Introduction
Bladder cancer has become the most common tumor of
the urinary system globally (1) with
almost 350,000-380,000 new cases diagnosed worldwide annually
(2). Although a great number of
bladder cancer patients have experienced effective treatment
including chemotherapy, radiation therapy and transurethral
resection, the prognosis for bladder cancer patients is still
unsatisfactory due to advanced stages with distal metastases and
frequent recurrences (2,3). Therefore, it is urgent to elucidate the
detailed molecular mechanism associated with the tumorigenesis of
bladder cancer.
To date, multiple histone methyltransferases and
demethylases have been revealed. The JMJD2 family recognizes and
demethylates di- and tri-methylated H3K9 and H3K36 (4). JMJD2A is the most studied member of the
JMJD2 family, and has been revealed to either activate or suppress
gene transcription through its demethylase activity (4–6). Notably,
JMJD2A has been revealed to be aberrantly expressed in multiple
types of cancer, including breast (7,8), bladder
(9,10), lung (10,11) and
colon cancer (12). The aim of this
study was to explore the function of JMJD2A in human bladder cancer
and detail its molecular mechanism.
Previous studies have revealed that
epithelial-mesenchymal transition (EMT) is essential for the
metastasis of cancers, such as bladder cancer (13). The main characteristic of EMT is
cadherin switching and the change of cytoskeleton and cell
polarity, which facilitates the motility of cells (14). Moreover, EMT has been demonstrated to
be associated with the emergence of chemoresistance in several
cancers (14). Additionally, several
studies have also revealed that intravesical recurrence is
associated with EMT (15,16).
In the present study, our findings revealed that
JMJD2A was upregulated in bladder cancer, and high expression of
JMJD2A was closely associated with advanced histological grade and
poor prognosis of bladder cancer. Moreover, it was revealed that
JMJD2A promoted cell migration and invasion through regulation of
EMT. Additionally, it was determined that JMJD2A transcriptionally
regulated SLUG.
Materials and methods
Cell culture
Human bladder cancer cell lines (T24 and 5637) and
human immortalized bladder urothelial cell line SV-HUC-1 were
purchased from the American Type Culture Collection (ATCC;
Manassas, VA, USA). T24 and 5637 cells were cultured in McCoy's 5A
modified medium (Gibco; Thermo Fisher Scientific, Inc., Waltham,
MA, USA) or RPMI-1640 medium (Gibco; Thermo Fisher Scientific,
Inc.) with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher
Scientific, Inc). SV-HUC-1 cells were cultured in F-12K medium
(Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS.
All cells were maintained in a humidified incubator at 37°C with 5%
CO2.
Patient tissue specimens
A total of 89 resected specimens from bladder cancer
patients were collected for this study. Bladder cancer specimens
were compared with paired normal bladder tissue from the same
patient. All specimens had been histologically and clinically
diagnosed at Tengzhou Central People's Hospital from August 2006 to
April 2014, independently, by three experienced pathologists. Prior
to surgery, none of the patients had undergone hormone therapy,
chemotherapy or radiotherapy. All patients provided written signed
informed consent. The study was approved and supervised by the
Ethics Committee of Tengzhou Central People's Hospital (Tengzhou,
China).
Real-time quantitative polymerase
chain reaction (RT-qPCR)
Total RNA was extracted from bladder cancer tissues
and cell lines using TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) following the manufacturer's instructions. An RNA
sample (2 µg) was utilized to synthesize first-strand cDNA using a
cDNA Reverse Transcription kit (Takara Bio, Inc., Otsu, Japan).
Subsequently, SYBR Green was used to perform a quantitative
real-time PCR (qPCR) assay on an Applied Biosystems 7300 Real-Time
PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.).
The primer sequences were as follows: JMJD2A forward,
5′-GAAGCCACGAGCATCCTATGA-3′ and reverse,
5′-GCGGAACTCTCGAACAGTCA-3′; E-cadherin forward,
5′-AAACATCATTGATGCAGACC-3′ and reverse,
5′-GATAGATTCTTGGGTTGGGTC-3′; N-cadherin forward,
5′-CAAAGCCTGGAACATATGTG-3′ and reverse, 5′-GTTTGAAAGGCCATATGTGG-3′;
SLUG forward, 5′-ACACATACAGTGATTATTTCCC-3′ and reverse,
5′-ACTGTAGTCTTTCCTCTTCAT-3′; SNAIL forward,
5′-TCTAATCCAGAGTTTACCTTCCAG-3′ and reverse,
5′-TGAAGTAGAGGAGAAGGACGA-3′; TWIST1 forward,
5′-GTACATCGACTTCCTCTACC-3′ and reverse,
5′-GAAACAATGACATCTAGGTCTC-3′; GAPDH forward,
5′-AGAAGGCTGGGGCTCATTTG-3′ and reverse, 5′-AGGGGCCATCCACAGTCTTC-3′.
GAPDH served as an internal control. The 2−ΔΔCq method
was utilized to calculate the relative expression of the target
gene (17). All experiments were
carried out at least three times.
Western blot analysis
Following transfection, cells were collected and
lysed in RIPA buffer (Beyotime Institute of Biotechnology, Haimen,
China) with protease and phosphatase inhibitors at 4°C for 45 min,
and the concentration of the samples was quantified using a BCA
protein assay kit (Pierce; Thermo Fisher Scientific, Inc.)
according to manufacturer's protocol. Western blot analysis was
performed according to a previously described procedure (18). A total of 40 µg protein was loaded in
10% SDS-PAGE. Subsequently, proteins were transferred to
polyvinylidene difluoride (PVDF) membranes and blocked in 5%
skimmed milk for 1 h at room temperature. The membranes were
incubated with indicated primary antibodies at 4°C overnight. After
washing with PBST three times, membranes were incubated with HRP
conjugated second antibodies at room temperature for 1 h. The
following antibodies were used: anti-JMJD2A (dilution 1:1,000; cat.
no. ab105953; Abcam, Cambridge, UK); anti-E-cadherin (dilution
1:1,000; cat. no. 14472; Cell Siganling Technology Danvers, MA,
USA) and anti-N-cadherin (dilution 1:1,000; cat. no. 13116; Cell
Siganling Technology); and anti-SLUG (dilution 1:2,000; cat. no.
SAB1305973; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany),
anti-TWIST1 (dilution 1:500; cat. no. SAB1409777; Sigma-Aldrich;
Merck KGaA), anti-SNAIL (dilution 1:2,000; cat. no. SAB1306281;
Sigma-Aldrich; Merck KGaA) and anti-β-actin (dilution 1:4,000; cat.
no. A1978; Sigma-Aldrich; Merck KGaA). The blots were visualized
using an enhanced chemiluminescent (ECL) kit (Thermo Fisher
Scientific, Inc.). All experiments were carried out at least three
times. The densitometry of the blots was quantified by ImageJ
(National Institutes of Health, Bethesda, MD, USA).
Wound healing assay
A wound healing assay was carried out to determine
the effect of JMJD2A on cell migration ability. Succinctly, a
straight wound was scratched using a 20-µl pipette tip when the
transfected cells reached ~85–95% confluence in 12-well plates. The
cells were washed with phosphate-buffered saline (PBS) to remove
the detached cells, and the cells were maintained at 37°C in a
humidified incubator containing 5% CO2. The images of
wound closure were captured at 0 and 48 h with a digital camera
system (Olympus Corp., Tokyo, Japan). All experiments were
performed at least three times.
Transwell invasion assay
A Transwell invasion assay was used to examine the
effect of JMJD2A on cell invasion ability. In brief, each Transwell
chamber (BD Biosciences, Franklin Lakes, NJ, USA) was first coated
with 80 µl Matrigel following the manufacturer's protocol.
Approximately 2×105 transfected cells were suspended in
500 µl serum-free McCoy's 5A modified medium or RPMI-1640 medium
and added into the top chamber. Subsequently, 500 µl McCoy's 5A
modified medium or RPMI-1640 medium containing 10% FBS was added
into the bottom chamber. After incubation for 36 h, the invaded
cells were fixed with formaldehyde at room temperature for 10 min
and stained with 0.1% crystal violet at room temperature for 10
min. The non-invaded cells were removed using a cotton swab. The
cells were counted in six different fields using a light microscope
(Olympus Corp., Tokyo, Japan). All experiments were performed at
least three times.
Chromatin immunoprecipitation (ChIP)
assay
A chromatin immunoprecipitation (ChIP) assay was
performed as previously described (19). Briefly, JMJD2A was overexpressed or
knocked down in T24 and 5637 cells, and after transfection for 48
h, the cells were harvested and crosslinked with 1% formaldehyde
for 20 min. Subsequently, the cells were lysed and then sonicated
to obtain DNA fragments (200–500 bp in size). Next, the samples
were immunoprecipitated with 2 µg ChIP-grade antibodies H3K9me2
(cat. no. ab1220; Abcam) overnight at 4°C, supplemented with
protein G beads. Reversing the cross-links was carried out at 65°C
overnight. DNA was purified with a Qiagen DNA extraction kit
(Qiagen, Inc., Valencia, CA, USA). The immunoprecipitated DNA was
purified for RT-qPCR analyses with primers as follows: SLUG
forward, 5′-CTGGATTATGCCTCTGTGAT-3′ and reverse,
5′-TGGTATTTATTTGCTGGTAG-3′. All experiments were performed at least
three times.
Colony formation assay
Approximately 5×103 infected T24 and 5637
cells were plated in 6-well culture plates and cultured with
serum-free DMEM. Following culture for 2 weeks at 37°C in a 5%
CO2 humidified incubator, the cells were washed with PBS
and stained with 0.1% crystal violet at room temperature for 10
min. Colonies (number of cells >50) were manually calculated
under a light microscope. All experiments were performed at least
three times.
Cell counting kit-8 (CCK-8) assay
CCK-8 (Beyotime Institute of Biotechnology) assays
were carried out to determine the effect of JMJD2A on cell
proliferation. Approximately 4×103 transfected cells
were suspended and placed in each well of 96-well plates and
incubated at 37°C in a humidifier incubator containing 5%
CO2. After incubation for 0, 24, 48 or 72 h, 20 µl of
CCK-8 solution was added into the cells of each well and cultured
at 37°C for 30 min. The optical density (OD) was determined at 450
nm using an ELISA microplate reader (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA). All experiments were carried out at least three
times.
Statistical analysis
All the data is presented as the mean ± standard
deviation (SD) and analyzed using GraphPad Prism 5.0 (GraphPad
Software, La Jolla, CA, USA). Statistical differences among groups
were examined using Student's t-test or one-way analysis of
variance (ANOVA) followed by Tukey's post hoc test. Kaplan-Meier
analysis and log-rank tests were carried out to assess overall
survival. P<0.05 was considered to indicate a statistically
significant difference.
Results
JMJD2A is upregulated in bladder
cancer tissues
A previous study had indicated that JMJD2A was
upregulated in bladder cancer (10).
To ascertain whether JMJD2A was upregulated in bladder cancer
tissues, RT-qPCR and western blot analysis were conducted in 89
pairs of bladder cancer tissue samples and matched adjacent normal
tissue samples. As revealed in Fig.
1A, the expression of JMJD2A was higher in the bladder cancer
tissue samples compared with the normal tissue samples (Fig. 1A, P<0.05). The relationship between
the expression of JMJD2A and the clinicopathological
characteristics of bladder cancer was then assessed. The mean value
of JMJD2A mRNA content in tumor cells was used as the standard.
Thus, higher values than the standard value were defined as high
expression, and lower values than the standard value were defined
as low expression. As revealed in Table
I, high expression of JMJD2A was positively associated with
tumor size and lymphatic invasion as well as high-grade bladder
cancer, suggesting that JMJD2A played an essential role in tumor
growth and metastasis in bladder cancer. In addition, the survival
curve demonstrated that high expression of JMJD2A was associated
with shorter overall survival and predicted a poor prognosis
(Fig. 1B). Subsequently, the
expression of JMJD2A in two bladder cancer cell lines (T24 and
5637) and a human immortalized bladder urothelial cell line
(SV-HUC-1), used as a control, was determined. As revealed in
Fig. 1C, both the mRNA and protein
expression levels of JMJD2A were high in bladder cancer cell lines,
T24 and 5637 compared with those in SV-HUC-1 (Fig. 1C, P=0.042). These data indicated that
JMJD2A may function as an oncogene in bladder cancer.
 | Table I.Clinicopathological variables in 89
bladder cancer patients. |
Table I.
Clinicopathological variables in 89
bladder cancer patients.
|
|
| JMJD2A protein
expression |
|
|---|
|
|
|
|
|
|---|
| Variables | No. (n=89) | Low (n=36) | High (n=53) | P-value |
|---|
| Age (years) |
|
|
| 0.709 |
|
<55 | 35 | 15 | 20 |
|
| ≥55 | 54 | 21 | 33 |
|
| Tumor size |
|
|
| 0.019 |
| Small
(≤1.5 cm) | 41 | 22 | 19 |
|
| Large
(>1.5 cm) | 48 | 14 | 34 |
|
| Lymphatic
invasion |
|
|
| 0.047 |
| Yes | 41 | 12 | 29 |
|
| No | 48 | 24 | 24 |
|
| TNM stage |
|
|
| 0.007 |
| I–II | 31 | 24 | 20 |
|
|
III–IV | 35 | 12 | 33 |
|
JMJD2A promotes bladder cancer cell
migration and invasion
To explore the effect of JMJD2A on bladder cancer
cells, JMJD2A was first overexpressed or knocked down in T24 and
5637 cells. After confirming the expression of JMJD2A (Fig. 2A), wound healing and Transwell
invasion assays were performed to detect whether JMJD2A affected
bladder cancer cell migration and invasion. Wound healing assays
revealed that the migratory ability of T24 and 5637 cells was
significantly enhanced when JMJD2A was overexpressed; whereas the
migratory ability of T24 and 5637 cells was decreased when JMJD2A
was knocked down (Fig. 2B).
Consistent with the wound healing assay result, in Transwell
invasion assays, the number of invaded cells in
JMJD2A-overexpressed T24 and 5637 was increased; however, the
number of invaded cells in JMJD2A-depleted T24 and 5637 was
decreased, compared to the control cell group (Fig. 2C). Overall, these results indicated
that JMJD2A promoted bladder cancer cell migration and
invasion.
Overexpression of JMJD2A facilitates
EMT in bladder cancer cells
EMT plays a crucial role in the metastasis of
bladder cancer cells (20). To
further determine whether JMJD2A promoted bladder cancer cell
migration and invasion through regulation of EMT, the expression of
EMT-associated genes, including E-cadherin, N-cadherin and vimentin
in bladder cancer cells, was detected by RT-qPCR and western
blotting. As revealed in Fig. 3A and
B, the expression of N-cadherin and vimentin was upregulated,
and in contrast, E-cadherin expression was significantly
downregulated upon JMJD2A overexpression in T24 and 5637 cells;
however, inhibition of JMJD2A resulted in an increased expression
of E-cadherin and a decreased expression of N-cadherin and vimentin
(Fig. 3C and D). These findings
demonstrated that JMJD2A promoted bladder cancer cell migration and
invasion by inactivating the EMT pathway.
JMJD2A transcriptionally regulates
SLUG in bladder cancer cells
To further decipher the detailed mechanism of JMJD2A
on the modulation of EMT, it was next examined whether JMJD2A
regulated the expression of EMT-related transcription factors,
including SLUG, Twist1, and SNAIL. Notably, it was revealed that
the overexpression/knockdown of JMJD2A had little effect on the
expression of TWIST1 and SNAIL but affected the expression of SLUG
(Fig. 4A and B). These results
revealed that JMJD2A regulated EMT possibly through the modulation
of SLUG. qChIP analysis was next performed to determine whether
JMJD2A regulated this gene by modulating H3K9me2 demethylation at
promoter regions. In order to verify this hypothesis, qChIP assay
was used with anti-H3K9me2 in JMJD2A overexpression or
JMJD2A-depleted T24 or 5637 cells. The results revealed that
H3K9me2 at the promoter of SLUG was significantly increased
when JMJD2A was knocked down in T24 and 5637 cells; however,
H3K9me2 at the promoter of SLUG was significantly decreased
when JMJD2A was overexpressed in T24 and 5637 cells (Fig. 4C). These results indicated that
SLUG was transcriptionally regulated by JMJD2A through its
H3K9 demethylase activity. The aforementioned results revealed that
JMJD2A transcriptionally activated SLUG expression in bladder
cancer.
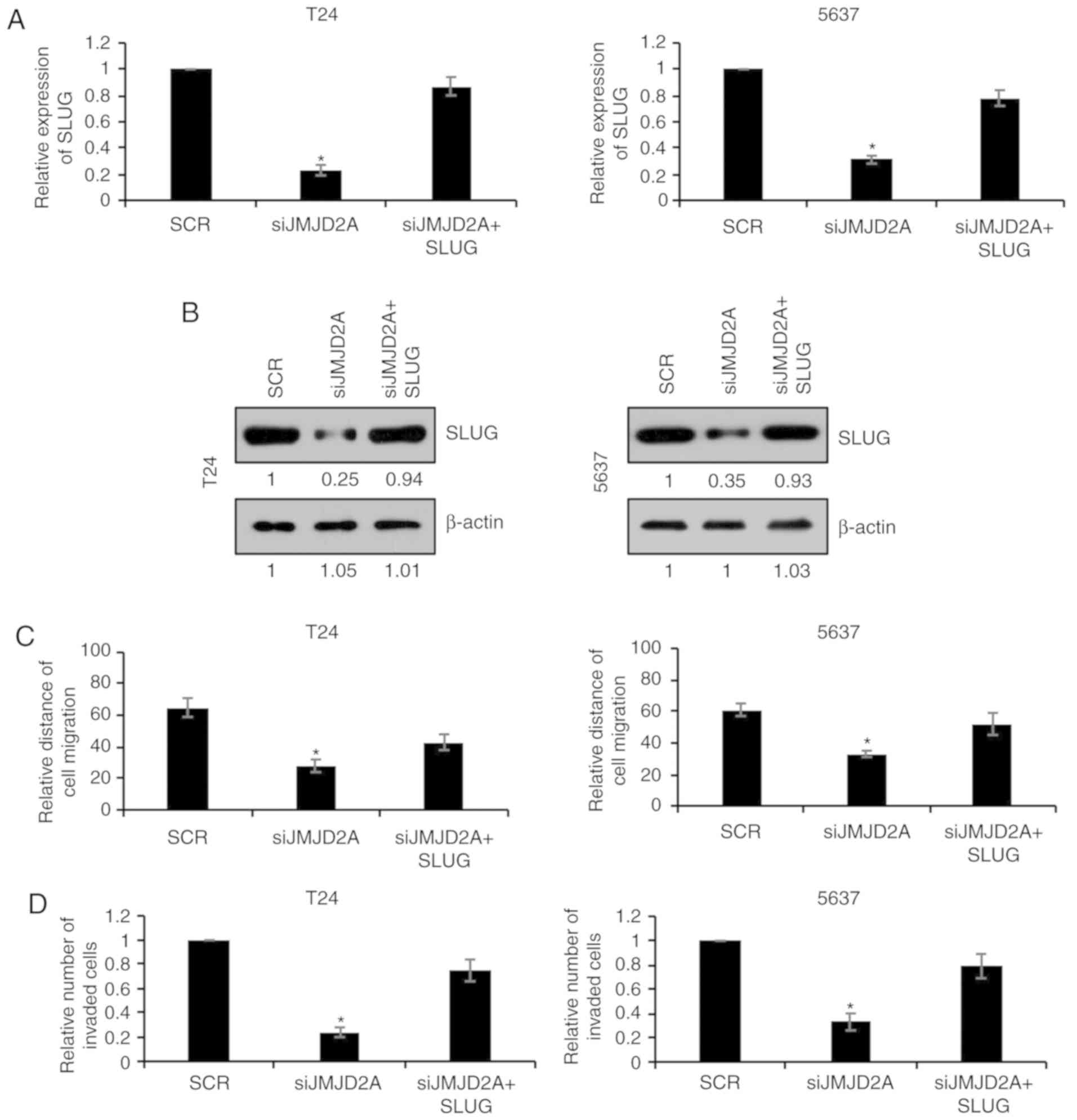
SLUG overexpression reverses the
effects of JMJD2A on migration and invasion in bladder cancer
cells
To further investigate whether SLUG was involved in
JMJD2A-mediated malignant phenotypes of bladder cancer cells, SLUG
was overexpressed in JMJD2A-depleted T24 or 5637 cells. The mRNA
and protein levels of SLUG were subsequently established using
RT-qPCR and western blot analyses, respectively. As revealed in
Fig. 5A and B, co-transfection with
JMJD2A siRNA and SLUG plasmids reversed the inhibitory effect on
SLUG expression in T24 or 5637 cells compared with transfection
with JMJD2A siRNA alone (Fig. 5A and
B). The migration and invasion of these two groups were also
compared. As revealed in Fig. 5C and
D, overexpression of SLUG reversed the inhibitory effects on
migration and invasion of JMJD2A-depleted T24 and 5637 cells
(Fig. 5C and D), revealing that SLUG
was in fact involved in JMJD2A-mediated malignant phenotypes of
bladder cancer cells.
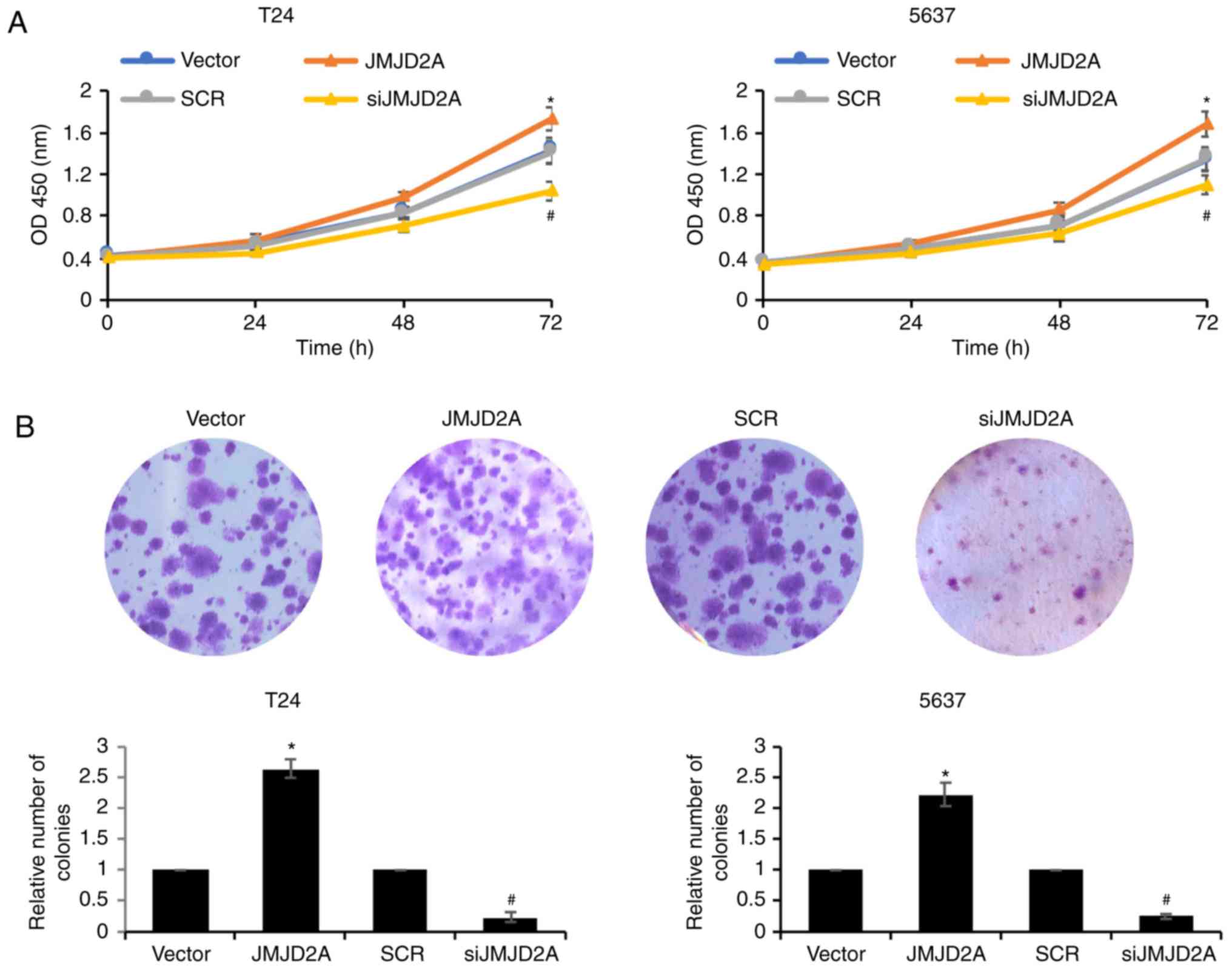
JMJD2A promotes bladder cancer cell
proliferation
To further confirm the biological function of
JMJD2A, CCK-8 and colony formation assays were performed to
identify the roles of JMJD2A on bladder cancer cell proliferation.
As revealed ectopic expression of JMJD2A promoted the cell
proliferation rate, compared with vector control group; however,
inhibition of JMJD2A suppressed the cell proliferation rate
compared with the SCR control group (Fig.
6). Consistent with the CCK-8 assay results, in the colony
formation assays, the ectopic expression of JMJD2A in T24 and 5637
cells was increased; whereas the colony formation of
JMJD2A-depleted T24 and 5637 cells was decreased (Fig. 6B). These findings indicated that
JMJD2A promoted the proliferation ability of bladder cancer
cells.
Discussion
JMJD2A has been revealed to be aberrantly-regulated
in various types of cancers, including breast (7,8), bladder
(9), lung (10,11) and
colon cancer (12). In the present
study, the results of RT-qPCR and western blot analyses revealed
that JMJD2A was upregulated in bladder cancer (BC) tissues and cell
lines, which was in contrast to a previous study (9). However, our research was consistent with
a study by Kogure et al (10).
The reason for the discrepancy in the results obtained may be due
to the difference in the number of specimens or geographical
regions of selected samples. In addition, the expression of JMJD2A
was also analyzed in a bladder database on cBioPortal (http://www.cbioportal.org; data not shown), suggesting
that JMJD2A is increased in bladder cancer. In order to further
decipher the roles of JMJD2A in bladder cancer, CCK-8, colony
formation, wound healing and Transwell invasion assays were
performed, which revealed that ectopic expression of JMJD2A
promoted T24 and 5637 cell proliferation, migration and invasion
while these effects were reversed when the expression of JMJD2A was
knocked down. The results demonstrated that JMJD2A could regulate
bladder cancer cell functions. Moreover, Kaplan-Meier curves
revealed that the patients with high expression of JMJD2A
experienced poor prognosis (P<0.05).
One noteworthy observation was that JMJD2A promoted
migration and invasion of bladder cancer cells. EMT is a
well-characterized process that results in invasion and metastatic
dissemination of human cancers (21,22).
Therefore, whether JMJD2A could modulate EMT of bladder cancer
cells was further examined. Gain- and loss-of-function analysis
revealed that JMJD2A overexpression decreased E-cadherin expression
and increased N-cadherin expression, while JMJD2A inhibition led to
the opposite results. These data indicated that JMJD2A may modulate
cell invasion by facilitating EMT in bladder cancer cells.
In the present study, our findings suggested that
JMJD2A transcriptionally activated SLUG. SLUG was upregulated by
JMJD2A at both the mRNA and protein level, and SLUG promoter
activity was significantly activated by JMJD2A, indicating a
transcriptional activation of SLUG by JMJD2A.
However, there are still some limitations in our
study. Our research revealed that JMJD2A expression was associated
with the stage of bladder cancer, and JMJD2A promoted cell
proliferation, migration and invasion in bladder cancer. However,
the upstream of JMJD2A in bladder cancer is still unknown. It would
be interesting to decipher whether the stages of bladder cancer can
also influence the expression of JMJD2A. We assume that there are
some proteins which regulate JMJD2A expression in different stages
of bladder cancer. In future, ATAC-seq and RNA-seq could be
performed using diverse stages of bladder cancer tissue samples and
potential transcription factors may be predicted. This may help us
to further gain insight on bladder cancer development. Moreover,
detection of the expression of JMJD2A in tissues using
immunohistochemical analysis should be performed. In addition, the
effect of JMJD2A in vivo should also be assessed for further
investigation.
In summary, this study indicated that JMJD2A
contributed to tumorigenesis in bladder cancer by regulating SLUG.
Additionally, high JMJD2A expression was associated with a poor
prognosis for bladder cancer patients. JMJD2A may act as an
oncogene in bladder cancer and may be a potential therapeutic
target for bladder cancer. Moreover, considering current changing
clinical treatment standards, it would be of high relevance to
explore the correlation between JMD2A and PD-L1 expression or other
immune markers and T-cell infiltration.
Acknowledgements
Not applicable.
Funding
Not applicable.
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
FW, YL and GC conceived and designed the study. FW,
QZ, LW, FS and BS performed the experiments. FW, QZ, FS and LW
wrote the paper. FW, YL and BS reviewed and edited the manuscript.
All authors read and approved the manuscript and agree to be
accountable for all aspects of the research in ensuring that the
accuracy or integrity of any part of the work are appropriately
investigated and resolved.
Ethics approval and consent to
participate
The Ethics Committee of Tengzhou Central People's
Hospital (Tengzhou, China) approved the present study. All patients
provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jin Y, Lu J, Wen J, Shen Y and Wen X:
Regulation of growth of human bladder cancer by miR-192. Tumour
Biol. 36:3791–3797. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Berry WL and Janknecht R: KDM4/JMJD2
histone demethylases: Epigenetic regulators in cancer cells. Cancer
Res. 73:2936–2942. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gray SG, Iglesias AH, Lizcano F,
Villanueva R, Camelo S, Jingu H, Teh BT, Koibuchi N, Chin WW,
Kokkotou E and Dangond F: Functional characterization of JMJD2A, a
histone deacetylase- and retinoblastoma-binding protein. J Biol
Chem. 280:28507–28518. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang D, Yoon HG and Wong J: JMJD2A is a
novel N-CoR-interacting protein and is involved in repression of
the human transcription factor achaete scute-like homologue 2
(ASCL2/Hash2). Mol Cell Biol. 25:6404–6414. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li BX, Zhang MC, Luo CL, Yang P, Li H, Xu
HM, Xu HF, Shen YW, Xue AM and Zhao ZQ: Effects of RNA
interference-mediated gene silencing of JMJD2A on human breast
cancer cell line MDA-MB-231 in vitro. J Exp Clin Cancer Res.
30:902011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Berry WL, Shin S, Lightfoot SA and
Janknecht R: Oncogenic features of the JMJD2A histone demethylase
in breast cancer. Int J Oncol. 41:1701–1706. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kauffman EC, Robinson BD, Downes MJ,
Powell LG, Lee MM, Scherr DS, Gudas LJ and Mongan NP: Role of
androgen receptor and associated lysine-demethylase coregulators,
LSD1 and JMJD2A, in localized and advanced human bladder cancer.
Mol Carcinog. 50:931–944. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kogure M, Takawa M, Cho HS, Toyokawa G,
Hayashi K, Tsunoda T, Kobayashi T, Daigo Y, Sugiyama M, Atomi Y, et
al: Deregulation of the histone demethylase JMJD2A is involved in
human carcinogenesis through regulation of the G(1)/S transition.
Cancer Lett. 336:76–84. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mallette FA and Richard S: JMJD2A promotes
cellular transformation by blocking cellular senescence through
transcriptional repression of the tumor suppressor CHD5. Cell Rep.
2:1233–1243. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kim TD, Shin S, Berry WL, Oh S and
Janknecht R: The JMJD2A demethylase regulates apoptosis and
proliferation in colon cancer cells. J Cell Biochem. 113:1368–1376.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bryan RT: Cell adhesion and urothelial
bladder cancer: The role of cadherin switching and related
phenomena. Philos Trans R Soc Lond B Biol Sci. 370:201400422015.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lamouille S, Xu J and Derynck R: Molecular
mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell
Biol. 15:178–196. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Muramaki M, Miyake H, Terakawa T, Kumano
M, Sakai I and Fujisawa M: Expression profile of E-cadherin and
N-cadherin in non-muscle-invasive bladder cancer as a novel
predictor of intravesical recurrence following transurethral
resection. Urol Oncol. 30:161–166. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Han B, Cui D, Jing Y, Hong Y and Xia S:
Estrogen receptor β (ERβ) is a novel prognostic marker of
recurrence survival in non-muscle-invasive bladder cancer
potentially by inhibiting cadherin switch. World J Urol.
32:149–155. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhi Y, Pan J, Shen W, He P, Zheng J, Zhou
X, Lu G, Chen Z and Zhou Z: Ginkgolide B inhibits human bladder
cancer cell migration and invasion through microRNA-223-3p. Cell
Physiol Biochem. 39:1787–1794. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wan J, Zhan J, Li S, Ma J, Xu W, Liu C,
Xue X, Xie Y, Fang W, Chin YE and Zhang H: PCAF-primed EZH2
acetylation regulates its stability and promotes lung
adenocarcinoma progression. Nucleic Acids Res. 43:3591–3604. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
McConkey DJ, Choi W, Marquis L, Martin F,
Williams MB, Shah J, Svatek R, Das A, Adam L, Kamat A, et al: Role
of epithelial-to-mesenchymal transition (EMT) in drug sensitivity
and metastasis in bladder cancer. Cancer Metastasis Rev.
28:335–344. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gonzalez DM and Medici D: Signaling
mechanisms of the epithelial-mesenchymal transition. Sci Signal.
7:re82014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yun SJ and Kim WJ: Role of the
epithelial-mesenchymal transition in bladder cancer: From prognosis
to therapeutic target. Korean J Urol. 54:645–650. 2013. View Article : Google Scholar : PubMed/NCBI
|