Introduction
Colon cancer is the most commonly diagnosed cancer
and one of the deadliest diseases worldwide (1). Despite extensive research into
strategies to treat colon cancer, conventional approaches have
shown little promise in improving prognosis (2). The overall survival rate of patients who
receive chemotherapy is only 10% (3).
Thus, it is crucial to study the mechanism of colon carcinogenesis
and to establish a theoretical basis for its treatment.
Transfer RNA (tRNA)-derived fragments (tRFs), a
novel type of small non-coding RNA that originates from tRNAs
(4–6),
are associated with several cellular activities (7–9).
Previously, small non-coding RNAs, including microRNAs (miRNAs),
tRFs, and piwi-interacting RNAs (piRNAs), have been implicated in
tumorigenesis (10,11). Among the tRFs, tRF-544 and tRF-315 are
potential prognostic biomarkers for prostate cancer (12). tRF-1001 is expressed highly in many
proliferating tumor cells and positively regulates cell
proliferation (6). High expression of
the tRFs ts-46 and ts-47 in lung cancer cell lines was identified
to suppress cell proliferation (13).
tRFs have been isolated as part of Argonaute (AGO) complexes, which
implies that tRFs play a functional role, similar to that of
miRNAs, and possess the ability to suppress mRNA translation in
human B cells (14). An association
with AGO implies that tRFs may be regulators of mRNA expression.
miR-2476, which is one base pair different from the 5′-tRF derived
from tRNA-Glu-CTC, is now not available due to its location near a
known tRNA (15). miR-3676, which is
in fact a tRF, targets the 3′ untranslated region (3′UTR) of the
oncogene TCL1 and downregulates the expression of TCL1 (13,16).
Collectively, these data suggest that tRFs play key roles in
various cancers and may be involved in tumorigenesis via
post-translational alteration of gene expression. However, the role
of tRFs in colon cancer remains unclear. Therefore, a comprehensive
analysis was conducted to explore the role of tRFs in colon
cancer.
In the present study, tRFs that were differentially
expressed (DE) between colon cancer and normal peritumor tissues
were identified by small RNA sequencing. The target genes of DE
tRFs and known colon cancer-associated miRNAs were predicted to
study the functional roles of tRFs in colon cancer by integrative
analysis of tRFs, mRNAs and miRNAs.
Materials and methods
Sample collection
Tumor and matched normal peritumor tissues were
collected from surgical specimens obtained from patients who
underwent surveillance colonoscopy or routine screening in
Department of Radiation Oncology, The Third Affiliated Hospital of
Kunming Medical University (Kunming, China) between July 2016 and
January 2018. A total of 30 patients who had not received any drug
treatment before surgery were included. Three pairs of samples were
used for small RNA sequencing and all 30 pairs of samples were used
for reverse transcription-quantitative polymerase chain reaction
(RT-qPCR) validation. All tissues were preserved in liquid nitrogen
immediately after resection. The present study was approved by the
Human Ethics Committee of Kunming Medical University. Written
informed consent was obtained from all patients. Subjects with
familial cancers, including familial adenomatous polyposis and
Lynch syndrome, history of prior colonic resections or inflammatory
bowel syndrome were excluded. The demographics and clinical
characteristics of patient samples with low or moderate/high
differentiation level according to histological characteristics
(17) are presented in Table I. Characteristics recorded include
sex, age, tumor size, location of hemicolon, TNM stage (18), lymph node and distant metastases.
 | Table I.Demographics and clinical
characteristics of patients with low (n=10) or moderate/high (n=20)
differentiation. |
Table I.
Demographics and clinical
characteristics of patients with low (n=10) or moderate/high (n=20)
differentiation.
| Characteristic | n | Low
differentiation, n (%) | Moderate/high
differentiation, n (%) | P-value |
|---|
| Sex |
|
|
| 0.698 |
|
Male | 15 | 5 (50) | 10 (50) |
|
|
Female | 15 | 5 (50) | 10 (50) |
|
| Age, years |
|
|
| 0.896 |
|
>60 | 17 | 6 (60) | 11 (55) |
|
|
≤60 | 13 | 4 (40) | 9 (45) |
|
| Tumor size, cm |
|
|
| 0.897 |
|
>12 | 16 | 6 (60) | 10 (50) |
|
|
≤12 | 14 | 4 (40) | 10 (50) |
|
| Location of
hemicolon |
|
|
| 0.896 |
|
Left | 17 | 6 (60) | 11 (55) |
|
|
Right | 13 | 4 (40) | 9 (45) |
|
| TNM stage |
|
|
| 0.897 |
|
I+II | 14 | 4 (40) | 10 (50) |
|
|
III+IV | 16 | 6 (60) | 10 (50) |
|
| Lymph node
metastasis |
|
|
| 0.897 |
|
Yes | 14 | 5 (50) | 9 (45) |
|
| No | 16 | 5 (50) | 11 (55) |
|
| Distant
metastasis |
|
|
| 0.519 |
|
Yes | 3 | 2 (20) | 1 (5) |
|
| No | 27 | 8 (80) | 19 (95) |
|
RNA isolation, construction of small
RNA library and sequencing
Total RNA was isolated from colon cancer and matched
peritumor tissues with TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA). The quantity and purity of the
total RNA were analyzed using NanoDrop (Thermo Fisher Scientific,
Inc.) and 1% agarose gel electrophoresis. Purified and quantified
RNA was used to construct small RNA libraries using the NEBNext
Multiplex Small RNA Library Prep kit for Illumina (New England
Biolabs, Inc., Ipswich, MA, USA) according to the manufacturer's
protocol. Finally, small RNA sequencing was performed on HiSeq 2500
(Illumina, Inc., San Diego, CA, USA).
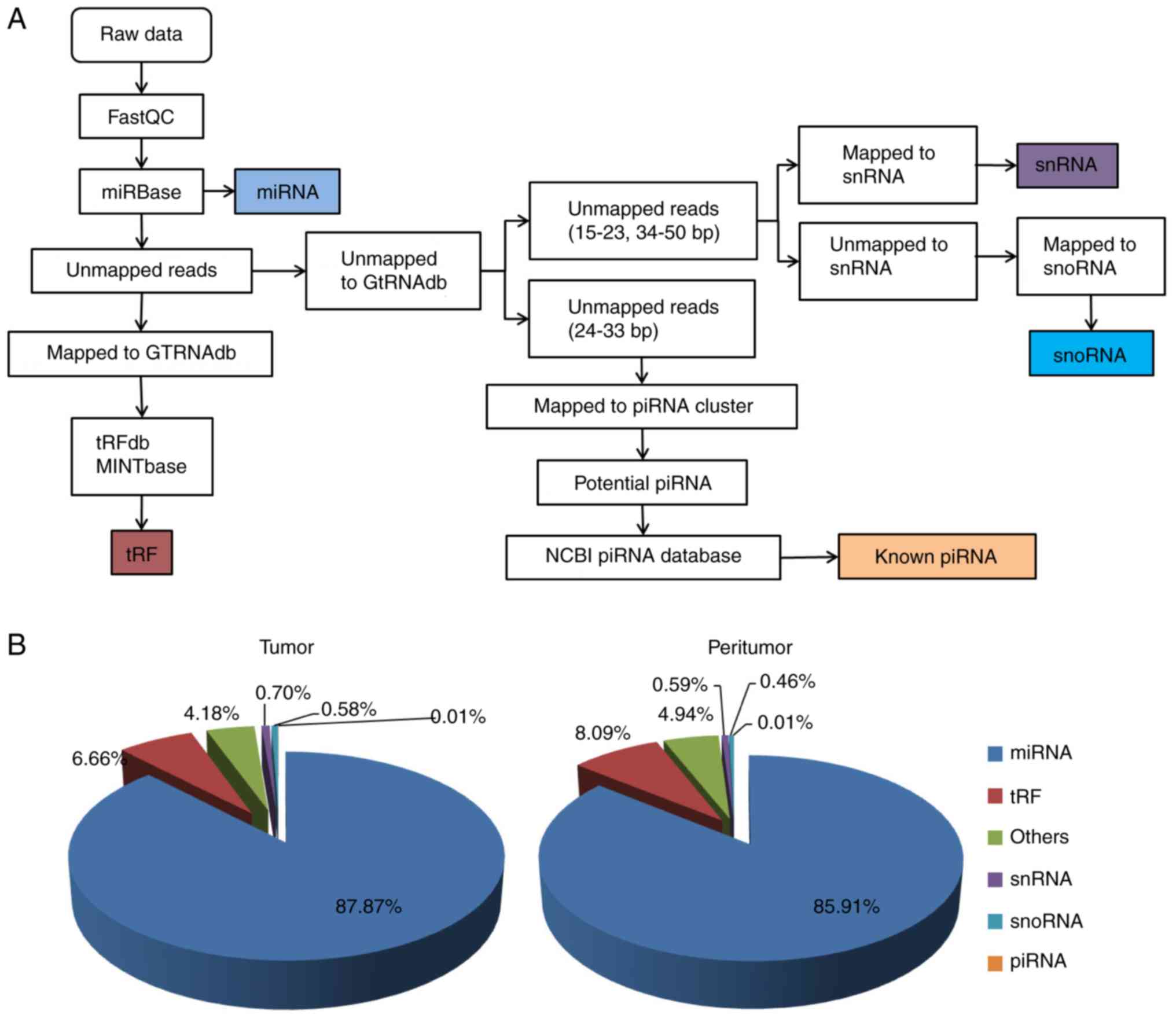
Data filtering and mapping
The raw sequencing data were refined with FastQC,
which included filtering out low-quality and short reads (<15
nucleotides). The clean small RNA reads were mapped using the
miRBase database (http://www.mirbase.org/) to identify known miRNAs, and
the unmapped reads were further aligned with sequences in the
Genomic tRNA Database (GtRNAdb, http://gtrnadb.ucsc.edu/). The reads mapped to GtRNAdb
sequences were then mapped to those in tRFdb (http://genome.bioch.virginia.edu/trfdb/)
and MINTbase (https://cm.jefferson.edu/MINTbase/) to identify tRFs.
Unmapped reads from the GtRNAdb analysis were aligned to the piRNA
sequences from the National Center for Biotechnology Information
(https://www.ncbi.nlm.nih.gov/) and small
nucleolar RNA (snoRNA) and small nuclear RNA (snRNA) sequences from
Rfam (http://rfam.xfam.org/) (19,20).
Screening of DE tRFs, miRNAs, and
mRNAs
DE tRFs and miRNAs between the colon cancer and
normal tissues were screened using the respective EBseq packages
(http://www.bioconductor.org/packages/devel/bioc/html/EBSeq.html).
The cut-off thresholds were set to |Log2(fold change)|
>1, and false-discovery rate (FDR) was set to <0.05. Gene
Expression Profiling Interactive Analysis (GEPIA; http://gepia.cancer-pku.cn/index.html)
(21), an interactive website based
on the expression profiles of tumor and normal samples from The
Cancer Genome Atlas and the Genotype-Tissue Expression projects,
provides tools for gene functional analysis, including tumor/normal
differential expression analysis according to cancer types or
pathological stages, correlation analysis, similar gene detection
and patient survival analysis. DE mRNAs between colon
adenocarcinoma and paired normal samples were identified using
limma on GEPIA, with the following parameters:
|Log2(fold change)| >1, FDR <0.05.
Prediction of target genes of DE tRFs
and key miRNAs
DE miRNAs implicated in colon cancer were selected
as key miRNAs. MiRanda (http://www.microrna.org/microrna/home.do) and
RNAhybrid (https://bibiserv.cebitec.uni-bielefeld.de/rnahybrid/)
were used to predict the binding of DE tRFs or key miRNAs to the
putative target genes. Genes with overlapping MiRanda (score ≥150
and energy ≤20) and RNAhybrid (energy ≤25) results were considered
as potential targets of tRFs or miRNAs. Next, the potential target
genes common to tRFs and miRNAs were compared with DE mRNAs between
colon adenocarcinoma and paired normal samples. Only the target
genes that showed significant DE between the colon cancer and
normal tissues and correlated negatively with the DE tRFs and
candidate miRNAs were screened for further analysis. An integrated
tRF/mRNA/miRNA network was then constructed using Cytoscape
software (version 3.5.1) (22).
Gene Ontology (GO) and pathway
analyses of predicted target genes
GO (http://www.geneontology.org/) analysis (23) was performed to analyze the functions
of the screened target genes (24,25).
Pathway enrichment analysis of the screened target genes was
performed using the Kyoto Encyclopedia of Genes and Genomes (KEGG;
http://www.kegg.jp/) (23). The cut-off threshold was set to
P<0.05.
RT-qPCR
Total RNAs from colon cancer tissues and normal
peritumor tissue, colon cell lines and the human colon epithelial
cell line were extracted with TRIzol reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) and reverse transcribed into cDNA using a
RevertAidAM First Strand cDNA Synthesis kit (#K1622; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. qPCR
was performed on the ABI Q6 detection system (Applied Biosystems;
Thermo Fisher Scientific, Inc.) using a Real Time SYBR Master Mix
kit (cat. no. 33041; Qiagen, Inc., Valencia, CA, USA). The
thermocycling conditions were: 95°C for 10 min for 1 cycle; 95°C
for 15 sec and 60°C for 60 sec for 45 cycles. Relative expression
was calculated using the comparative threshold (Ct) cycle method
(2−ΔΔCq) (26). U6 was
used as an internal reference for the relative DE miRNA and tRF
expression. Primers (Yingbiotech Co., Shanghai, China) and
sequences are presented in Table
II.
 | Table II.Primer sequences for quantitative
polymerase chain reaction. |
Table II.
Primer sequences for quantitative
polymerase chain reaction.
| Primer | Sequence
(5′-3′) |
|---|
| U6-F |
CGATACAGAGAAGATTAGCATGGC |
| U6-R |
AACGCTTCACGAATTTGCGT |
|
hsa-miR-378a-3pF |
CGCAGACTGGACTTGGAGTCA |
|
hsa-miR-424-3pF |
GCTGCAAAACGTGAGGCG |
|
hsa-miR-483-3pF |
GGGCAAATCACTCCTCTCCTC |
|
hsa-miR-615-3pF |
GCAGTCCGAGCCTGGGTCT |
|
tRF-24-NMEH623K25F |
CAGCTGTCACGCGGGAGA |
|
tRF-30-XSXMSL73VL4YF |
TGCCGTGATCGTATAGTGGTTAG |
|
tRF-29-QU7BPN6ISBJOF |
GCTGAGTGAAGCATTGGACTGTAA |
|
tRF-27-Q99P9P9NH5NF |
GCAGGCTTCTGTAGTGTAGTGGTTA |
| miR/tRF-R |
AGTGCGTGTCGTGGAGTCG |
Cell culture
Human colon cancer cell lines SW480, SW620, HCT116,
HT29 and human colon epithelial cell line NCM460 were purchased
from the Type Culture Collection of the Chinese Academy of Sciences
(Shanghai, China). HCT116 and SW480 cells were maintained by serial
passage in RPMI-1640 medium (Corning, Inc., Corning, NY, USA)
supplemented with 10% fetal bovine serum (FBS; Thermo Fisher
Scientific, Inc.), 10 U/ml penicillin and 10 µg/ml streptomycin
(P/S; Thermo Fisher Scientific, Inc.), and cultured in a 5%
CO2, humidified atmosphere at 37°C. HT29 cells were
maintained in McCoy's 5A medium (Thermo Fisher Scientific, Inc.)
with 10% FBS and 1% P/S and cultured in a 5% CO2,
humidified atmosphere at 37°C. SW620 cells were cultured in L15
medium (Beijing Solarbio Science & Technology Co., Ltd.,
Beijing, China) with 10% FBS and 1% P/S and cultured in 100% air at
37°C. Dulbecco's modified Eagle's medium (Thermo Fisher Scientific,
Inc.) and F12 medium (Thermo Fisher Scientific, Inc.) were used to
culture the human colon epithelial cell line NCM460 with 10% FBS
and 1% P/S, in a 5% CO2, humidified atmosphere at
37°C.
Statistical analysis
Statistical analysis was performed on SPSS version
12.0 statistical software (SPSS, Inc., Chicago, IL, USA) and the
data were presented using the software GraphPad Prism 5.0 (GraphPad
Software, Inc., La Jolla, CA, USA). Associations between
demographics and differentiation were determined statistically
using Pearson's Chi-square test. Measurement data are presented as
the mean ± standard deviation and were analyzed by paired Student's
t-test for comparisons of two groups or one-way analysis of
variance followed by Tukey's post hoc test for comparisons among
multiple groups. P<0.05 was considered to indicate a
statistically significant difference.
Results
Associations between demographic and
clinical characteristics of patients and differentiation of
cancer
The demographic and clinical characteristics of the
30 patients are presented in Table I.
Characteristics included sex, age and tumor size, location of
hemicolon, TNM stage, lymph node metastasis and distant metastasis.
There were no significant differences between the low and
moderate/high differentiation groups.
Data mapping and results
After filtering with FastQC, the clean reads were
mapped to different small RNA databases or sequences (Fig. 1A). Of all mapped reads from colon
cancer tissues, 87.87% were miRNAs, 6.66% tRFs, 0.70% snRNAs, 0.58%
snoRNAs and 0.01% known piRNAs (Fig.
1B). The various small RNAs in peritumor tissues were 85.91%
miRNAs, 8.09% tRFs, 0.59% snRNAs, 0.46% snoRNAs and 0.01% piRNAs
(Fig. 1B).
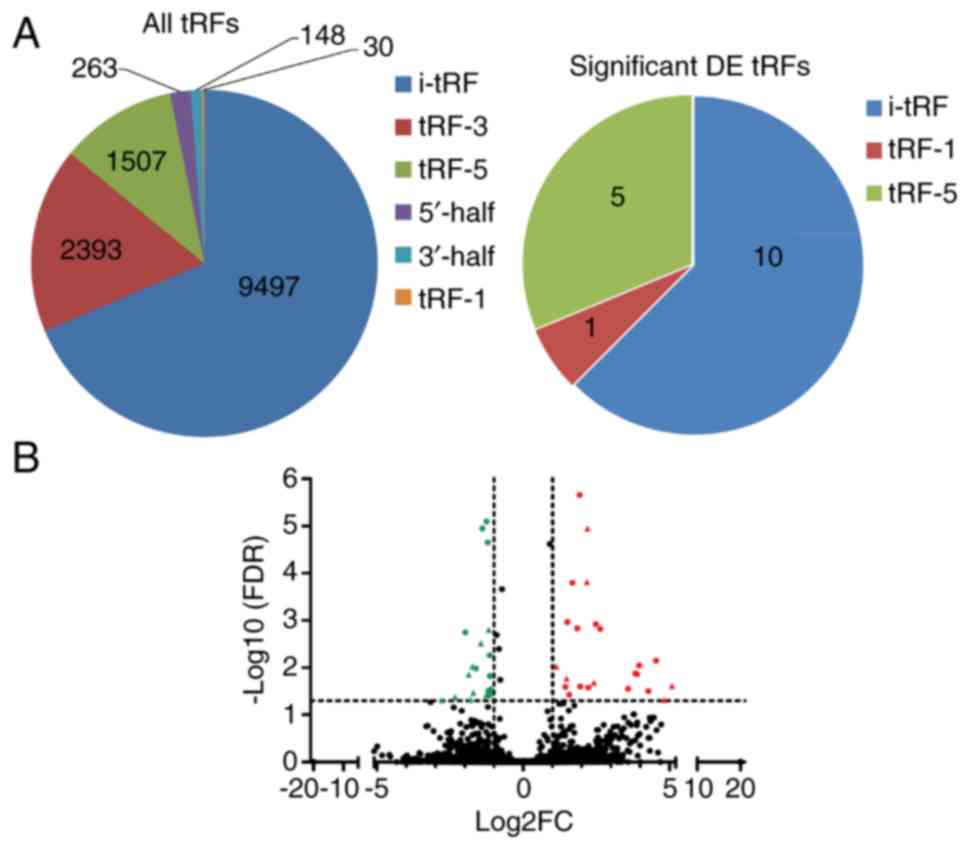
DE tRFs and miRNAs
As shown in Table
III, a total of 13,838 tRFs, 2,588 miRNAs and 167 known piRNAs
were identified in the six samples. The identified tRFs were of six
types: 3′-half, 5′-half, i-tRF, tRF-1, tRF-3 and tRF-5 (Fig. 2A). In total, 16 tRFs (Table IV) and 26 miRNAs exhibited
significant differential expression between the colon cancer and
normal tissues with |Log2(fold change)| >1 and
FDR<0.05 (Fig. 2B). No
significantly DE known piRNAs were detected.
 | Table III.Numbers of detected small non-coding
RNAs. |
Table III.
Numbers of detected small non-coding
RNAs.
| Type | Total | Differentially
expressed |
|---|
| miRNA | 2,588 | 26 |
| tRF | 13,838 | 16 |
| piRNA | 167 | 0 |
 | Table IV.Significantly differentially
expressed tRFs between colon cancer tissues and peritumor
tissues. |
Table IV.
Significantly differentially
expressed tRFs between colon cancer tissues and peritumor
tissues.
| Database ID |
Log2FC | FDR | Fragment
sequence | Type | Anticodon |
|---|
|
tRF-28-6Y1UFQ9D9ODD | 4.82 | 0.048029 |
GGCTGAGTGAAGCATTGGACTGTAAATC | i-tRF | TyrGTA |
|
tRF-25-6YYDLBRY73 | −2.78 | 0.048997 |
GGCTGTTAACCGAAAGGTTGGTGGT | i-tRF | AsnGTT |
| tRF-16-9L5HMVE | −1.18 | 0.001581 |
TGGTAGAGCGCGTGCT | i-tRF | AlaCGC, AlaAGC |
|
tRF-22-9LON4VN11 | −1.26 | 0.038332 |
TGGTAGAATTCTCGCCTGCCAC | i-tRF | GlyGCC |
|
tRF-25-P940KK5Y93 | −1.44 | 0.003051 |
GCCTCCTAAGCCAGGGATTGTGGGT | i-tRF | ArgCCT |
|
tRF-24-PY8HM2OSIZ | −1.71 | 0.033956 |
GCCTGTCACGCGGGAGACCGGGGT | i-tRF | AspGTC |
| tRF-38-YDLBRY73W0
K5KKDR | −1.80 | 0.047935 |
TTAACCGAAAGGTTGGTGGTTCGAGCCCACCCAGGGAC | i-tRF | AsnGTT |
|
tRF-29-P27JPJ60MVJY | −2.33 | 0.041736 |
GCAGAGTGGCGCAGCGGAAGCGTGCTGGG | i-tRF | MetCAT |
|
tRF-24-NMEH623K25 | −1.73 | 0.009462 |
CTGTCACGCGGGAGACCGGGGTTC | i-tRF | AspGTC |
|
tRF-29-QU7BPN6ISBJO | 5.08 | 0.024387 |
GCTGAGTGAAGCATTGGACTGTAAATCTA | i-tRF | TyrGTA |
| tsRNA-1020 | −1.87 | 0.014078 |
GAGAGCGCTCGGTTTTT | tRF-1 | PheGAA |
|
tRF-27-Q99P9P9NH5N | 2.17 |
1.53×10−4 |
GCTTCTGTAGTGTAGTGGTTATCACGT | tRF-5 | ValCAC |
|
tRF-24-Q99P9P9NF2 | 2.41 | 0.020747 |
GCTTCTGTAGTGTAGTGGTTATCA | tRF-5 | ValCAC |
|
tRF-30-6SXMSL73VL4Y | 2.19 |
1.13×10−5 |
GGCCGTGATCGTATAGTGGTTAGTACTCTG | tRF-5 | HisGTG |
|
tRF-26-P4R8YP9LOND | 1.13 | 0.009501 |
GCATGGGTGGTTCAGTGGTAGAATTC | tRF-5 | GlyGCC |
|
tRF-30-XSXMSL73VL4Y | 1.48 | 0.016991 |
TGCCGTGATCGTATAGTGGTTAGTACTCTG | tRF-5 | HisGTG |
DE mRNA and function analysis
From the GEPIA, 5,372 DE mRNAs were identified
between colon adenocarcinoma and paired normal samples. Of these,
2,692 were significantly upregulated and 2,680 were significantly
downregulated (Fig. 3A). The DE mRNAs
were further investigated by GO and KEGG pathway enrichment
analyses. The DE mRNAs were mainly enriched in cell
proliferation-related GO terms, including extracellular matrix
organization, cell adhesion, mitotic cell cycle, mitotic nuclear
division, cell cycle, negative regulation of cell proliferation,
G1/S transition of mitotic cell cycle, regulation of cell
proliferation, positive regulation of cell proliferation, DNA
replication and DNA strand elongation involved in DNA replication
(Fig. 3B). In the KEGG pathway
enrichment analysis, the DE mRNAs were primarily enriched in cell
proliferation-related (including cell cycle, ECM-receptor
interaction and DNA replication) and cancer-related (including p53
signaling replication, small cell lung cancer, pathways in cancer
and PI3K-Akt signaling pathway) KEGG terms (Fig. 3C).
Integrated tRF/mRNA/miRNA
analysis
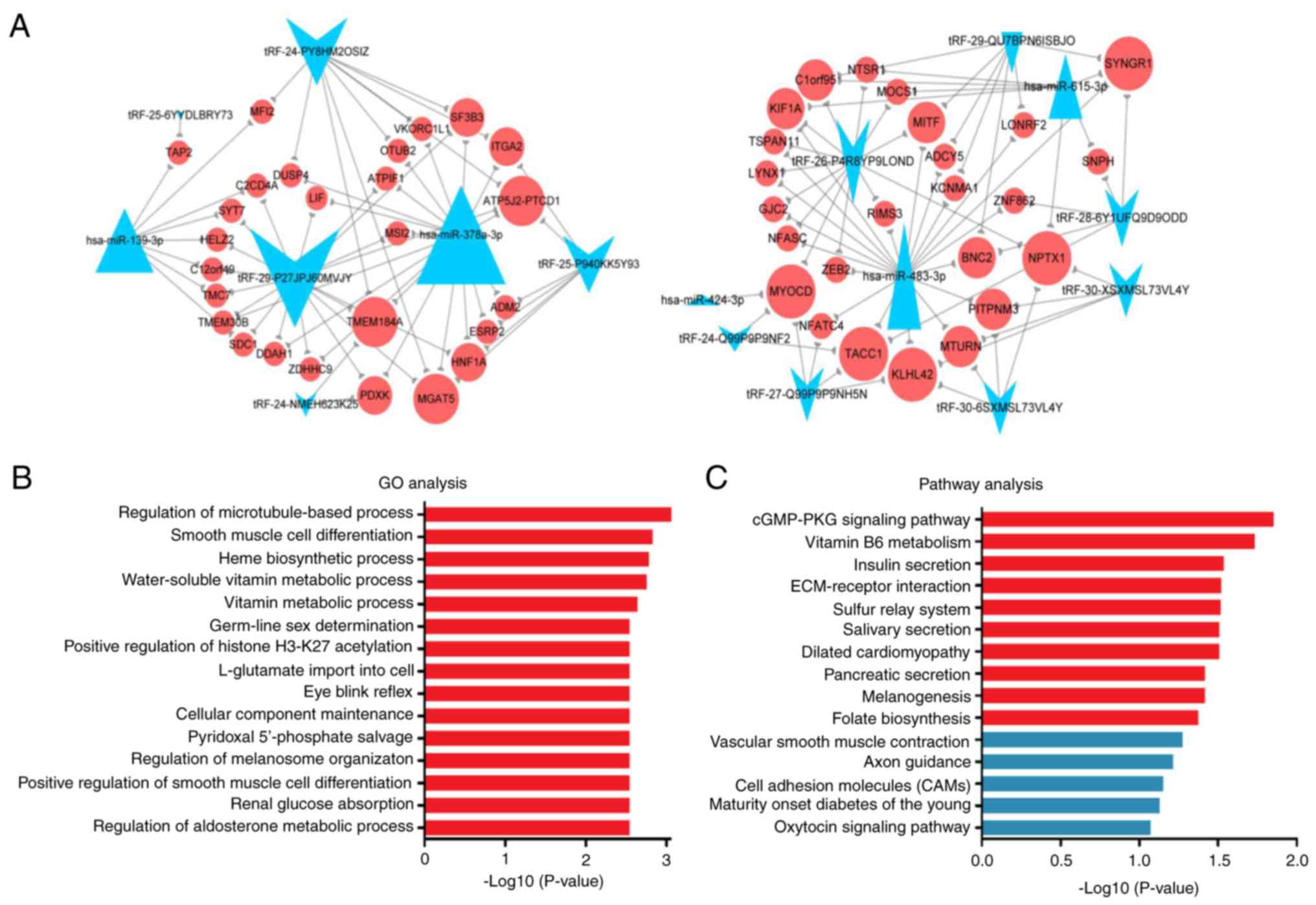
Through literature mining, five of the 26 DE miRNAs
(hsa-miR-139-3p, hsa-miR-378a-3p, hsa-miR-424-3p, hsa-miR-483-3p
and hsa-miR-615-3p) were selected as key miRNAs. The DE tRFs and
five key miRNAs were common to and negatively correlated with 55
target genes. An integrated tRF-mRNA-miRNA network was constructed
(Fig. 4A). Next, the 55 potential
targets of DE tRFs and key miRNAs were subjected to GO and pathway
enrichment analyses. These potential targets were mainly enriched
in the GO terms regulation of microtubule-based process, smooth
muscle cell differentiation, heme biosynthetic process,
water-soluble vitamin metabolic process and vitamin metabolic
process (Fig. 4B). In the KEGG
analyses, the potential targets were enriched in the terms cGMP-PKG
signaling pathway, vitamin B6 metabolism, insulin secretion,
ECM-receptor interaction and sulfur relay system (Fig. 4C).
The target genes MOCS1 and PDXK were
enriched in the GO terms water-soluble vitamin metabolic process
and vitamin metabolic process (Table
V). PDXK was enriched in the pathway term vitamin B6
metabolism. KCNMA1, ADCY5 and NFATC4 were
significantly enriched in the pathway term cGMP-PKG signaling
pathway. From the integrated tRF-mRNA-miRNA network (Fig. 4A), tRF-26-P4R8YP9LOND was associated
with MOCS1, tRF-24-NMEH623K25 and tRF-29-P27JPJ60MVJY were
associated with PDXK, tRF-29-QU7BPN6ISBJO was associated
with KCNMA1 and ADCY5, and tRF-27-Q99P9P9NH5N was
associated with NFATC4.
 | Table V.GO and pathway terms and enriched
genes. |
Table V.
GO and pathway terms and enriched
genes.
| ID | Term | Genes | P-value | Enrichment |
|---|
| GO:0032886 | Regulation of
microtubule-based process | TACC1, KLHL42 | 0.0005298 | 57.690476 |
| GO:0051145 | Smooth muscle cell
differentiation | MYOCD, NFATC4 | 0.0015027 | 34.614286 |
| GO:0006783 | Heme biosynthetic
process | ATPIF1, HNF1A | 0.0016579 | 32.965986 |
| GO:0006767 | Water-soluble
vitamin metabolic process | MOCS1, GSTO2,
PDXK | 0.00177 | 12.511188 |
| GO:0006766 | Vitamin metabolic
process | MOCS1, GSTO2,
PDXK | 0.0023028 | 11.411303 |
| GO:0018992 | Germ-line sex
determination | TMEM184A | 0.002889 | 346.14286 |
| GO:1901676 | Positive regulation
of histone H3-K27 acetylation | LIF | 0.002889 | 346.14286 |
| GO:1990123 | L-glutamate import
into cell | NTSR1 | 0.002889 | 346.14286 |
| GO:0060082 | Eye blink
reflex | KCNMA1 | 0.002889 | 346.14286 |
| GO:0043954 | Cellular component
maintenance | MYOCD | 0.002889 | 346.14286 |
| GO:0009443 | Pyridoxal
5′-phosphate salvage | PDXK | 0.002889 | 346.14286 |
| GO:1903056 | Regulation of
melanosome organization | ZEB2 | 0.002889 | 346.14286 |
| GO:2000724 | Positive regulation
of cardiac vascular smooth muscle cell differentiation | MYOCD | 0.002889 | 346.14286 |
| GO:0035623 | Renal glucose
absorption | HNF1A | 0.002889 | 346.14286 |
| GO:0032344 | Regulation of
aldosterone metabolic process | KCNMA1 | 0.002889 | 346.14286 |
| PATH:04022 | cGMP-PKG signaling
pathway | KCNMA1, ADCY5,
NFATC4 | 0.0140781 | 5.8015398 |
| PATH:00750 | Vitamin B6
metabolism | PDXK | 0.0184421 | 53.825397 |
| PATH:04911 | Insulin
secretion | KCNMA1, ADCY5 | 0.0291694 | 7.4241927 |
| PATH:04512 | ECM-receptor
interaction | ITGA11, SDC1 | 0.0304212 | 7.2573569 |
| PATH:04122 | Sulfur relay
system | MOCS1, KCNMA1,
ADCY5 | 0.0305564 | 32.295238 |
| PATH:05414 | Dilated
cardiomyopathy | ITGA11, ADCY5 | 0.0310552 | 7.1767196 |
| PATH:04972 | Pancreatic
secretion | KCNMA1, ADCY5 | 0.0383707 | 6.3950967 |
| PATH:04916 | Melanogenesis | MITF, ADCY5 | 0.0383707 | 6.3950967 |
| PATH:00790 | Folate
biosynthesis | MOCS1 | 0.0425282 | 23.068027 |
Validation of key miRNAs and candidate
tRF expression by RT-qPCR
Four key DE miRNAs, of which one was downregulated
(hsa-miR-378a-3p) and three were upregulated (hsa-miR-424-3p,
hsa-miR-483-3p, hsa-miR-615-3p) were verified by RT-qPCR in colon
cancer tissue and adjacent normal mucosa (n=30; Fig. 5A). Four DE tRFs (tRF-24-NMEH623K25,
tRF-30-XSXMSL73VL4Y, tRF-29-QU7BPN6ISBJO and tRF-27-Q99P9P9NH5N)
were significantly upregulated in colon cancer tissue compared with
adjacent normal mucosa (Fig. 5B).
Notably, DE tRF expression increased with tumor differentiation
(Fig. 5C). Furthermore,
tRF-29-QU7BPN6ISBJO and tRF-27-Q99P9P9NH5N were overexpressed in
colon cancer cell lines (SW480, SW620, HCT116 and HT29) compared
with a colon epithelial cell line (NCM460), as verified by RT-qPCR
(Fig. 5D).
Discussion
In the present study, 16 DE tRFs and 26 DE miRNAs
were identified between colon cancer and normal peritumor tissues
by small RNA sequencing. In addition, 5,327 DE mRNAs between colon
cancer and normal tissues were screened by GEPIA. In the GO and
pathway analyses, the DE mRNAs were enriched in cell
proliferation-related GO and pathway terms. Recent evidence
suggests that tRFs exert their effects in a similar way to miRNAs
(27). Therefore the subsequent
analysis and experiments were based on the hypothesis that tRFs and
miRNA target the same DE mRNAs and perform similar biological
functions in the development of colon cancer.
Among the 26 DE miRNAs between colon cancer and
paired normal tissues, five key miRNAs (hsa-miR-139-3p,
hsa-miR-378a-3p, hsa-miR-424-3p, hsa-miR-483-3p and hsa-miR-615-3p)
have been implicated in colon cancer. For example, low expression
of miR-139a-3p and miR-378a-3p is associated with poor survival of
colorectal cancer patients (28–30).
Overexpression of miR-378a-3p suppresses colon cancer cell
proliferation and induces apoptosis (30). miR-424-3p is upregulated in primary
and metastatic cancers and colorectal cancer cell lines, and its
overexpression is associated with poor outcomes in patients with
colorectal cancer (31). miR-483-3p
was identified to be significantly upregulated in tissue and blood
samples of colorectal cancer patients, and to promote the
proliferation of colorectal cancer cells and inhibit DLC-1
expression by targeting its 3′UTR. Furthermore, transfection with
an miR-483-3p antagomir suppressed the growth of colorectal cancer
cells (32,33). High expression of miR-615-3p was
reported in right-sided colon cancer (34). In the present study, hsa-miR-139-3p
and hsa-miR-378a-3p were identified to be downregulated in colon
cancer tissues, whereas hsa-miR-424-3p, hsa-miR-483-3p and
hsa-miR-615-3p were identified to be upregulated. These results are
consistent with previous reports. Thus, these key miRNAs were
selected to analyze the roles of tRFs in colon cancer.
To explore the roles of tRFs in colon cancer, an
integrated tRF/mRNA/miRNA analysis was performed. The mRNAs in the
constructed network were enriched in the GO terms regulation of
microtubule-based process, smooth muscle cell differentiation, heme
biosynthetic process, water-soluble vitamin metabolic process and
vitamin metabolic process. The top five pathway terms were cGMP-PKG
signaling pathway, vitamin B6 metabolism, insulin secretion,
ECM-receptor interaction and sulfur relay system. Several studies
have reported that vitamins play important roles in colon cancer.
Dietary vitamin B6 and D inhibited oncogenesis of colon cancer in a
murine model and human colon cancer cells (35–37). Low
intake of folate and vitamin B6, along with p53 overexpression,
increased the risk of colon cancer development (38). Increased vitamin B6 altered expression
of genes in the colon and inhibited the development and progression
of colon cancer by suppressing cancer cell proliferation (39,40). Thus,
vitamin B6 plays an important role in colon cancer by activating
p53 and increasing the expression of p21 (41). Vitamin K1 treatment inhibited the
proliferation and induced apoptosis of human colon cancer cells
(42). These reports suggest that
vitamin metabolic processes are intensively associated with colon
tumorigenesis. In the present study, the target genes of
tRF-26-P4R8YP9LOND, tRF-25-P940KK5Y93, tRF-30-XSXMSL73VL4Y,
tRF-24-NMEH623K25 and tRF-29-P27JPJ60MVJY were associated with
vitamin metabolism-related processes and pathways (water-soluble
vitamin metabolic process, vitamin metabolic process and vitamin B6
metabolism). In summary, these findings indicate that tRFs,
particularly tRF-26-P4R8YP9LOND, tRF-25-P940KK5Y93,
tRF-30-XSXMSL73VL4Y, tRF-24-NMEH623K25 and tRF-29-P27JPJ60MVJY, may
regulate vitamin levels and contribute to the development and
progression of colon cancer.
In addition, the cGMP-PKG signaling pathway can be
activated by the non-steroidal anti-inflammatory drug sulindac
sulfide, which is known to inhibit tumorigenesis by blocking the
proliferation and inducing apoptosis of colon cancer cells
(43,44). In the present study, the target genes
of tRF-29-QU7BPN6ISBJO and tRF-27-Q99P9P9NH5N were identified to be
enriched in the pathway term cGMP-PKG signaling pathway. This
result suggests that tRF-29-QU7BPN6ISBJO and tRF-27-Q99P9P9NH5N may
be involved in the development of colon cancer via the cGMP-PKG
signaling pathway.
In conclusion, the present study identified several
tRFs that may be involved in the development and progression of
colon cancer via vitamin metabolism-related processes and pathways
and the cGMP-PKG signaling pathway. However, the study has certain
limitations that must be considered. First, only three samples were
studied. Second, the expression pattern of the identified DE tRFs
and their association with the identified target genes require
further verification. The interaction between tRFs and their target
genes could be detected using the AGO2-RIP method and a
dual-luciferase reporter gene system. Finally, the specific role of
tRFs in colon cancer should be analyzed further. tRF expression
should be evaluated following exogenous expression or interference
of target genes, and the effect of tRFs and their target mRNAs on
the occurrence and development of tumor cells should be verified
through cell function experiments, including cell proliferation,
migration and invasion assays.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant no. 81301825) and the
Training Program for Medical Reserve Talents of the Health and
Family Planning Commission of Yunnan Province (grant nos. H-201641
and H-201624).
Availability of data and materials
The datasets used in the present study are available
from the corresponding author upon reasonable request.
Authors' contributions
YuL, JQ and WX (first author) conceived and design
the study. WX (first author), XW, XC, WX (fourth author), YaL, CL
and QL performed the experiments. WX (first author) wrote the
manuscript. XW, JQ and YuL reviewed and edited the manuscript. All
authors read and approved the manuscript and agree to be
accountable for all aspects of the research and for ensuring that
the accuracy or integrity of all parts of this work are
appropriately investigated and resolved.
Ethics approval and consent to
participate
All experimental protocols were approved by the
Human Ethics Committee of Kunming Medical University (Kunming,
China). Written informed consent was obtained from all
patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jaganathan SK, Vellayappan MV, Narasimhan
G and Supriyanto E: Role of pomegranate and citrus fruit juices in
colon cancer prevention. World J Gastroenterol. 20:4618–4625. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen J, Elfiky A, Han M, Chen C and Saif
MW: The role of Src in colon cancer and its therapeutic
implications. Clin Colorectal Cancer. 13:5–13. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kawaji H, Nakamura M, Takahashi Y,
Sandelin A, Katayama S, Fukuda S, Daub CO, Kai C, Kawai J, Yasuda
J, et al: Hidden layers of human small RNAs. BMC Genomics.
9:1572008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cole C, Sobala A, Lu C, Thatcher SR,
Bowman A, Brown JW, Green PJ, Barton GJ and Hutvagner G: Filtering
of deep sequencing data reveals the existence of abundant
Dicer-dependent small RNAs derived from tRNAs. RNA. 15:2147–2160.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lee YS, Shibata Y, Malhotra A and Dutta A:
A novel class of small RNAs: TRNA-derived RNA fragments (tRFs).
Genes Dev. 23:2639–2649. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Thompson DM and Parker R: Stressing out
over tRNA cleavage. Cell. 138:215–219. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Martens-Uzunova ES, Olvedy M and Jenster
G: Beyond microRNA - novel RNAs derived from small non-coding RNA
and their implication in cancer. Cancer Lett. 340:201–211. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kumar P, Anaya J, Mudunuri SB and Dutta A:
Meta-analysis of tRNA derived RNA fragments reveals that they are
evolutionarily conserved and associate with AGO proteins to
recognize specific RNA targets. BMC Biol. 12:782014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Muller S, Raulefs S, Bruns P, Afonso-Grunz
F, Plötner A, Thermann R, Jäger C, Schlitter AM, Kong B, Regel I,
et al: Next-generation sequencing reveals novel differentially
regulated mRNAs, lncRNAs, miRNAs, sdRNAs and a piRNA in pancreatic
cancer. Mol Cancer. 14:942015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Olvedy M, Scaravilli M, Hoogstrate Y,
Visakorpi T, Jenster G and Martens-Uzunova ES: A comprehensive
repertoire of tRNA-derived fragments in prostate cancer.
Oncotarget. 7:24766–24777. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Balatti V, Nigita G, Veneziano D, Drusco
A, Stein GS, Messier TL, Farina NH, Lian JB, Tomasello L, Liu CG,
et al: tsRNA signatures in cancer. Proc Natl Acad Sci U S A.
114:8071–8076. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Maute RL, Schneider C, Sumazin P, Holmes
A, Califano A, Basso K and Dalla-Favera R: tRNA-derived microRNA
modulates proliferation and the DNA damage response and is
down-regulated in B cell lymphoma. Proc Natl Acad Sci U S A.
110:1404–1409. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kozomara A and Griffiths-Jones S: miRBase:
Annotating high confidence microRNAs using deep sequencing data.
Nucleic Acids Res. 42:D68–D73. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Balatti V, Rizzotto L, Miller C,
Palamarchuk A, Fadda P, Pandolfo R, Rassenti LZ, Hertlein E,
Ruppert AS, Lozanski A, et al: TCL1 targeting
miR-3676 is codeleted with tumor protein p53 in chronic
lymphocytic leukemia. Proc Natl Acad Sci USA. 112:2169–2174. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Qi L and Ding Y: Screening of
differentiation-specific molecular biomarkers for colon cancer.
Cell Physiol Biochem. 46:2543–2550. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yang L, Wang S, Zhou Y, Lai S, Xiao G,
Gazdar A and Xie Y: Evaluation of the 7th and 8th editions of the
AJCC/UICC TNM staging systems for lung cancer in a large North
American cohort. Oncotarget. 8:66784–66795. 2017.PubMed/NCBI
|
|
19
|
Nawrocki EP, Burge SW, Bateman A, Daub J,
Eberhardt RY, Eddy SR, Floden EW, Gardner PP, Jones TA, Tate J, et
al: Rfam 12.0: Updates to the RNA families database. Nucleic Acids
Res. 43:D130–D137. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kalvari I, Argasinska J, Quinones-Olvera
N, Nawrocki EP, Rivas E, Eddy SR, Bateman A, Finn RD and Petrov AI:
Rfam 13.0: Shifting to a genome-centric resource for non-coding RNA
families. Nucleic Acids Res. 46:D335–D342. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tang Z, Li C, Kang B, Gao G and Zhang Z:
GEPIA: A web server for cancer and normal gene expression profiling
and interactive analyses. Nucleic Acids Res. 45:W98–W102. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hulsegge I, Kommadath A and Smits MA:
Globaltest and GOEAST: Two different approaches for gene ontology
analysis. BMC Proc. 4 (Suppl 3):S102009. View Article : Google Scholar
|
|
25
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The Gene
Ontology Consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCT method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kuscu C, Kumar P, Kiran M, Su Z, Malik A
and Dutta A: tRNA fragments (tRFs) guide Ago to regulate gene
expression post-transcriptionally in a Dicer independent manner.
RNA. 24:1093–1105. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Liu X, Duan B, Dong Y, He C, Zhou H, Sheng
H, Gao H and Zhang X: MicroRNA-139-3p indicates a poor prognosis of
colon cancer. Int J Clin Exp Pathol. 7:8046–8052. 2014.PubMed/NCBI
|
|
29
|
Ng L, Wan TM, Man JH, Chow AK, Iyer D,
Chen G, Yau TC, Lo OS, Foo DC, Poon JT, et al: Identification of
serum miR-139-3p as a non-invasive biomarker for colorectal cancer.
Oncotarget. 8:27393–27400. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li H, Dai S, Zhen T, Shi H, Zhang F, Yang
Y, Kang L, Liang Y and Han A: Clinical and biological significance
of miR-378a-3p and miR-378a-5p in colorectal cancer. Eur J Cancer.
50:1207–1221. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Torres S, Garcia-Palmero I, Bartolome RA,
Fernandez-Aceñero MJ, Molina E, Calviño E, Segura MF and Casal JI:
Combined miRNA profiling and proteomics demonstrates that different
miRNAs target a common set of proteins to promote colorectal cancer
metastasis. J Pathol. 242:39–51. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yong FL, Law CW and Wang CW: Potentiality
of a triple microRNA classifier: MiR-193a-3p, miR-23a and
miR-338-5p for early detection of colorectal cancer. BMC cancer.
13:2802013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Cui H, Liu Y, Jiang J, Liu Y, Yang Z, Wu
S, Cao W, Cui IH and Yu C: IGF2-derived miR-483 mediated
oncofunction by suppressing DLC-1 and associated with colorectal
cancer. Oncotarget. 7:48456–48466. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Schee K, Lorenz S, Worren MM, Günther CC,
Holden M, Hovig E, Fodstad O, Meza-Zepeda LA and Flatmark K: Deep
sequencing the MicroRNA transcriptome in colorectal Cancer. PloS
One. 8:e661652013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Komatsu S, Yanaka N, Matsubara K and Kato
N: Antitumor effect of vitamin B6 and its mechanisms. Biochim
Biophys Acta. 1647:127–130. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Meeker S, Seamons A, Paik J, Treuting PM,
Brabb T, Grady WM and Maggio-Price L: Increased dietary vitamin D
suppresses MAPK signaling, colitis, and colon cancer. Cancer Res.
74:4398–4408. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Pereira F, Larriba MJ and Munoz A: Vitamin
D and colon cancer. Endocr Relat Cancer. 19:R51–R71. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Schernhammer ES, Ogino S and Fuchs CS:
Folate and vitamin B6 intake and risk of colon cancer in
relation to p53 expression. Gastroenterology. 135:770–780. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Toya K, Hirata A, Ohata T, Sanada Y, Kato
N and Yanaka N: Regulation of colon gene expression by vitamin B6
supplementation. Mol Nutr Food Res. 56:641–652. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Komatsu SI, Watanabe H, Oka T, Tsuge H,
Nii H and Kato N: Vitamin B-6-supplemented diets compared with a
low vitamin B-6 diet suppress azoxymethane-induced colon
tumorigenesis in mice by reducing cell proliferation. J Nutr.
131:2204–2207. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhang P, Suidasari S, Hasegawa T, Yanaka N
and Kato N: Vitamin B6 activates p53 and elevates p21
gene expression in cancer cells and the mouse colon. Oncol Rep.
31:2371–2376. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Orlando A, Linsalata M, Tutino V, D'Attoma
B, Notarnicola M and Russo F: Vitamin K1 exerts antiproliferative
effects and induces apoptosis in three differently graded human
colon cancer cell lines. Biomed Res Int. 2015:2967212015.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Li N, Xi Y, Tinsley HN, Gurpinar E, Gary
BD, Zhu B, Li Y, Chen X, Keeton AB, Abadi AH, et al: Sulindac
selectively inhibits colon tumor cell growth by activating the
cGMP/PKG pathway to suppress Wnt/β-catenin signaling.
Mol Cancer Ther. 12:1848–1859. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Li N, Chen X, Zhu B, Ramírez-Alcántara V,
Canzoneri JC, Lee K, Sigler S, Gary B, Li Y, Zhang W, et al:
Suppression of β-catenin/TCF transcriptional activity
and colon tumor cell growth by dual inhibition of PDE5 and 10.
Oncotarget. 6:27403–27415. 2015.PubMed/NCBI
|