Introduction
Breast cancer (BC) is the most commonly diagnosed,
and the second most common cause of cancer-related deaths in women
(1). There were ~2.1 million new
diagnoses of female BC cases worldwide in 2018, accounting for
almost 1 in 4 cancer deaths (2).
Triple-negative breast cancer (TNBC), one of the BC subtypes has
been revealed to be immunonegative for estrogen receptor,
progesterone receptor and human epidermal growth factor receptor-2.
TNBC is a heterogeneous type of tumor characterized by several
clinical features including its aggressive nature, higher rates of
relapse and shorter overall survival in comparison with other
subtypes of BC (3). In addition, TNBC
more often metastasizes to the lungs, bone, liver, and brain than
any other of the subtypes of BC (4–6). Although
the biomarkers of human epidermal growth factor receptor 2,
progesterone receptor and estrogen receptor have provided
significant prognostic value for TNBC patients (7), development of new biomarkers is still
necessary to obtain valuable information concerning diagnosis and
treatment, and to further exploit effective therapeutic strategies
and improve survival.
MicroRNAs (miRNAs), composed of 19–24 nucleotides,
play crucial roles in a variety of biological processes, including
cell proliferation, differentiation, migration, invasion, and
apoptosis (8–10). Dysregulation of miRNA expression often
appears in many cancers such as lung cancer, BC and cervical
cancer, and is directly associated with tumor initiation,
progression, and metastases (11–14). Human
miR-126-3p is an endothelial-specific miRNA, encoding gene located
in the intron of the EGFL7 gene at chromosome 9q34.3. This
miRNA plays an important role during embryonic development and in
angiogenesis (15). Previously it was
revealed that the expression level of miR-126-3p in many aggressive
tumors was significantly downregulated, such as osteosarcoma,
colorectal cancer, prostate cancer as well as in other cancers
(16–20), suggesting miR-126-3p to be a novel
biomarker for cancer treatment.
Blood supply supports tumor growth and distant
dissemination. It has caused the widespread concern of how
angiogenesis occurs and functions, since vasculogenic mimicry (VM)
was firstly demonstrated as a phenomenon of highly aggressive and
metastatic melanoma cells forming highly patterned vascular
channels in vitro in 1999. VM could be composed of a
basement membrane in the absence of endothelial cells and
fibroblasts (21), supported further
by numerous underlying molecular pathway studies (22). Among these mechanisms, miRNAs may be
involved. For example, vascular integrity may be affected by
miR-126-3p, resulting in a disorganized tumor vasculature (23). Increasing evidence has indicated that
the alternative microcirculation pathway was established by
vasculogenic mimicry (VM) channels which exert a fatal effect in
the early growth, metastasis and relapse of invasive BC (24–26).
Hence, in the present study, the relationship between miR-126 and
vascular mimicry was investigated, which may provide the
theoretical basis for anti-vascular therapy in TNBC.
As a G-protein signaling regulator, regulator of
G-protein signaling (RGS3) is characterized by a homologous
sequence of a core RGS domain and switches off receptor-activated
cellular signal transduction (27,28). RGS3
is associated with tumor cell proliferation and migration (29,30). In
addition, ingenuity pathway analysis indicates that the RGS3 mRNA
interacts with miR-126 (31,32). Previous studies further revealed that
there is a direct 3′-untranslated region binding site between RGS3
mRNA and miRNA-126, indicating that overexpressed RGS3 regulated by
miRNA-126 through post-transcriptional modulation, is significantly
associated with a poor prognosis of cancer patients (33).
Therefore, to explore new biomarkers for TNBC
treatment, miR-126 expression in human MDA-MB-231 and HCC1937 cell
lines was firstly investigated. Then, the effect of miR-126
expression on proliferation, migration, invasion, and angiogenesis
of human TNBC was determined. Subsequently, RGS3 as the potential
target gene of miR-126 was analyzed and validated by bioinformatics
and luciferase activity assay. Finally, how miR-126-3p regulates
target-gene expression was also investigated and the underlying
mechanisms of TNBC progression were elucidated.
Materials and methods
Cell culture
The TNBC cell lines, MDA-MB-231 and HCC1937, and
normal breast cell line MCF-10A were obtained from the Cell
Resource Center of the Shanghai Institutes for Biological Sciences.
The base medium used in the present study was L-15 Medium (cat. no.
JCML1514500; Nalgene) for MDA-MB-231 cells, RPMI-1640 (Invitrogen;
Thermo Fisher Scientific, Inc.) for HCC1937 cells, and the complete
growth medium was supplemented with 10% fetal bovine serum, 1%
penicillin (100 U/ml) and 1% streptomycin (100 U/ml). MCF-10A cells
were cultured in complete growth medium consisting of 1:1 mixture
of Dulbecco's modified Eagle's medium and Ham's F12 medium
supplemented with 5% (v/v) horse serum, 10 µg/ml insulin, 100 ng/ml
cholera toxin, 20 ng/ml recombinant human epidermal growth factor,
0.5 µg/ml hydrocortisone and 1 unit/ml penicillin/streptomycin. The
cells were all incubated in a humidified incubator at 37°C with 5%
CO2.
Cell transfection
miR-126-3p mimic, miR-126-3p inhibitor, negative
control of miR-126-3p (miRNA-NC) which is defined as the cells
transfected with the plasmids without miR-126-3p mimic or
inhibitor, and RGS3-specific small interfering RNA (siRNA) were all
synthesized by GenePharma Co., Ltd. According to the manufacturer's
protocols, Lipofectamine 2000 (cat. no. 52887; Invitrogen; Thermo
Fisher Scientific, Inc.) was used to transfect the synthesized
miRNAs aforementioned or expression-ready RGS3 ORF construct and NC
construct into the investigated MDA-MB-231 and HCC1937 cells. After
48 h of transfection, the cells were collected for the following
experiments.
Cell counting kit-8 (CCK-8) assay
Cell proliferation was assessed by a Cell Counting
Kit-8 (Dojindo Molecular Technologies). Cells were seeded at a
density of 1×105 cells/well to a 96-well plate.
Forty-eight hours after transfection, CCK-8 fluids were added to
the cells with a final concentration of 10% and maintained at 37°C
for 4 h. The optical density was assessed at 450 nm using a
Microplate Reader (Bio-Rad Laboratories, Inc.).
Migration and invasion assays
The Transwell assay kit (product no. 356234; Corning
Incorporated) was used to evaluate the invasion of the transfected
cells. These cells supplemented with DMEM were seeded to the upper
chamber. DMEM supplemented with FBS was added to the lower chamber.
The cells were maintained at 37°C for 48 h and were then fixed by
methanol. Non-migrated cells on the upper surface of the membrane
were removed by a cotton swab. The migrated cells on the lower side
of the membrane were stained using 0.1% crystal violet for 5 min at
room temperature, and imaged. ImageJ v1.52 software (National
Institutes of Health) was used to count the number of invading
cells. The invasion assay was performed in accordance with
migration assay except that the Transwell membrane was pre-coated
with Matrigel (BD Biosciences).
Colony formation
Cells transfected with synthetic miRNAs were seeded
to a 6-well plate at a density of 500 cells/well and incubated in
DMEM with 10% FBS at 37°C for 2 weeks. Then, the cells were fixed
with methanol, washed with PBS and then stained with 0.1% crystal
violet solution (Beyotime Institute of Biotechnology). The number
of colonies with >50 cells were counted.
VM formation assays
To assess the effect of miR-126-3p and RGS3 on the
VM of TNBC cells, 2×104 transfected cells overexpressing
plasmid or a control Matrigel (100 µl) were placed on a 48-well
plate, and incubated at 37°C. The number of tubes (complete
circular structures) in each well were captured and counted using
an inverted microscope (Nikon Corp.). The three readings of each
well were averaged to be the final reading.
Quantitative real-time PCR
(qRT-PCR)
Total RNA was extracted from the cultured cells
using Trizol reagent (Invitrogen; Thermo Fisher Scientific, Inc.)
and quantified using Ultra-micro UV analyzer Q6000UV (Quawell
Technology, Inc.). The sequence of stem-loop structures used in the
present study was:
5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCGCATTAT-3′. The sequence of
microRNA-126-3p was 5′-CGTACCGTGAGTAATAATGCG-3′. The underlined
part was the combined part of the stem-loop and target sequence.
Thus, the first cDNA strand specific to miRNA-126-3p was
synthesized from total RNA using Bestar™ qPCR RT Kit (cat. no.
2220; DBI Bioscience) on a PCR Amplifier (Product no. K960;
Hangzhou Jingge Scientific Instrument Co., Ltd.). Real-time PCR was
conducted using Stratagene Mx3000P (Agilent Technologies, Inc.) and
by applying Bestar™ qPCR Master Mix (cat. no. 2043; DBI
Bioscience). U6 snRNA was used as an endogenous control and
analyzed with the 2−ΔΔCq method (34). The primers are listed in Table I.
 | Table I.Primer sequences used in RT-qPCR. |
Table I.
Primer sequences used in RT-qPCR.
| Item | Sequences
(5′-3′) |
|---|
| GAPDH F |
TGTTCGTCATGGGTGTGAAC |
| GAPDH R |
ATGGCATGGACTGTGGTCAT |
| U6 F |
CTCGCTTCGGCAGCACA |
| U6 R |
AACGCTTCACGAATTTGCGT |
| All R |
CTCAACTGGTGTCGTGGA |
| hsa-miR-126-3p
RT |
CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCGCATTAT |
| hsa-miR-126-3p
F |
ACACTCCAGCTGGGTCGTACCGTGAGTAATAA |
| miR-126-3p NC |
CAGUACUUUUGUGUAGUACAA |
| miR-126-3p
mimic |
UCGUACCGUGAGUAAUAAUGCG |
| miR-126-3p
inhibitor |
CGCAUUAUUACUCACGGUACGA |
| RGS3 F |
TCAGAGGAAGCCCTCAAGTG |
| RGS3 R |
TGGATGCCATCTTGGACTGT |
Dual luciferase activity assay
The targets of miR-126-3p were predicted by
bioinformatics analysis using algorithm TargetScan (http://www.targetscan.org/vert_72/), and the
TargetScan score was investigated for has-miR-126-3p binding to
RGS3 in miRDB (http://mirdb.org/cgi-bin/search.cgi). Then, the wild
3′UTR fragment of RGS3 carrying the putative miR-126-binding site
was amplified and sub-cloned into the psiCHECK-2 vector (Bioneer
Co., Ltd.) to construct pMiR-wild-type (pMiR-WT). The isolated
plasmid was sequenced and mutated using Directed Mutagenesis System
(Invitrogen; Thermo Fisher scientific, Inc.), to produce
pMiR-mutant-plasmid (pMiR-Mut) with seven point mutations: CTGCCC
C(CtoA) G(GtoT) G(GtoA) T(TtoC) A(AtoT) C(CtoG) G(GtoA) AGGGGGC.
The sites of the mutation in RGS3 sequence are C in 152438, G in
152439, G in 152440, T in 152441, A in 152442, C in 152443 and G in
152444, which is based on the sequence provided in the NCBI GenBank
(https://www.ncbi.nlm.nih.gov/nuccore/NG_029512.1?from=4999&to=158013&report=genbank).
Sunsequently, the manufacturer's instructions were followed and
Lipofectamine 2000 (cat. no. 52887; Invitrogen; Thermo fisher
Scientific, Inc.) was used to transfect plasmids containing the
3′UTR fragment (pMiR-WT) or mutant (pMiR-Mut) respectively into
MDB-MA-231 and HCC1937 cells to express miRNA-NC or pcDNA3.0-NC or
miR-126-3p mimic. Forty-eight hours after transfection, the cells
were lysed and a reporter assay was performed using the
Dual-luciferase assay system (Promega Corp.).
Statistical analysis
All the experiments were repeated three times. The
results were expressed as the mean ± standard deviation (SD). A
Student's t-test was used to analyze the differences between two
groups. One-way analysis of variance and Tukey's post hoc test were
used to analyze the differences among three or more groups.
GraphPad Prism 6 (GraphPad Software, Inc.) was used for data
analysis. P<0.05 was considered to indicate a statistically
significant difference.
Results
Overexpression of miR-126-3p inhibits
proliferation, migration, invasion, and angiogenesis of TNBC cell
lines whereas silencing of miR-126-3p promotes these processes
The endogenous expression of miR-126-3p in MCF-10A,
MDA-MB-231 and HCC1937 cells are presented in Fig. 1A, indicating that the expression of
miR-126-3p in TNBC cells was significantly lower than that in
normal breast cells (P<0.001, respectively). To investigate the
effect of miR-126-3p on human TNBC cells, the MDA-MB-231 and
HCC1937 cells were transfected with miR-126-3p mimic or miR-126-3p
inhibitor. Non-transfected cells were used as the control group and
cells transfected with miRNA-NC were used as the negative control.
The expression of miR-126-3p in the miR-126-3p mimic group was
increased by 2.2-fold in MDA-MB-231 cells and 2.4-fold in HCC1937
cells, respectively, compared with that in the miRNA-NC groups
(P<0.001). The expression level of miR-126-3p in miR-126-3p
inhibitor group was decreased by 2.3-fold in MDA-MB-231 cells and
3.8-fold in HCC1937 cells respectively compared with that in
miRNA-NC group (P<0.001), indicating that overexpressing or
silencing of miR-126-3p were achieved by transfection of miR-126-3p
mimic or inhibitor respectively (Fig.
1A). The viability of MDA-MB-231 and HCC1937 cells was
significantly increased at the 72-h time-point by miR-126-3p
inhibition compared with that in the miRNA-NC group (2.47±0.09 vs.
1.57±0.03 in MDA-MB-231 cells and 3.66±0.05 vs. 2.81±0.05 in
HCC1937 cells, P<0.001, normalized to levels at 24 h,
respectively), and was decreased at the 72-h time-point by
miR-126-3p overexpression compared with that in the miRNA-NC group
(1.28±0.05 vs. 1.57±0.03 in MDA-MB-231 cells and 2.22±0.06 vs.
2.81±0.05 in HCC1937 cells, P<0.05, normalized to levels at 24
h, respectively, Fig. 1B). The
migration of MDA-MB-231 and HCC1937 cells was significantly
enhanced by miR-126-3p inhibition compared with that in the
miRNA-NC group (138.33±1.53 vs. 68±6.56 in MDA-MB-231 cells and
81.67±1.53 vs. 40.33±2.52 in HCC1937 cells, P<0.001,
respectively), and was reduced by miR-126-3p overexpression
compared with that in the miRNA-NC group (37.33±4.04 vs. 68±6.56 in
MDA-MB-231 cells and 14.33±1.53 vs. 40.33±2.52 in HCC1937 cells,
P<0.001, respectively, Fig. 1C and
D). Similarly, MDA-MB-231 cell invasion was significantly
enhanced by silencing of miR-126-3p compared with that in the
miRNA-NC group (132±2 vs. 72.67±5.69, P<0.001), and was
decreased by overexpression of miR-126-3p compared with that in the
miRNA-NC group (32±1.73 vs. 72.67±5.69, P<0.001, Fig. 1C and E). The number of colonies
exhibited a 50% reduction in MDA-MB-231 cells overexpressing
miR-126-3p and a 36% enhancement in MDA-MB-231 cells with
miR-126-3p silencing when compared to those in the miRNA-NC
respectively (63.67±4.04 and 172.67±8.50 vs. 126.67±5.86,
P<0.001, respectively, Fig. 1C and
F). The VM assay revealed that tube formation was reduced 70%
in the group of MDA-MB-231 cells overexpressing miR-126-3p when
compared to that in the miRNA-NC group (P<0.001) while it was
increased 3.7-fold in MDA-MB-231 cells with miR-126-3p silencing
when compared to the miRNA-NC group (P<0.001) (Fig. 1G and H), indicating that tube
formation capacity was inhibited by the miR-126-3p mimic and
enhanced by the miR-126-3p inhibitor. There was no difference
between the control group and miRNA-NC group in terms of cell
viability, migration, invasion, colony formation, and tube
formation.
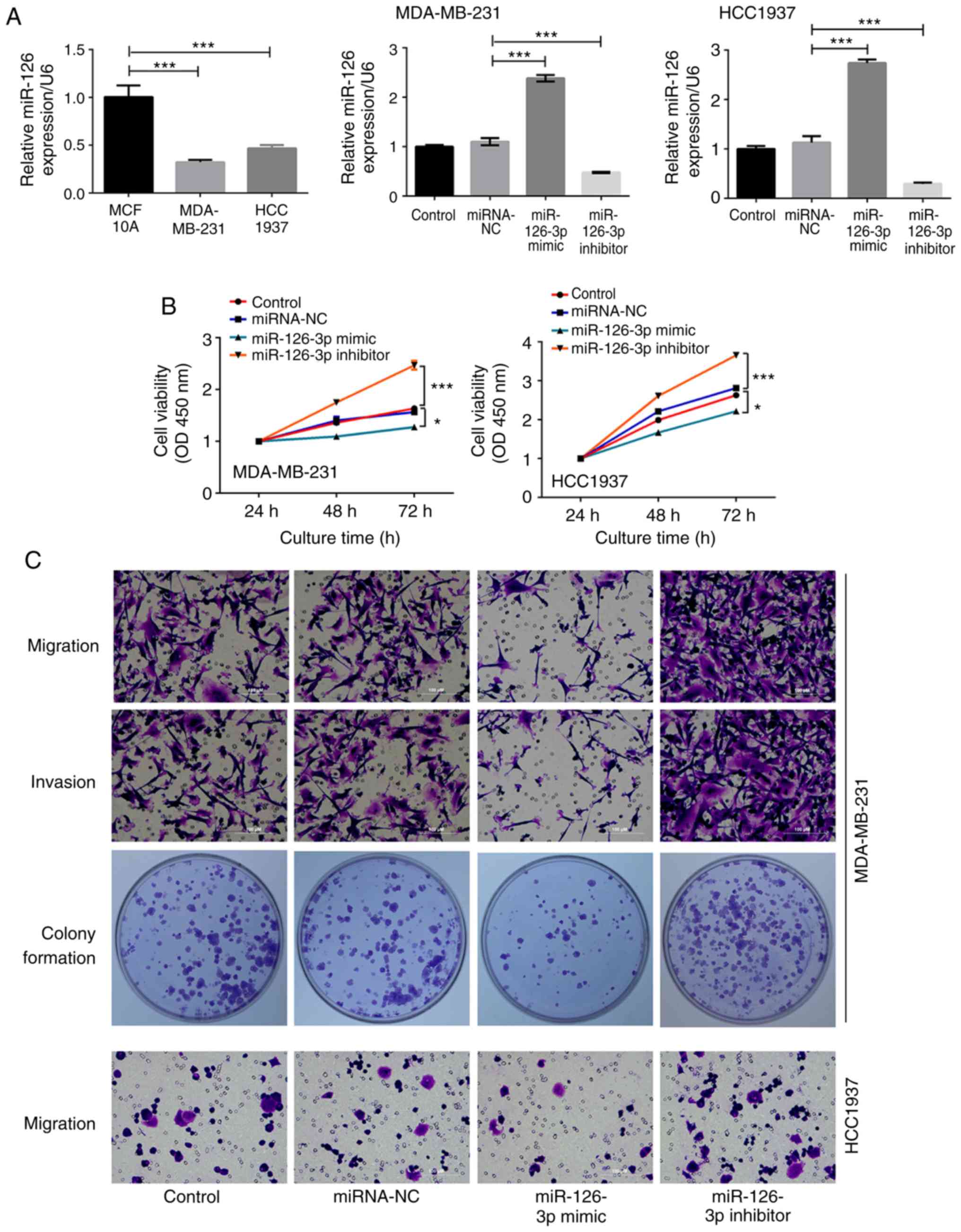 | Figure 1.miR-126-3p regulates MDA-MB-231 and
HCC1937 cell activities. (A) The expression of miR-126 in MCF-10A,
MDA-MB-231, HCC1937 cell lines, and in the control, miRNA-NC,
miR-126 mimic and miR-126 inhibitor groups of MDA-MB-231 and
HCC1937 cells were assessed by qRT-PCR. (B) The cell proliferation
in MDA-MB-231 and HCC1937 cell lines. (C) Images of cell migration,
invasion, and colony formation in MDA-MB-231 and HCC1937 cell
lines. Data presented were the averaged mean of at least three
independent experiments. Error bars indicate the standard
deviation. *P<0.05, and ***P<0.001. miR-126-3p regulates
MDA-MB-231 and HCC1937 cell activities. (D) The cell migration in
MDA-MB-231 and HCC1937 cell lines. (E) The cell invasion in the
MDA-MB-231 cell line. (F) The colony formation number in the
MDA-MB-231 cell line. (G) The VM in the control, miRNA-NC, miR-126
mimic, and miR-126 inhibitor groups of MDA-MB-231 cells are
presented (magnification, ×100). (H) The relative tube formation in
the MDA-MB-231 cell line were assessed after miRNA transfection.
Data presented were the averaged mean of at least three independent
experiments. Error bars indicate the standard deviation.
*P<0.05, and ***P<0.001. VM, vasculogenic mimicry. |
RGS3 is a target gene of
miR-126-3p
The TargetScan algorithm predicted that miR-126-3p
binded to 3′UTR of RGS3, and the TargetScan score was 67 for
has-miR-126-3p binding to RGS3 in miRDB. The DNA sequencing result
of pRGS3-Mut finally confirmed the same mutation sites as human
pRGS3-WT from the GenBank sequence, which validated the recombinant
pcDNA3.0-RGS3 plasmid. Therefore, the pRGS3-WT or pRGS3-Mut with
mimic control or miR-126-3p mimic were co-transfected into
MDA-MB-231 and HCC1937 cells, and luciferase activity was detected.
Luciferase activity in cells transfected with pRGS3-WT and
miR-126-3p mimic was reduced 2.2-fold compared with that in cells
transfected with pRGS3-WT and mimic control (P<0.001) (Fig. S1), however, the difference between
co-transfection with pRGS3-Mut and the miR-126-3p mimic was not
significant.
Ectopic expression of RGS3 affects
cell proliferation, migration, invasion, and angiogenesis
The endogenous expression of RGS3 in MCF-10A,
MDA-MB-231 and HCC1937 cells presented in Fig. 2A, revealed that the expression of RGS3
in TNBC cells was significantly higher than that in normal breast
cells (P<0.001, respectively). pcDNA3.0-NC, pcDNA3.0-RGS3 alone
or accompanied by si-RGS3 were respectively transfected into
MDA-MB-231 and HCC1937 cells. Non-transfected cells were used as
the control group. The expression of RGS3 in the pcDNA3.0-RGS3
group was increased by 3.1-fold in MDA-MB-231 cells and 3.4-fold in
HCC1937 cells compared with that in the pcDNA3.0-NC groups
(P<0.001, respectively). The expression level of RGS3 in the
pcDNA3.0+si-RGS3 group was decreased by 2.8-fold in MDA-MB-231
cells and 3.6-fold in HCC1937 cells compared with that in the
pcDNA3.0-NC group (P<0.001, respectively), indicating that
overexpression or silencing of RGS3 were achieved by transfection
of pcDNA3.0-RGS3 or pcDNA3.0+si-RGS3, respectively (Fig. 2A). The viability of MDA-MB-231 cells
and HCC1937 cells was significanlty increased at the 72-h
time-point in the transfected pcDNA3.0-RGS3 group compared with
that in the pcDNA3.0-NC group (2.52±0.05 vs. 1.74±0.05 in
MDA-MB-231 cells and 3.80±0.03 vs. 2.35±0.04 in HCC1937 cells,
P<0.001, normalized to levels at 24 h, respectively), and was
decreased at the 72-h time-point in the transfected
pcDNA3.0+si-RGS3 group compared with that in the pcDNA3.0-NC group
(1.42±0.07 vs. 1.74±0.05 in MDA-MB-231 cells and 1.66±0.05 vs.
2.35±0.04 in HCC1937 cells, P<0.05, normalized to levels at 24
h, respectively, Fig. 2B). In
Fig. 2C-E, migration, and invasion of
the MDA-MB-231 cell transfected by pcDNA3.0-RGS3 were enhanced
compared with those in the pcDNA3.0-NC group (100 and 77% increase
in cell migration and invasion, respectively; P<0.001). In
addition, MDA-MB-231 cell migration and invasion were attenuated by
pcDNA3.0+si-RGS3 in comparison with the pcDNA3.0-NC group (45% and
52% decline in cell migration and invasion, respectively;
P<0.001). The number of colonies presented a 37% increase in the
pcDNA3.0-RGS3 MDA-MB-231 cells and a 44% abatement in the
pcDNA3.0+si-RGS3 MDA-MB-231 cells when compared to pcDNA3.0-NC
respectively (P<0.001, Fig. 2C and
F). Tube formation presented in the VM assay was increased
3.2-fold in the pcDNA3.0-RGS3 MDA-MB-231 cells when compared to
that in the pcDNA3.0-NC group (P<0.001), while it was reduced
61% in the si-RGS3 MDA-MB-231 cells when compared to the
pcDNA3.0-NC group (P<0.05) (Fig. 2G
and H). There was no difference between the control group and
the pcDNA3.0-NC group regarding cell viability, migration,
invasion, colony formation, and tube formation.
 | Figure 2.Ectopic expression of RGS3 affects
miR-126-induced regulation of MDA-MB-231 cells activities. (A) The
expression of RGS in MCF-10A, MDA-MB-231, HCC1937 cell lines, and
in the control, miRNA-NC, miR-126 mimic and miR-126 inhibitor
groups of MDA-MB-231 and HCC1937 cells were assessed by qRT-PCR.
(B) The cell proliferation in MDA-MB-231 and HCC1937 cell lines.
(C) Images of cell migration, invasion, and colony formation in the
MDA-MB-231 cell line. Data presented were the averaged mean of at
least three independent experiments. Error bars indicate the
standard deviation. *P<0.05 and ***P<0.001. Ectopic
expression of RGS3 affects miR-126-induced regulation of MDA-MB-231
cells activities. (D) The cell migration, (E) the cell invasion,
(F) the colony formation number, in the control, pcDNA3.0-NC,
pcDNA3.0-RGS3, and pcDNA3.0+si-RGS3 groups of MDA-MB-231 cells are
presented. (G) The VM in the control, pcDNA3.0-NC, pcDNA3.0-RGS3,
and pcDNA3.0+si-RGS3 groups of MDA-MB-231 cells are presented
(magnification, ×100). (H) The relative tube formation in the
MDA-MB-231 cell line transfected with pcDNA3.0-NC, pcDNA3.0-RGS3
alone or combined with RGS3-specific si-RGS3 were assessed.
*P<0.05 and ***P<0.001. RGS3, regulator of G-protein
signaling 3; VM, vasculogenic mimicry. |
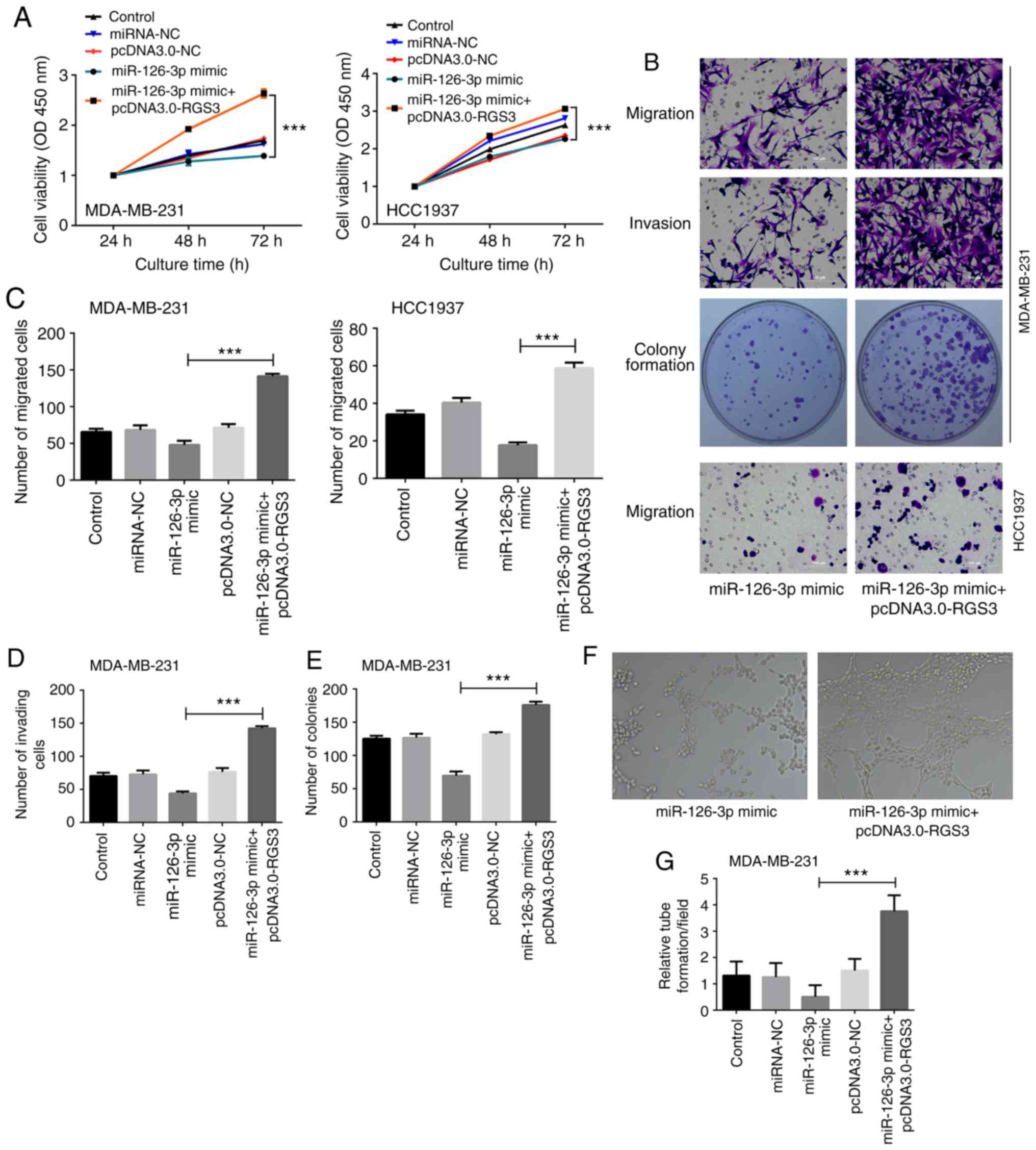
Restoration of RGS3 reverses the
inhibitory effects of miR-126-3p in TNBC cells
Two groups of miR-126-3p mimic alone or with
pcDNA3.0-RGS3 were set up to further validate whether the
restoration of RGS3 can reverse the inhibitory effects of
miR-126-3p in TNBC cells. Results in Fig.
3A-D revealed that the viability, migration, and invasion of
MDA-MB-231 cells were all increased by 1.5- (at the 72-h,
time-point normalized to the levels at 24 h), 3.0- and 3.2-fold
respectively by co-transfection of miR-126-3p mimic and
pcDNA3.0-RGS3 compared with cells only transfected with miR-126-3p
mimic (P<0.001). Similarly, the viability and migration of
HCC1937 cells were all increased by 1.4- (at the 72-h time-point,
normalized to levels at 24 h) and 3.3-fold respectively by
co-transfection of miR-126-3p mimic and pcDNA3.0-RGS3 compared with
cells only transfected with miR-126-3p mimic (P<0.001).
Moreover, there was a 1.5-fold enhancement of the number of
colonies in MDA-MB-231 cells co-transfected with miR-126-3p mimic
and pcDNA3.0-RGS3 when compared to that in the cells transfected
with the miR-126-3p mimic (P<0.001, Fig. 3B and E). In the VM assay, tube
formation was increased by 6.5-fold in the MDA-MB-231 cells
co-transfected with the miR-126-3p mimic and pcDNA3.0-RGS3 compared
with that in the cells transfected with the miR-126-3p mimic
(P<0.001, Fig. 3F-G).
Discussion
Increasing evidence has revealed that specific
microRNA clusters may play an important role in the biology of
TNBC, and may be of clinical relevance in TNBC, knowledge which
should assist disease prognostication and therapy (35). The present study revealed that
miR-126-3p was decreased in TNBC cells and miR-126-3p
overexpression inhibited cell proliferation, migration, and
invasion. RGS3 was predicted to be a target gene of miR-126-3p by
the TargetScan algorithm and validated by luciferase activity
assay. Furthermore, RGS3 silencing reversed the effect of
miR-126-3p knockdown. Finally, it was determined that miR-126-3p
knockdown activated VM, which could be reversed by silencing of
RGS3.
Multiple miRNAs are associated with the metastasis
and prognosis of BC (36–39). For instance, overexpression of
miR-30c-2-3p inhibited the migration and invasion of BC cells,
while low or underexpression of miR-30c-2-3p was correlated with
poor overall survival (40). An
increasing number of studies has confirmed that miRNA expression
profiles are dysregulated in TNBC patients compared to healthy
controls. For example, miR-190a, miR-136-5p, miR-126-5p,
miR-135b-5p and miR-182-5p may be associated with the development
and progression of TNBC (41).
Although miR-126-3p has been reported as an oncogenic miRNA,
promoting proliferation in gastric cancer cells (42), it has also been revealed to be a tumor
suppressor due to its inhibition in the growth of colon cancer cell
lines, HEK293 and MCF-7 cells by targeting p85b and IRS-1,
respectively (43,44). However, in a study of miRNA expression
profiles of 24 triple-negative breast cancers and 14 adjacent
normal tissues using deep sequencing technology, miR-126-3p was
revealed to be significantly downregulated (P<0.05; fold change
1.8–2.6) in TNBC (45). Additionally,
restoring miR-126-3p in MDA-MB-231 cells reduced cell proliferation
and the levels of miR-126-3p in tumor tissues predicted TNBC
outcomes (46). Novel prognostic and
predictive miRNA targets of TNBC, include a miRNA signature that
predicts patient response to anthracycline-based chemotherapy,
which may improve clinical management and/or lead to the
development of novel therapies (47).
Notably, it was surmised that miR-126-3p acts as a tumor-suppressor
miRNA in TNBC cancer cells in the present study, which further
extends the current knowledge of microRNA regulatory
characteristics and network in TNBC.
Notably, previous results indicated that a
combination treatment with miRNAs, in particular miR-126-3p,
enhanced the activity of specific BC drugs in vitro, even in
the most aggressive BC subtype, TNBC (48). In addition, whole transcriptome
studies, coupled with Gene Ontology and pathway analysis revealed a
notable landscape for miR-126-3p activity in BC, such as
involvement in cell cycle regulation of BC, in particular the M
phase (48). Whether miR-126-3p
overexpression affects apoptosis and the cell cycle in TNBC as well
as whether a combination treatment with miR-126-3p affects TNBC
cell function are our next goals in future research.
RGS3 was predicted to be a potential target of
miR-126-3p by TargetScan algorithm due to their complementary
binding which was further validated through a luciferase activity
reporter assay. Notably, it was revealed that targeting and
combination of transfected exogenous RGS3, which lacks the 3′UTR
region for miR-126-3p, could revert the inhibitory effects of
miR-126-3p on TNBC cell behaviors, indicating that RGS3 was
directly involved in miR-126-3p-induced inhibition of cell
behaviors.
VM is important for disease progression of many
aggressive tumors such as hepatocellular carcinoma, as well as
prostate, ovarian and lung cancer (49–52). VM
has been reported to be related to the metastasis and poor
prognosis of TNBC (24,53–54). The
present study revealed that angiogenesis could be induced by
miR-126-3p silencing and suppressed by miR-126-3p overexpression,
while it was promoted by overexpression of RGS3 and inhibited by
RGS3 silencing. Moreover, the overexpression of RGS3 could recover
the inhibitory effect of miR-126-3p on VM of TNBC cells, indicating
that RGS3 directly contributed to miR-126-3p-induced VM inhibition
processes which provides a new target for anti-angiogenesis
therapy. Therefore, it was deduced that miR-126-3p/RGS3 has the
potential to be a new biomarker for TNBC treatment.
Collectively, the tumor suppressive role of the
miR-126-3p/RGS3 axis on cell proliferation, colony formation,
migration, invasion, and VM in TNBC was verified, which may be a
potential target for cancer therapy treatment.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present research was supported by the Youth
Scientific Research Project of Fujian Provincial Health and Family
Planning Commission, PRC (no. 2016-1-91).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
ZH performed the majority of the study and wrote the
manuscript. CH conducted the vasculogenic mimicry formation assays
and data analysis and BM performed the dual luciferase activity
assay and data analysis. QW, XZ and LL helped with the cell
function assays and data analysis. CW and DC designed the
experiments and edited the manuscript. All authors read and
approved the final manuscript and agree to be accountable for all
aspects of the research in ensuring that the accuracy or integrity
of any part of the work are appropriately investigated and
resolved.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global Cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yuan N, Meng M, Liu C, Feng L, Hou L, Ning
Q, Xin G, Pei L, Gu S, Li X and Zhao X: Clinical characteristics
and prognostic analysis of triple-negative breast cancer patients.
Mol Clin Oncol. 2:245–251. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lehmann BD, Bauer JA, Chen X, Sanders ME,
Chakravarthy AB, Shyr Y and Pietenpol JA: Identification of human
triple-negative breast cancer subtypes and preclinical models for
selection of targeted therapies. J Clin Invest. 121:2750̄–2767.
2011. View
Article : Google Scholar
|
|
5
|
Gerratana L, Fanotto V, Bonotto M,
Bolzonello S, Minisini AM, Fasola G and Puglisi F: Pattern of
metastasis and outcome in patients with breast cancer. Clin Exp
Metastasis. 32:125–133. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tsai CH, Chiu JH, Yang CW, Wang JY, Tsai
YF, Tseng LM, Chen WS and Shyr YM: Molecular characteristics of
recurrent triple-negative breast cancer. Mol Med Rep. 12:7326–7334.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
M Braden A, V Stankowski R, M Engel J and
A Onitilo A: Breast cancer biomarkers: Risk assessment, diagnosis,
prognosis, prediction of treatment efficacy and toxicity, and
recurrence. Curr Pharm Des. 20:4879–4898. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yang C, Hou C, Zhang H, Wang D, Ma Y,
Zhang Y, Xu X, Bi Z and Geng S: miR-126 functions as a tumor
suppressor in osteosarcoma by targeting Sox2. Int J Mol Sci.
15:423–437. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jones KB, Salah Z, Del Mare S, Galasso M,
Gaudio E, Nuovo GJ, Lovat F, LeBlanc K, Palatini J, Randall RL, et
al: miRNA signatures associate with pathogenesis and progression of
osteosarcoma. Cancer Res. 72:1865–1877. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Najafi Z, Sharifi M and Javadi G:
Degradation of miR-21 induces apoptosis and inhibits cell
proliferation in human hepatocellular carcinoma. Cancer Gene Ther.
22:530–535. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sandhu R, Rivenbark AG, Mackler RM, Livasy
CA and Coleman WB: Dysregulation of microRNA expression drives
aberrant DNA hypermethylation in basal-like breast cancer. Int J
Oncol. 44:563–572. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhu X, Li H, Long L, Hui L, Chen H, Wang
X, Shen H and Xu W: miR-126 enhances the sensitivity of non-small
cell lung cancer cells to anticancer agents by targeting vascular
endothelial growth factor A. Acta Biochim Biophys Sin (Shanghai).
44:519–526. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yu Q, Liu SL, Wang H, Shi G, Yang P and
Chen X: miR-126 suppresses the proliferation of cervical cancer
cells and alters cell sensitivity to the chemotherapeutic drug
bleomycin. Asian Pac J Cancer Prev. 14:6569–6572. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Akao Y, Noguchi S, Iio A, Kojima K, Takagi
T and Naoe T: Dysregulation of microRNA-34a expression causes
drug-resistance to 5-FU in human colon cancer DLD-1 cells. Cancer
Lett. 300:197–204. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kuhnert F, Mancuso MR, Hampton J,
Stankunas K, Asano T, Chen C and Kuo CJ: Attribution of vascular
phenotypes of the murine Egfl7 locus to the microRNA miR-126.
Development. 135:3989–3993. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liu W, Zhao ZY, Shi L and Yuan WD: Tissue
microRNA-126 expression level predicts outcome in human
osteosarcoma. Diagn Pathol. 10:1162015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hansen TF, Nielsen BS, Jakobsen A and
Sørensen FB: Intra-tumoural vessel area estimated by expression of
epidermal growth factor-like domain 7 and microRNA-126 in primary
tumours and metastases of patients with colorectal cancer: A
descriptive study. J Transl Med. 13:102015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Fujii T, Shimada K, Tatsumi Y, Fujimoto K
and Konishi N: Syndecan-1 responsive microRNA-126 and 149 regulate
cell proliferation in prostate cancer. Biochem Biophys Res Commun.
456:183–189. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liu LY, Wang W, Zhao LY, Guo B, Yang J,
Zhao XG, Hou N, Ni L, Wang AY, Song TS, et al: MiR-126 inhibits
growth of SGC-7901 cells by synergistically targeting the oncogenes
PI3KR2 and Crk, and the tumor suppressor PLK2. Int J Oncol.
45:1257–1265. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yanaihara N, Caplen N, Bowman E, Seike M,
Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, et
al: Unique microRNA molecular profiles in lung cancer diagnosis and
prognosis. Cancer Cell. 9:189–198. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Maniotis AJ, Folberg R, Hess A, Seftor EA,
Gardner LM, Pe'er J, Trent JM, Meltzer PS and Hendrix MJ: Vascular
channel formation by human melanoma cells in vivo and in vitro:
Vasculogenic mimicry. Am J Pathol. 155:739–752. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kirschmann DA, Seftor EA, Hardy KM, Seftor
RE and Hendrix MJ: Molecular pathways: Vasculogenic mimicry in
tumor cells: Diagnostic and therapeutic implications. Clin Cancer
Res. 18:2726–2732. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Meister J and Schmidt MHH: miR-126 and
miR-126*: New players in cancer. ScientificWorldJournal.
10:2090–2100. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liu T, Sun B, Zhao X, Li Y, Gu Q, Dong X
and Liu F: OCT4 expression and vasculogenic mimicry formation
positively correlate with poor prognosis in human breast cancer.
Int J Mol Sci. 15:19634–19649. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu T, Sun B, Zhao X, Gu Q, Dong X, Yao Z,
Zhao N, Chi J, Liu N, Sun R and Ma Y: HER2/neu expression
correlates with vasculogenic mimicry in invasive breast carcinoma.
J Cell Mol Med. 17:116–122. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Shirakawa K, Kobayashi H, Sobajima J,
Hashimoto D, Shimizu A and Wakasugi H: Inflammatory breast cancer:
Vasculogenic mimicry and its hemodynamics of an inflammatory breast
cancer xenograft model. Breast Cancer Res. 5:136–139. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Dulin NO, Pratt P, Tiruppathi C, Niu J,
Voyno-Yasenetskaya T and Dunn MJ: Regulator of G protein signaling
RGS3T is localized to the nucleus and induces apoptosis. J Biol
Chem. 275:21317–21323. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Eusemann TN, Willmroth F, Fiebich B, Biber
K and van Calker D: Adenosine receptors differentially regulate the
expression of regulators of G-protein signalling (RGS) 2, 3 and 4
in astrocyte-like cells. PLoS One. 10:e01349342015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sethakorn N and Dulin NO: RGS expression
in cancer: Oncomining the cancer microarray data. J Recept Signal
Transduct Res. 33:166–171. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Tatenhorst L, Senner V, Püttmann S and
Paulus W: Regulators of G-protein signaling 3 and 4 (RGS3, RGS4)
are associated with glioma cell motility. J Neuropathol Exp Neurol.
63:210–222. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Feng R, Chen X, Yu Y, Su L, Yu B, Li J,
Cai Q, Yan M, Liu B and Zhu Z: miR-126 functions as a tumour
suppressor in human gastric cancer. Cancer Lett. 298:50–63. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang J, Chen X, Li P, Su L, Yu B, Cai Q,
Li J, Yu Y, Liu B and Zhu Z: CRKL promotes cell proliferation in
gastric cancer and is negatively regulated by miR-126. Chem Biol
Interact. 206:230–238. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wang J, Zhou Y, Fei X, Chen X and Zhu Z:
Regulator of G-protein signaling 3 targeted by miR-126 correlates
with poor prognosis in gastric cancer patients. Anticancer Drugs.
28:161–169. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Piasecka D, Braun M, Kordek R, Sadej R and
Romanska H: MicroRNAs in regulation of triple-negative breast
cancer progression. J Cancer Res Clin Oncol. 144:1401–1411. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhang L, Xu Y, Jin X, Wang Z, Wu Y, Zhao
D, Chen G, Li D, Wang X, Cao H, et al: A circulating miRNA
signature as a diagnostic biomarker for non-invasive early
detection of breast cancer. Breast Cancer Res Treat. 154:423–434.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang B, Li J, Sun M, Sun L and Zhang X:
miRNA expression in breast cancer varies with lymph node metastasis
and other clinicopathologic features. IUBMB Life. 66:371–377. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Gomes BC, Martins M, Lopes P, Morujão I,
Oliveira M, Araújo A, Rueff J and Rodrigues AS: Prognostic value of
microRNA-203a expression in breast cancer. Oncol Rep. 36:1748–1756.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wang Y, Zhang Z and Wang J: MicroRNA-384
inhibits the progression of breast cancer by targeting ACVR1. Oncol
Rep. 39:2563–2574. 2018.PubMed/NCBI
|
|
40
|
Zhang HD, Jiang LH, Hou JC, Zhou SY, Zhong
SL, Zhu LP, Wang DD, Yang SJ, He YJ, Mao CF, et al: Circular RNA
has_circ_0072995 promotes breast cancer cell migration and invasion
through sponge for miR-30c-2-3p. Epigenomics. 10:1229–1242. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Paszek S, Gabło N, Barnaś E, Szybka M,
Morawiec J, Kołacińska A and Zawlik I: Dysregulation of microRNAs
in triple-negative breast cancer. Ginekol Pol. 88:530–536. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Otsubo T, Akiyama Y, Hashimoto Y, Shimada
S, Goto K and Yuasa Y: MicroRNA-126 inhibits SOX2 expression and
contributes to gastric carcinogenesis. PLoS One. 6:e166172011.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Guo C, Sah JF, Beard L, Willson JK,
Markowitz SD and Guda K: The noncoding RNA, miR-126, suppresses the
growth of neoplastic cells by targeting phosphatidylinositol
3-kinase signaling and is frequently lost in colon cancers. Genes
Chromosomes Cancer. 47:939–946. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zhang J, Du YY, Lin YF, Chen YT, Yang L,
Wang HJ and Ma D: The cell growth suppressor, miR-126, targets
IRS-1. Biochem Biophys Res Commun. 377:136–140. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Chang YY, Kuo WH, Hung JH, Lee CY, Lee YH,
Chang YC, Lin WC, Shen CY, Huang CS, Hseih FJ, et al: Deregulated
microRNAs in triple-negative breast cancer revealed by deep
sequencing. Mol Cancer. 14:362015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Liu Y, Cai Q, Bao PP, Su Y, Cai H, Wu J,
Ye F, Guo X, Zheng W, Zheng Y and Shu XO: Tumor tissue microRNA
expression in association with triple-negative breast cancer
outcomes. Breast Cancer Res Treat. 152:183–191. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Turashvili G, Lightbody ED, Tyryshkin K,
SenGupta SK, Elliott BE, Madarnas Y, Ghaffari A, Day A and Nicol
CJB: Novel prognostic and predictive microRNA targets for
triple-negative breast cancer. FASEB J. 29:fj201800120R2018.
|
|
48
|
Baldassari F, Zerbinati C, Galasso M,
Corrà F, Minotti L, Agnoletto C, Previati M, Croce CM and Volinia
S: Screen for microRNA and drug interactions in breast cancer cell
lines points to miR-126 as a modulator of CDK4/6 and PIK3CA
inhibitors. Front Genet. 9:1742018. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Sun T, Zhao N, Zhao XL, Gu Q, Zhang SW,
Che N, Wang XH, Du J, Liu YX and Sun BC: Expression and functional
significance of Twist1 in hepatocellular carcinoma: Its role in
vasculogenic mimicry. Hepatology. 51:545–556. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Liu R, Yang K, Meng C, Zhang Z and Xu Y:
Vasculogenic mimicry is a marker of poor prognosis in prostate
cancer. Cancer Biol Ther. 13:527–533. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Wang JY, Sun T, Zhao XL, Zhang SW, Zhang
DF, Gu Q, Wang XH, Zhao N, Qie S and Sun BC: Functional
significance of VEGF-a in human ovarian carcinoma: Role in
vasculogenic mimicry. Cancer Biol Ther. 7:758–766. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Li Y, Sun B, Zhao X, Zhang D, Wang X, Zhu
D, Yang Z, Qiu Z and Ban X: Subpopulations of uPAR+ contribute to
vasculogenic mimicry and metastasis in large cell lung cancer. Exp
Mol Pathol. 98:136–144. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Zhang D, Sun B, Zhao X, Ma Y, Ji R, Gu Q,
Dong X, Li J, Liu F, Jia X, et al: Twist1 expression induced by
sunitinib accelerates tumor cell vasculogenic mimicry by increasing
the population of CD133+ cells in triple-negative breast cancer.
Mol Cancer. 13:2072014. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Wagenblast E, Soto M, Gutiérrez-Ángel S,
Hartl CA, Gable AL, Maceli AR, Erard N, Williams AM, Kim SY,
Dickopf S, et al: A model of breast cancer heterogeneity reveals
vascular mimicry as a driver of metastasis. Nature. 520:358–362.
2015. View Article : Google Scholar : PubMed/NCBI
|