly
shared protein domains with other molecules, such as CSTA, CSTB and
FETUB (Fig. 1A). Furthermore,
protein-protein interaction networks by STRING indicated that
cystatin proteins may be associated with immune proteins, such as
human lymphocyte antigen (Fig. 1B).
GO term enrichment analysis revealed that the cystatin family
encoded products which play roles in the extracellular region and
contains inhibitory activity against the peptidase and
endopeptidase of the enzyme (Fig.
1C).
Expression of the CST gene family in
tissues
The results of MERAV revealed that the expression of
the cystatin gene family was different between pancreas tumor and
paraneoplastic tissues (Fig. 2A).
Expression of CST1, CST2, CST4 and CST6 was upregulated in pancreas
tumors. The median CST7 mRNA level was increased in tumor tissues
but the difference was not statistically significant. From another
perspective, CST3 and CST5 mRNA levels were downregulated in
pancreas tumor tissues. Moreover, in the GTEx analysis of various
normal tissues, CST3 expression was higher than that of other
cystatin isoforms (Fig. 2B). CST7
is partly upregulated in whole blood and spleen. Co-expression
relationships between CST genes in tumor tissues, as evaluated
using Pearson's correlation, revealed that CST4 and CST5 had a
positive correlation (Fig.
S1).
Survival analysis of the cystatin gene
family in TCGA early-stage PDAC patients
The relationship between clinicopathological
features and prognosis of patients with early-stage PDAC in TCGA is
presented in Table SI. All
patients were divided into groups according to median values or
stages of the clinicopathological features. In the univariate
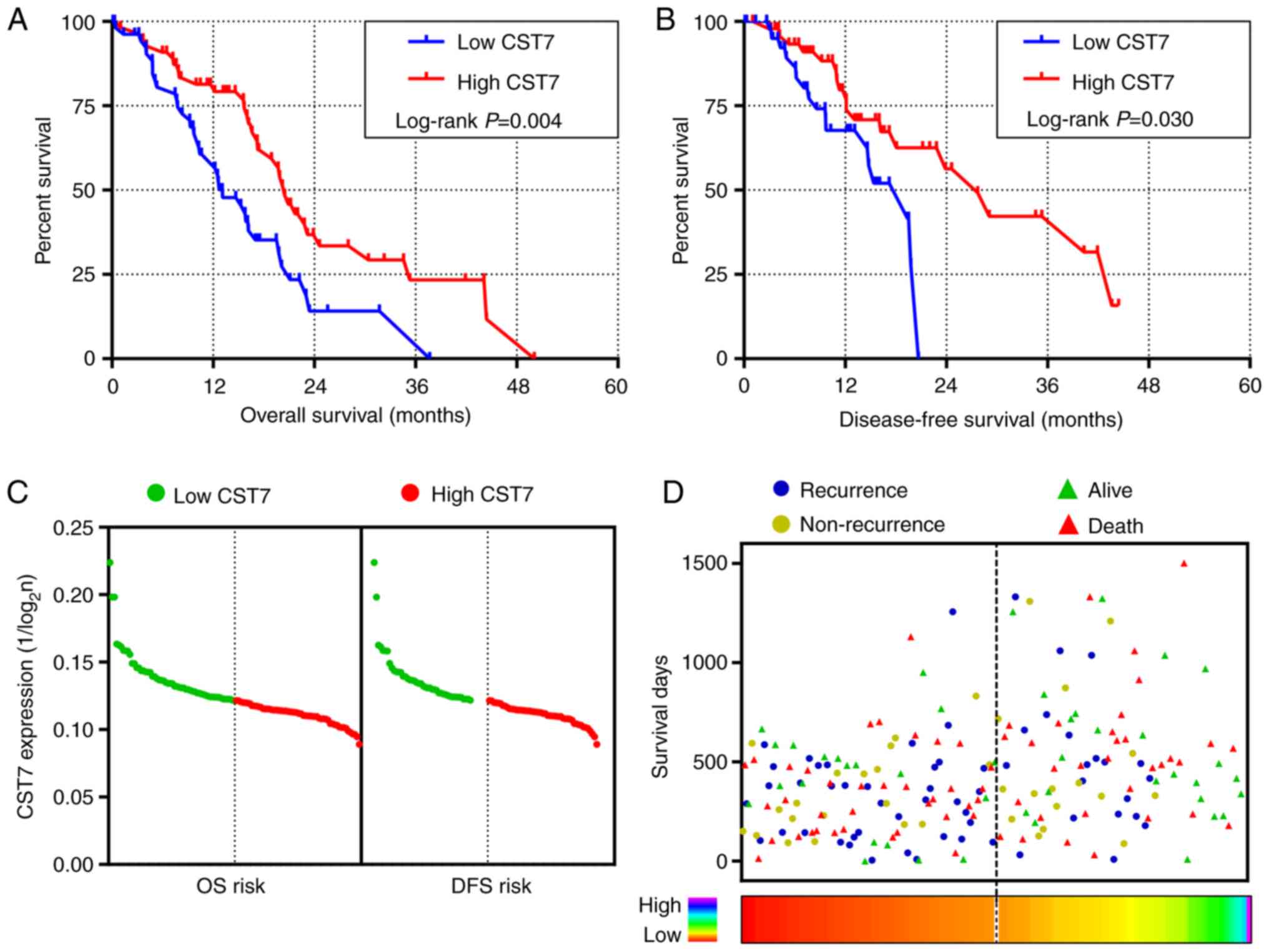
survival analysis, high CST7 expression was related to low-risk in
the OS and DFS of PDAC (Fig. 3A and
B and Table I). Based on the
regression coefficient (β=−0.709) in univariate Cox proportional
hazards regression, prognosis risk had a negative correlation with
CST7 expression level (Fig. 3C and
Table I). Patients diagnosed with
PDAC had longer OS and DFS when they harbored higher CST7
expression (Fig. 3D). Adjusted
pathologic stage T, pathologic stage N, histologic grade, radical
resection and targeted molecular therapy in the multivariate
survival analysis, CST2, CST3 and CST7 were associated with the OS
of PDAC patients. Higher CST2, CST3, and CS7 expression level have
a lower risk in OS of PDAC patients (HR=0.46, 0.50 and 0.44,
respectively; Table I). However,
none of the cystatin gene family members were associated with DFS
in early-stage PDAC. In this procedure, CST7 was selected as a key
cystatin isoform.
 | Table I.Survival analysis of the cystatin
gene family in patients with early-stage PDAC. |
Table I.
Survival analysis of the cystatin
gene family in patients with early-stage PDAC.
| Variables |
| MST (days) | P-value | Crude HR (95%
CI) | P-value | βa | Adjusted
HRb (95% CI) | P-value | MRT (days) | P-value | Crude HR (95%
CI) | P-value | βa | Adjusted
HRb (95% CI) | P-value |
|---|
| CST1 | Low | 596 | 0.210 |
|
|
|
|
| 593 | 0.698 |
|
|
|
|
|
|
| High | 481 |
| 1.36 | 0.212 | 0.310 | 1.14 | 0.690 | 620 |
| 1.15 | 0.699 | 0.137 | 1.76 | 0.202 |
|
|
|
|
| (0.84-2.22) |
|
| (0.61-2.14) |
|
|
| (0.57-2.29) |
|
| (0.74-4.18) |
|
| CST2 | Low | 473 | 0.079 |
|
|
|
|
| 831 | 0.421 |
|
|
|
|
|
|
| High | 607 |
| 0.65 | 0.082 | −0.430 | 0.46 | 0.008 | 581 |
| 1.33 | 0.422 | 0.286 | 1.20 | 0.649 |
|
|
|
|
| (0.40-1.06) |
|
|
(0.26-0.813) |
|
|
| (0.66-2.68) |
|
| (0.54-2.67) |
|
| CST3 | Low | 486 | 0.351 |
|
|
|
|
| 620 | 0.674 |
|
|
|
|
|
|
| High | 568 |
| 0.79 | 0.352 | −0.233 | 0.50 | 0.015 | 593 |
| 0.87 | 0.675 | −0.145 | 0.91 | 0.810 |
|
|
|
|
| (0.49-1.29) |
|
|
(0.28-0.88) |
|
|
| (0.44-1.70) |
|
| (0.42-1.98) |
|
| CST4 | Low | 596 | 0.270 |
|
|
|
|
| 593 | 0.986 |
|
|
|
|
|
|
| High | 481 |
| 1.31 | 0.272 | 0.271 | 1.37 | 0.276 | 716 |
| 0.99 | 0.986 | −0.006 | 1.45 | 0.358 |
|
|
|
|
| (0.81-2.13) |
|
| (0.78-2.43) |
|
|
| (0.51-1.94) |
|
| (0.66-3.18) |
|
| CST5 | Low | 517 | 0.667 |
|
|
|
|
| 831 | 0.092 |
|
|
|
|
|
|
| High | 518 |
| 1.11 | 0.667 | 0.106 | 1.31 | 0.354 | 486 |
| 1.78 | 0.096 | 0.577 | 1.45 | 0.348 |
|
|
|
|
| (0.69-1.80) |
|
| (0.74-2.31) |
|
|
| (0.90-3.51) |
|
| (0.67-3.15) |
|
| CST6 | Low | 517 | 0.895 |
|
|
|
|
| 620 | 0.295 |
|
|
|
|
|
|
| High | 518 |
| 0.97 | 0.896 | −0.032 | 1.17 | 0.572 | 593 |
| 1.45 | 0.298 | 0.369 | 2.15 | 0.078 |
|
|
|
|
| (0.60-1.56) |
|
| (0.68-2.04) |
|
|
| (0.72-2.90) |
|
| (0.92-5.06) |
|
| CST7 | Low | 393 | 0.004 |
|
|
|
|
| 581 | 0.030 |
|
|
|
|
|
|
| High | 614 |
| 0.49 | 0.005 | −0.709 | 0.44 | 0.004 | 831 |
| 0.45 | 0.034 | −0.800 | 0.56 | 0.204 |
|
|
|
|
|
(0.30-0.80) |
|
|
(0.25-0.77) |
|
|
|
(0.21-0.94) |
|
| (0.23-1.37) |
|
To investigate the correlation between CTS7
expression and clinical features in the prognosis of PDAC patients,
we performed a combined analysis of CST7 mRNA level with age, sex,
tumor dimension and residual tumor status. The results revealed
that high CST7 expression in groups with age ≤60 years, females,
tumor dimension and radical resection had a favorable outcome in
PDAC patients (all P<0.05; Fig.
4). Adjusting for number of positive lymph nodes, histologic
grade, radiation therapy and targeted molecular therapy in
multivariate regression analysis, groups of high CST7 expression
and age ≤60 years, male/female, dimension >3 cm and radical
resection had a lower risk in the OS of PDAC patients (all
P<0.05; Table II). In the DFS
of PDAC patients, HRs for high CST7 expression and age >60
years, male/female, dimension ≤3 cm and radical resection were
0.14, 0.17/0.24, 0.23 and 0.17, respectively (Table II).
 | Figure 4.Combined survival analysis of CST7
expression and clinicopathological features in patients with
early-stage PDAC. (A-D) Kaplan-Meier survival curves of OS for high
and low CST7 expression combined with age, sex, tumor dimension,
and residual tumor status, respectively. (E-H) Kaplan-Meier
survival curves of DFS for high and low CST7 expression combined
with age, sex, tumor dimension, and residual tumor status,
respectively. CST7, cystatin F; PDAC, pancreatic ductal
adenocarcinoma; OS, overall survival; DFS, disease-free
survival. |
 | Table II.Combined survival analysis of CST7
expression in patients with early-stage PDAC in TCGA. |
Table II.
Combined survival analysis of CST7
expression in patients with early-stage PDAC in TCGA.
| Variables | Patients
(n=112) | MST (months) | P-value | Crude HR (95%
CI) | Adjusted
HRa (95% CI) | Adjusted
P-value | MRT (months) | P-value | Crude HR (95%
CI) | Adjusted
HRa (95% CI) | Adjusted
P-value |
|---|
| Age+CST7 group |
| Age
≤60+low CST7 | 14 | 12.2 | 0.010 |
|
| 0.012 | 14.63 | 0.078 |
|
| 0.016 |
| Age
≤60+high CST7 | 24 | 22.8 |
| 0.29
(0.12-0.71) | 0.31
(0.11-0.93) | 0.037 | 27.7 |
| 0.35
(0.13-1.01) | 0.35
(0.12-1.07) | 0.065 |
| Age
>60+low CST7 | 42 | 15.27 |
| 0.92
(0.44-1.90) | 1.09
(0.48-2.47) | 0.841 | 19.37 |
| 0.58
(0.21-1.58) | 0.44
(0.16-1.27) | 0.129 |
| Age
>60+high CST7 | 32 | 18.93 |
| 0.62
(0.29-1.32) | 0.52
(0.22-1.24) | 0.139 | >44.4 |
| 0.26
(0.08-0.80) | 0.14
(0.04-0.47) | 0.002 |
| Sex+CST7 group |
|
Female+low CST7 | 24 | 12.6 | 0.020 |
|
| 0.003 | 9.7 | 0.013 |
|
| 0.012 |
|
Female+high CST7 | 29 | 19.87 |
| 0.44
(0.22-0.88) | 0.25
(0.11-0.55) | 0.001 | 29.07 |
| 0.20
(0.07-0.56) | 0.17
(0.05-0.53) | 0.002 |
|
Male+low CST7 | 32 | 15.77 |
| 0.72
(0.37-1.39) | 0.54
(0.27-1.08) | 0.080 | 19.77 |
| 0.42
(0.16-1.11) | 0.37
(1.37-1.00) | 0.050 |
|
Male+high CST7 | 27 | 21.73 |
| 0.37
(0.17-0.76) | 0.31
(0.14-0.71) | 0.006 | 18.07 |
| 0.37
(0.15-0.92) | 0.24
(0.08-0.67) | 0.011 |
| Dimension+CST7
group |
| D≤3+low
CST7 | 20 | 15.87 | 0.046 |
|
| 0.012 | 19.37 | 0.105 |
|
| 0.108 |
|
D≤3+high CST7 | 21 | 19.87 |
| 0.44
(0.19-1.04) | 0.53
(0.21-1.37) | 0.191 | >44.4 |
| 0.24
(0.06-0.94) | 0.23
(0.06-0.93) | 0.040 |
|
D>3+low CST7 | 35 | 12.5 |
| 1.10
(0.56-2.18) | 1.22
(0.58-2.58) | 0.607 | 14.76 |
| 0.98
(0.36-2.68) | 0.87
(0.31-2.46) | 0.795 |
|
D>3+high CST7 | 34 | 17.23 |
| 0.60
(0.30-1.21) | 0.44
(0.20-0.97) | 0.041 | 23.87 |
| 0.58
(0.21-1.60) | 0.44
(0.15-1.26) | 0.126 |
| Residual tumor
status+CST7 group |
| R0+low
CST7 | 27 | 15.77 | 0.002 |
|
| 0.006 | 14.63 | 0.002 |
|
| 0.006 |
| R0+high
CST7 | 39 | 23.17 |
| 0.37
(0.19-0.72) | 0.36
(0.18-0.72) | 0.004 | 40.33 |
| 0.19
(0.06-0.58) | 0.17
(0.05-0.55) | 0.003 |
|
R1/Rx+low CST7 | 28 | 12.2 |
| 1.16
(0.61-2.23) | 1.13
(0.56-2.29) | 0.730 | 19.77 |
| 0.77
(0.29-2.07) | 0.77
(0.28-2.10) | 0.611 |
|
R1/Rx+high CST7 | 16 | 17.27 |
| 0.95
(0.44-2.08) | 0.57
(0.22-1.47) | 0.244 | 11.33 |
| 1.32
(0.48-3.67) | 1.36
(0.47-3.98) | 0.570 |
Prognosis model for CST7 expression in
early-stage PDAC
To predict the clinical outcome risk in different
CST7 mRNA levels and clinicopathological features, we constructed a
prognosis nomogram for OS and DFS of PDAC patients (C-index=0.799
and 0.772, respectively). CST7 expression, histological grade,
lymph node metastasis, resection status, therapy methods were
associated with the OS of PDAC patients (Fig. 5A). Moreover, histological grade
played an important role in the recurrence of PDAC and had a
synergism with CST7 expression, lymph node metastasis, resection
status, and tumor stages (Fig.
5B).
GSEA for CST7 in PDAC
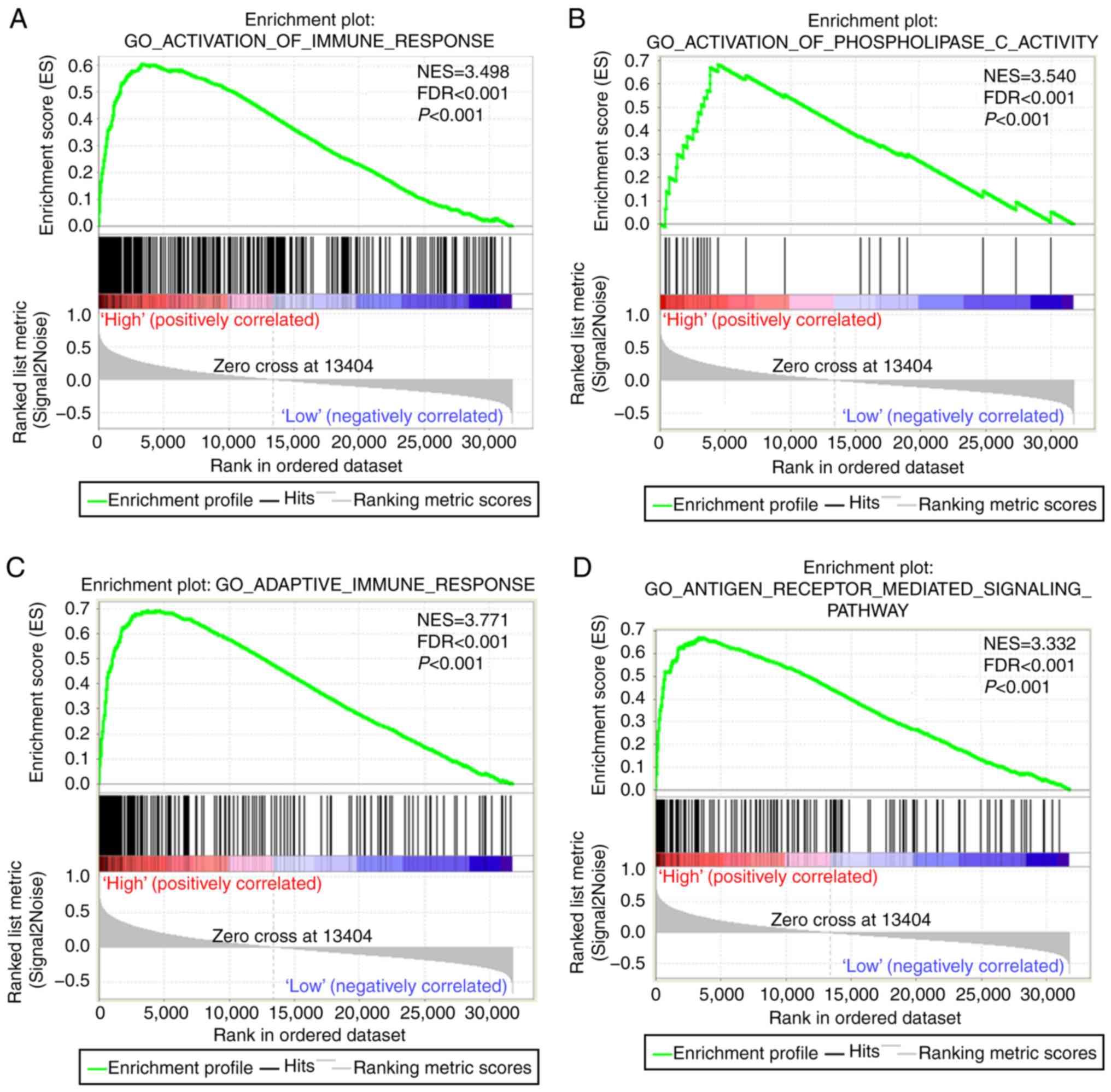
In the GSEA, we identified that CST7 may be involved
in immunomodulation, immune response, and cellular immune
regulation (Fig. 6A-I). In terms of
the biological process of PDAC patients, CST7 was associated with
cell adhesion molecule CAMS and played a role in cell adhesion
(Fig. 6J-L).
Discussion
In the present study, we analyzed the biological
functions of family 2 cystatins using bioinformatics analysis. Gene
expression levels of family 2 cystatins differed between
paraneoplastic and pancreas tumor tissues. Furthermore, we
identified CST7 as a key cystatin subunit in patients with
early-stage PDAC in TCGA. DFS and OS of patients with surgically
resected PDACs were significantly prolonged in the high-CST7
expression subgroup compared with the low-CST7 expression subgroup.
Moreover, we investigated the prognostic value of CST7 using
combined analysis and constructed a prognostic model for PDAC
patients after pancreatectomy. The result of the GSEA demonstrated
that CST7 may be involved in immune regulation.
As the most commonly used tumor biomarker for
detecting PDAC, the main limitations of CA 19-9 include its
frequent elevations associated with non-malignant diseases such as
pancreatitis and obstructive jaundice, and the inability to detect
many early-stage tumors (23). CA
19-9 is also not suitable for estimating 5-10% of patients who are
carriers of the Lewis-negative genotype and advance malignancies
that do not express antigen (24).
Thus, it is extremely important and necessary to screen additional
key genes associated with PDAC that may act as diagnostic,
prognostic or therapeutic biomarkers for improving the diagnostic
ability of CA19-9.
After screening, CST7 was considered to be a key
cystatin subunit in early-stage PDAC. Using GSEA, we identified
that CST7 was related to the regulation of immune cell activity.
Previous studies demonstrated that CST7 expression may play roles
in regulating the activation of natural killer cells,
differentiation of monocytes to macrophages and influenza vaccine
responses (25–27). CST7 encodes a glycosylated cysteine
protease inhibitor with a putative role in immune regulation
through inhibition of a unique target in the hematopoietic system.
It has been reported that CST7 is mainly expressed by immune cells
(28,29) where it is often elevated when these
cells differentiate or activate from a stationary precursor.
Cystatins are considered to be typical emergency inhibitors,
trapping proteases that escape from endosomes/lysosomes or cells,
and converting them into proteolytically inactive complexes
(30). However, the amino acids
present in the N-terminal region of the protease-binding loop and
CST7 differ from the amino acids of other family members,
indicating that CST7 may bind to different protease targets.
Compared with other cystatins, core sugars of the N-linked
glycosylation sites of CST7 are highly structured, and conformation
and interactions with the combined proteins indicate that unique
features of CST7 may modulate its inhibitory properties though
structural reconstruction (31).
In the present study, GO term annotations revealed
that the cystatin family plays roles in the extracellular region
and demonstrates inhibitory activity against biologically active
proteins. A recent study demonstrated that CST7 regulated
intracellular, but not extracellular, protease activity by
targeting cathepsin C (32). CST7
plays an important role in activation of various serine protease
zymogens in secretory granules of tumor-related immune cells, such
as cytotoxic T lymphocytes, mast cells, NK cells, and neutrophils
(33–35). Although destruction of the CST7
dimer is enough to produce an endopeptidase inhibitor such as
cathepsin L, a proteolytic cleavage event is applied for converting
inactive CST7 into an active cathepsin C inhibitor. Cathepsin C is
essential for the activation of cytokines, such as TNF-α and IL-1β.
CST7 was revealed specifically to bind and inhibit cathepsin C in
immune cells via regulation of split anergy (32,36).
Moreover, expression of CST7 has been observed in
various human cancer cell lines established from malignant tumors.
As the only family 2 cystatin able to enter endosomal/lysosomal
vesicles and to regulate directly the activity of intracellular
cysteine cathepsins, CST7 is highly upregulated in promonocytic
U937 and promyeloblast HL-60 cells (26). Pierre and Mellman (37) reported that the apparent
distribution of smaller vesicles/particles in U937 cells indicates
that CST7 is partially localized to organelles that resemble fusion
lysosomes/endosomes, wherein antigen and constant chain processing
occurs in antigen-presenting cells. Immunocytochemical staining of
CST7 in human promonocyte U937 cells displays a vesicular pattern
(38).
Furthermore, recent studies have revealed that CST7
expression level was associated with tumor progression and served
as an independent factor for liver metastasis in colorectal cancer
patients (39,40). The prognosis of the patients with a
higher expression of CST7 was significantly worse than those with a
lower expression. However, our results indicated that high CST7
expression level may be a protective factor for patients with
early-stage PDAC. Using the Human Protein Atlas project (HPA;
http://www.proteinatlas.org/), an online
visualization website for detecting the relationship between the
protein level and clinical outcomes based on TCGA data, we
determined that the protein level of CST7 was not associated with
the prognosis of PDAC patients but would be a potential prognostic
marker in renal cancer, endometrial, head and neck, breast and
cervical cancer. We speculate that this gene has tissue differences
in tumor prognosis and prognosis evaluation. CST7 may serve as a
tumor suppressor gene in PDAC patients, and high CST7 expression
promotes autoimmune cell monitoring. For our prognostic model
constructed using a nomogram, CST7 harbored a high predictive
effect on the clinical outcomes of PDAC and could assist clinicians
to judge disease outcomes and treatment. Considering the lack of
independent data and some missing clinical data which are
limitations of the present study, an additional large verification
cohort to validate the results is necessary.
In conclusion, the results of the present study
indicated that CST7 could be a useful biomarker for the prognostic
prediction of early-stage PDAC.
Supplementary Material
Supporting Data
Acknowledgements
We are grateful to TCGA group for providing relevant
data.
Funding
The present study was supported in part by the
National Natural Science Foundation of China (nos. 81560535,
81802874, 81072321, 30760243, 30460143 and 30560133), Natural
Science Foundation of Guangxi Province of China (grant no.
2018GXNSFBA138013 and 2018GXNSFAA050119), 2009 Program for New
Century Excellent Talents in University (NCET), Guangxi Natural
Sciences Foundation (no. GuiKeGong 1104003A-7), and Guangxi Health
Ministry Medicine Grant (Key-Scientific Research-Grant Z201018).
This study was also partly supported by the Scientific Research
Fund of the Health and Family Planning Commission of Guangxi Zhuang
Autonomous Region (Z2016318), Key Laboratory of
High-Incidence-Tumor Prevention and Treatment (Guangxi Medical
University), Ministry of Education (GKE2018-01), the Guangxi Key
R&D Program (GKEAB18221019), The Basic Ability Improvement
Project for Middle-aged and Young Teachers in Colleges and
Universities in Guangxi (2018KY0110), Innovation Project of Guangxi
Graduate Education (JGY2018037), and 2018 Innovation Project of
Guangxi Graduate Education (YCBZ2018036). Furthermore, the present
study was also partly supported by the Research Institute of
Innovative Think-tank in Guangxi Medical University (The
gene-environment interaction in hepatocarcinogenesis in Guangxi
HCCs and its translational applications in the HCC prevention). We
would also acknowledge the support of the National Key Clinical
Specialty Programs (General Surgery and Oncology) and the Key
Laboratory of Early Prevention and Treatment for Regional
High-Incidence-Tumor (Guangxi Medical University), Ministry of
Education, China.
Availability of data and materials
All data generated or analyzed during the present
study are included in this published article.
Authors' contributions
CY and TP designed the study; CY, CH, XY and TY
performed research; XY, XW, XL, ZL and TY provided sample
collection and clinical support; GZ, TY and WQ contributed to data
interpretation. CY and ZL wrote the manuscript, and XL and TP
critically revised the manuscript and participated in the analysis
and interpretation of the data. All authors reviewed, edited and
approved the final version of the manuscript and agree to be
accountable for all aspects of the research in ensuring that the
accuracy or integrity of any part of the work are appropriately
investigated and resolved.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of The First Affiliated Hospital of Guangxi Medical
University (Guangxi, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
PDAC
|
pancreatic ductal adenocarcinoma
|
|
CST7
|
cystatin F
|
|
STRING
|
Search Tool for the Retrieval of
Interacting Genes/Proteins
|
|
GO
|
Gene Ontology
|
|
DAVID
|
Database for Annotation, Visualization
and Integrated Discovery
|
|
MERAV
|
Metabolic gEne RApid Visualizer
|
|
GTEx
|
Genotype-Tissue Expression
|
|
TCGA
|
The Cancer Genome Atlas
|
|
AJCC
|
American Joint Committee on Cancer
|
|
OS
|
overall survival
|
|
DFS
|
disease-free survival
|
|
HR
|
hazard ratio
|
|
CI
|
confidence interval
|
|
C-index
|
concordance index
|
|
GSEA
|
gene set enrichment analysis
|
|
FDR
|
false discovery rate
|
References
|
1
|
Li D, Xie K, Wolff R and Abbruzzese JL:
Pancreatic cancer. Lancet. 363:1049–1057. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2012. CA Cancer J Clin. 62:10–29. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jemal A, Siegel R, Ward E, Murray T, Xu J
and Thun MJ: Cancer statistics, 2007. CA Cancer J Clin. 57:43–66.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Conrad C and Lillemoe KD: Surgical
palliation of pancreatic cancer. Cancer J. 18:577–583. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Vincent A, Herman J, Schulick R, Hruban RH
and Goggins M: Pancreatic cancer. Lancet. 378:607–620. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Choi EH, Kim JT, Kim JH, Kim SY, Song EY,
Kim JW, Kim SY, Yeom YI, Kim IH and Lee HG: Upregulation of the
cysteine protease inhibitor, cystatin SN, contributes to cell
proliferation and cathepsin inhibition in gastric cancer. Clin Chim
Acta. 406:45–51. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Travis J and Potempa J: Bacterial
proteinases as targets for the development of second-generation
antibiotics. Biochim Biophys Acta. 1477:35–50. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Koblinski JE, Ahram M and Sloane BF:
Unraveling the role of proteases in cancer. Clin Chim Acta.
291:113–135. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kos J and Lah TT: Cysteine proteinases and
their endogenous inhibitors: Target proteins for prognosis,
diagnosis and therapy in cancer (review). Oncol Rep. 5:1349–1410.
1998.PubMed/NCBI
|
|
11
|
Verbovšek U, Van Noorden CJ and Lah TT:
Complexity of cancer protease biology: Cathepsin K expression and
function in cancer progression. Semin Cancer Biol. 35:71–84. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cox J: Cystatins as regulators of cancer.
Med Res Arch. 5:2017. View Article : Google Scholar
|
|
13
|
Barrett AJ: The cystatins: A new class of
peptidase inhibitors. Trends Biochem Sci. 12:193–196. 1987.
View Article : Google Scholar
|
|
14
|
Henskens YM, Veerman EC and Nieuw
Amerongen AV: Cystatins in health and disease. Biol Chem Hoppe
Seyler. 377:71–86. 1996.PubMed/NCBI
|
|
15
|
Abrahamson M, Alvarez-Fernandez M and
Nathanson CM: Cystatins. Biochem Soc Symp. 179–199. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chimonidou M, Tzitzira A, Strati A,
Sotiropoulou G, Sfikas C, Malamos N, Georgoulias V and Lianidou E:
CST6 promoter methylation in circulating cell-free DNA of breast
cancer patients. Clin Biochem. 46:235–240. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cao X, Li Y, Luo RZ, Zhang L, Zhang SL,
Zeng J, Han YJ and Wen ZS: Expression of Cystatin SN significantly
correlates with recurrence, metastasis, and survival duration in
surgically resected non-small cell lung cancer patients. Sci Rep.
5:82302015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Alvarez-Díaz S, Valle N, García JM, Peña
C, Freije JM, Quesada V, Astudillo A, Bonilla F, López-Otín C and
Muñoz A: Cystatin D is a candidate tumor suppressor gene induced by
vitamin D in human colon cancer cells. J Clin Invest.
119:2343–2358. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hünten S and Hermeking H: p53 directly
activates cystatin D/CST5 to mediate mesenchymal-epithelial
transition: A possible link to tumor suppression by vitamin D3.
Oncotarget. 6:15842–15856. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liao X, Huang K, Huang R, Liu X, Han C, Yu
L, Yu T, Yang C, Wang X and Peng T: Genome-scale analysis to
identify prognostic markers in patients with early-stage pancreatic
ductal adenocarcinoma after pancreaticoduodenectomy. Onco Targets
Ther. 10:4493–4506. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Harrell FE Jr: rms: Regression modeling
strategies. R package version 4.0-0. City. 2013.
|
|
22
|
Harrell FE Jr, Califf RM, Pryor DB, Lee KL
and Rosati RA: Evaluating the yield of medical tests. JAMA.
247:2543–2546. 1982. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Koopmann J, Rosenzweig CN, Zhang Z, Canto
MI, Brown DA, Hunter M, Yeo C, Chan DW, Breit SN and Goggins M:
Serum markers in patients with resectable pancreatic
adenocarcinoma: Macrophage inhibitory cytokine 1 versus CA19-9.
Clin Cancer Res. 12:442–446. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Locker GY, Hamilton S, Harris J, Jessup
JM, Kemeny N, Macdonald JS, Somerfield MR, Hayes DF and Bast RC Jr;
ASCO: ASCO 2006 update of recommendations for the use of tumor
markers in gastrointestinal cancer. J Clin Oncol. 24:5313–5327.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Maher K, Konjar S, Watts C, Turk B and
Kopitar-Jerala N: Cystatin F regulates proteinase activity in
IL-2-activated natural killer cells. Protein Pept Lett. 21:957–965.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Dautović E, Perišić Nanut M, Softić A and
Kos J: The transcription factor C/EBPα controls the role of
cystatin F during the differentiation of monocytes to macrophages.
Eur J Cell Biol. 97:463–473. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Voigt EA, Grill DE, Zimmermann MT, Simon
WL, Ovsyannikova IG, Kennedy RB and Poland GA: Transcriptomic
signatures of cellular and humoral immune responses in older adults
after seasonal influenza vaccination identified by data-driven
clustering. Sci Rep. 8:7392018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Halfon S, Ford J, Foster J, Dowling L,
Lucian L, Sterling M, Xu Y, Weiss M, Ikeda M, Liggett D, et al:
Leukocystatin, a new class II cystatin expressed selectively by
hematopoietic cells. J Biol Chem. 273:16400–16408. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ni J, Fernandez MA, Danielsson L,
Chillakuru RA, Zhang J, Grubb A, Su J, Gentz R and Abrahamson M:
Cystatin F is a glycosylated human low molecular weight cysteine
proteinase inhibitor. J Biol Chem. 273:24797–24804. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Turk B, Turk D and Salvesen GS: Regulating
cysteine protease activity: Essential role of protease inhibitors
as guardians and regulators. Curr Pharm Des. 8:1623–1637. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Schüttelkopf AW, Hamilton G, Watts C and
van Aalten DM: Structural basis of reduction-dependent activation
of human cystatin F. J Biol Chem. 281:16570–16575. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hamilton G, Colbert JD, Schuettelkopf AW
and Watts C: Cystatin F is a cathepsin C-directed protease
inhibitor regulated by proteolysis. EMBO J. 27:499–508. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Pham CT and Ley TJ: Dipeptidyl peptidase I
is required for the processing and activation of granzymes A and B
in vivo. Proc Natl Acad Sci USA. 96:8627–8632. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wolters PJ, Pham CT, Muilenburg DJ, Ley TJ
and Caughey GH: Dipeptidyl peptidase I is essential for activation
of mast cell chymases, but not tryptases, in mice. J Biol Chem.
276:18551–18556. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Adkison AM, Raptis SZ, Kelley DG and Pham
CT: Dipeptidyl peptidase I activates neutrophil-derived serine
proteases and regulates the development of acute experimental
arthritis. J Clin Invest. 109:363–371. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Magister Š, Tseng HC, Bui VT, Kos J and
Jewett A: Regulation of split anergy in natural killer cells by
inhibition of cathepsins C and H and cystatin F. Oncotarget.
6:22310–22327. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Pierre P and Mellman I: Developmental
regulation of invariant chain proteolysis controls MHC class II
trafficking in mouse dendritic cells. Cell. 93:1135–1145. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Nathanson CM, Wassélius J, Wallin H and
Abrahamson M: Regulated expression and intracellular localization
of cystatin F in human U937 cells. FEBS J Biochem. 269:5502–5511.
2002. View Article : Google Scholar
|
|
39
|
Georgieva M, Krasteva M, Angelova E,
Ralchev K, Dimitrov V, Bozhimirov S, Georgieva E and Berger MR:
Analysis of the K-ras/B-raf/Erk signal cascade, p53 and CMAP as
markers for tumor progression in colorectal cancer patients. Oncol
Rep. 20:3–11. 2008.PubMed/NCBI
|
|
40
|
Utsunomiya T, Hara Y, Kataoka A, Morita M,
Arakawa H, Mori M and Nishimura S: Cystatin-like
metastasis-associated protein mRNA expression in human colorectal
cancer is associated with both liver metastasis and patient
survival. Clin Cancer Res. 8:2591–2594. 2002.PubMed/NCBI
|