Introduction
Approximately 1.3 million new cases of breast cancer
are diagnosed annually worldwide, and an estimated 450,000 patients
die from this disease each year (1).
The incidence is 3–4% per year, and patients are being diagnosed at
younger ages each year (2). In China,
nearly 200,000 women are diagnosed with breast cancer every year,
and breast cancer ranks first among the five most commonly
diagnosed malignancies in women (3).
Moreover, since 2013 it has been the fourth leading cause of
cancer-related death among women. Although the precise causes of
breast cancer remain unclear, the primary risk factors include
environmental factors, hormonal effects, and importantly, genetic
factors (4–8). Epidemiological studies have found that
5–10% of breast cancer cases are hereditary and associated with
inherited genetic mutations (9). At
present, breast cancer treatment is primarily based on the
diagnosis of clinical stage, pathological type and molecular
subtype, which includes the expression status of estrogen receptor
(ER), progesterone receptor (PR), and human epidermal growth factor
2 receptor (HER2) as well as the proliferation index Ki67 (10,11).
However, conventional clinicopathological diagnoses have limited
accuracy and specificity, making precise classification of
individual breast cancer patients difficult. The effectiveness of
specific treatments can vary among patients classified as having
breast cancer of the same clinical stage, pathological
classification and molecular subtype. Therefore, personalized and
precise treatments must be developed based on the heterogeneity of
breast cancer (12,13).
In recent years, targeted cancer therapy has
developed rapidly, resulting in the availability of precision
medicine. Basic research and clinical studies have gradually
identified and validated many gene mutations associated with
targeted cancer therapy. The study of gene mutations relies on
advancements in gene detection techniques, such as Sanger
sequencing, pyrosequencing, and real-time fluorescent polymerase
chain reaction (PCR), which typically permit detection of mutations
in individual genes or exons (14).
By comparison, next-generation sequencing (NGS) technology utilizes
massively parallel sequencing (MPS) to sequence millions or even
billions of DNA fragments in parallel, which enables the
identification of as many as hundreds of tumor-associated genes
through examination of all exons and the whole genome (15,16). NGS
has broad potential for the detection of tumor-related gene
mutations (17,18).
Ion Torrent sequencing is an NGS technology based on
semiconductor chip technology and uses the same principle of
complementary base pairing. This approach uses an ion sensor to
detect local pH changes caused by the release of hydrogen ions
(H+) during DNA polymerization. This chemically encoded
information is transferred directly into digital information;
therefore, bases are interpreted in real time, allowing sequences
of DNA fragments to be obtained rapidly. Compared to other
sequencing technologies, Ion Torrent sequencing does not require
fluorescent labeling, reducing the need for expensive optics and
offering incomparable advantages in terms of equipment costs,
sequencing speed, and sequencing costs (19–21). In
the present study, we utilized the Ion Torrent sequencing platform
and the PrimBio Breast Cancer Panel, which is a NGS gene panel
targeting breast cancer-related genes, including i) BRCA1/2,
the most studied genes in inherited breast cancer ii), other
candidate genes for inherited breast cancer, and iii)
susceptibility genes that may be sporadically mutated in breast
cancer. With coverage of 31 genes, the PrimBio Breast Cancer Panel
is the most comprehensive breast cancer gene panel available and
provides rapid target selection within formalin-fixed,
paraffin-embedded samples.
As the development and progression of breast cancer
are highly variable, drug responses can differ among various tumor
cells of the same patient. The purpose of the present study was to
obtain the multigene mutation spectra of female patients with
breast cancer in Northeast China, in order to explore the
correlation between mutations and clinicopathological
characteristics and to identify genetic mutations that might be
useful for predicting the prognosis and survival of breast cancer
patients. We used the Ion Torrent sequencing platform (PrimBio
Medical Laboratory, PrimBio Genes Biotechnology, Wuhan, China) to
detect mutations in all coding exons of 31 genes in 621 specimens
from 286 breast cancer patients, and 91 blood samples from the
enrolled patients were collected as a control group. The
harmful/pathogenic gene mutations (somatic nonsynonymous mutations)
in each tissue were identified, and their correlations with
clinicopathological data and survival outcomes were analyzed. The
ultimate purpose of this study was to identify genetic mutations
that can be used to predict the prognosis and survival of breast
cancer patients.
Materials and methods
Sample collection
All enrolled breast cancer patients were treated at
the China-Japan Union Hospital of Jilin University from October
2010 to April 2013 and did not receive any neoadjuvant therapy
before surgery. The specimens were all obtained from the hospital
tissue bank. We analyzed breast cancer tissues from 286 patients
and blood samples from 91 patients. For 72 patients, four tissue
samples were collected at the same time, including cancerous
tissue, paracancerous tissue (within 3 cm of the cancerous tissue),
normal tissue (normal tissue located more than 3 cm from the
cancerous tissue), and blood. All experiments were approved by the
Ethics Committee of the China-Japan Union Hospital of Jilin
University, and an informed consent form was signed by each
enrolled patient. The clinicopathological data for the 286 patients
enrolled in this study are presented in Table I.
 | Table I.Clinicopathological data of the 286
breast cancer patients enrolled in this study. |
Table I.
Clinicopathological data of the 286
breast cancer patients enrolled in this study.
|
Characteristics | Data n (%) |
|---|
| Age (years) |
|
|
≤35 | 21 (7.3) |
|
35-50 | 115 (40.2) |
|
>50 | 150 (52.4) |
| Anti-estrogen
therapy |
|
|
Yes | 100 (35.0) |
| No | 186 (65.0) |
| Number of
births |
|
| 0 | 8 (2.8) |
| 1 | 141 (49.3) |
| 2 | 84 (29.4) |
| ≥3 | 53 (18.5) |
| Number of
abortions |
|
| 0 | 110 (38.5) |
| 1 | 82 (28.7) |
| 2 | 66 (23.1) |
| ≥3 | 28 (9.8) |
| Tumor size |
|
| Primary
cancer | 6 (2.1) |
| ≤2
cm | 148 (51.7) |
| 2-5
cm | 114 (39.9) |
| >5
cm | 14 (4.9) |
| Skin
involvement | 4 (1.4) |
| Lymph node
metastasis |
|
| 0 | 140 (49.0) |
| ≤3 | 61 (21.3) |
|
4-9 | 40 (14.0) |
|
≥10 | 45 (15.7) |
| Type of
pathology |
|
| Ductal
carcinoma | 254 (88.9) |
| Lobular
carcinoma | 5 (1.7) |
|
Mucinous carcinoma | 6 (2.1) |
|
Hybrid | 21 (7.3) |
| Vascular
involvement |
|
|
Yes | 124 (43.4) |
| No | 162 (56.6) |
| Expression of
P53 |
|
|
Yes | 171 (59.8) |
| No | 115 (40.2) |
| Ki67 |
|
|
<14% | 103 (36.0) |
|
≥14% | 183 (64.0) |
| Molecular subtype
(except primary cancer) |
|
| Luminal
A | 76 (26.6) |
| Luminal
B | 120 (42.0) |
| Her2
overexpression | 29 (10.1) |
|
Triple-negative | 55 (19.2) |
| Clinical stage |
|
| Primary
cancer | 6 (2.1) |
| 1 | 99 (34.6) |
| 2 | 92 (32.2) |
| 3 | 81 (28.3) |
| 4 | 8 (2.8) |
| Recurrence
risk |
|
|
Low | 9 (3.1) |
|
Moderate | 168 (58.7) |
|
High | 109 (38.1) |
Sequencing and analysis
Genomic DNA was extracted from tissue or blood
samples from the breast cancer patients, and a library was
generated using the PrimBio breast cancer gene panel, which covers
the complete coding regions of 31 genes associated with breast
cancer. Samples were sequenced using an Ion Proton sequencing
platform, and the obtained sequencing results were analyzed using
Human Genome Build 19 (Hg19, The Genome Reference Consortium Human
Genome Build 37 (GRCh 37)) as a reference. Breast cancer-specific
mutations were identified using PrimBio by comparing DNA sequences
in tumor tissue to those in blood from the same patient, and then,
tumor-specific mutations were identified using StrandNGS software
(https://www.strand-ngs.com), which can
detect harmful/pathogenic mutations by comprehensively evaluating
scores predicted by the Scale-Invariant Feature Transform (SIFT),
Polyphen2, Likelihood Ratio Test (LRT), Mutation Taster, Mutation
Assessor, and Functional Analysis Through Hidden Markov Models
(FATHMM) features. After synonymous mutations were identified, all
the other mutations were defined as nonsynonymous mutations. By
examining four samples from each patient, mutations identified in
blood samples were defined as germline mutations, while those found
only in the other three tissues were defined as somatic mutations.
All somatic mutations were annotated using the Catalogue of Somatic
Mutations in Cancer (COSMIC) database (http://cancer.sanger.ac.uk/cosmic).
Data analysis and processing
Significant differences in the obtained data were
identified by one-way analysis of variance (ANOVA), and mutations
associated with the survival of breast cancer patients were
identified by Kaplan-Meier survival analysis with a log rank test.
Finally, multivariate analyses were performed using a Cox
proportional hazard regression model to identify gene mutations
independently associated with breast cancer prognosis. P<0.05
indicated statistical significance.
Results
Gene mutations identified in the
breast cancer patients
A total of 179 somatic nonsynonymous mutations in 11
harmful/pathogenic genes were detected (Table II). Approximately 54.2 and 5.6% of
the patients carried one and multiple gene mutations, respectively.
The two most frequently mutated genes were PIK3CA (39.2%)
and TP53 (12.9%). In the PIK3CA gene, 117 pathogenic
mutations (somatic nonsynonymous mutations) were detected in 112
breast cancer patients, and the distribution of these mutations is
presented in Table III.
 | Table II.Gene mutations according to the
different histologic subtypes of the breast carcinoma cases. |
Table II.
Gene mutations according to the
different histologic subtypes of the breast carcinoma cases.
| Mutation
(number) | Ductal (n=254) | Lobular (n=5) | Mucinous (n=6) | Mixed (n=21) |
|---|
| PIK3CA
(117) | 105 | 3 | 2 | 7 |
| TP53
(37) | 34 | 1 | 0 | 2 |
| AKT1
(14) | 14 | 0 | 0 | 0 |
| PTEN
(4) | 4 | 0 | 0 | 0 |
| GATA3
(1) | 0 | 0 | 0 | 1 |
| ATM (1) | 1 | 0 | 0 | 0 |
| BRCA2
(1) | 1 | 0 | 0 | 0 |
| BRCA1
(1) | 1 | 0 | 0 | 0 |
| PALB2
(1) | 1 | 0 | 0 | 0 |
| RAD51D
(1) | 1 | 0 | 0 | 0 |
| CHEK2
(1) | 1 | 0 | 0 | 0 |
 | Table III.Mutation distribution among the exons
of PIK3CA. |
Table III.
Mutation distribution among the exons
of PIK3CA.
| mut_pos | Case Freq, n (%)
N=286 | dbSNP | 1,000-genome MAF
(%) |
|---|
|
c.1258T>C:p.C420R | 1 (0.3) | rs121913272 | 0 |
|
c.1624G>A:p.E542K | 13 (4.5) | rs121913273 | 0 |
|
c.1633G>A:p.E545K | 23 (8.0) | rs104886003 | 0 |
|
c.1634A>G:p.E545G | 1 (0.3) | rs121913274 | 0 |
|
c.1635G>T:p.E545D | 2 (0.7) | rs121913275 | 0 |
|
c.1636C>A:p.Q546K | 1 (0.3) | rs121913286 | 0 |
|
c.1637A>G:p.Q546R | 1 (0.3) | rs397517201 | 0 |
|
c.3140A>G:p.H1047R/L | 75 (26.2) | rs121913279 | 0 |
Distribution of mutation frequencies
according to recurrence risk
According to the risk of recurrence, the patients
were divided into low-risk (n=9), moderate-risk (n=168), and
high-risk (n=109) groups. The mutation distributions for the three
groups are shown in Fig. 1. More
genetic mutations were detected in the moderate- and high-risk
groups, while only one mutation site in the AKT1 gene was
detected in the low-risk group.
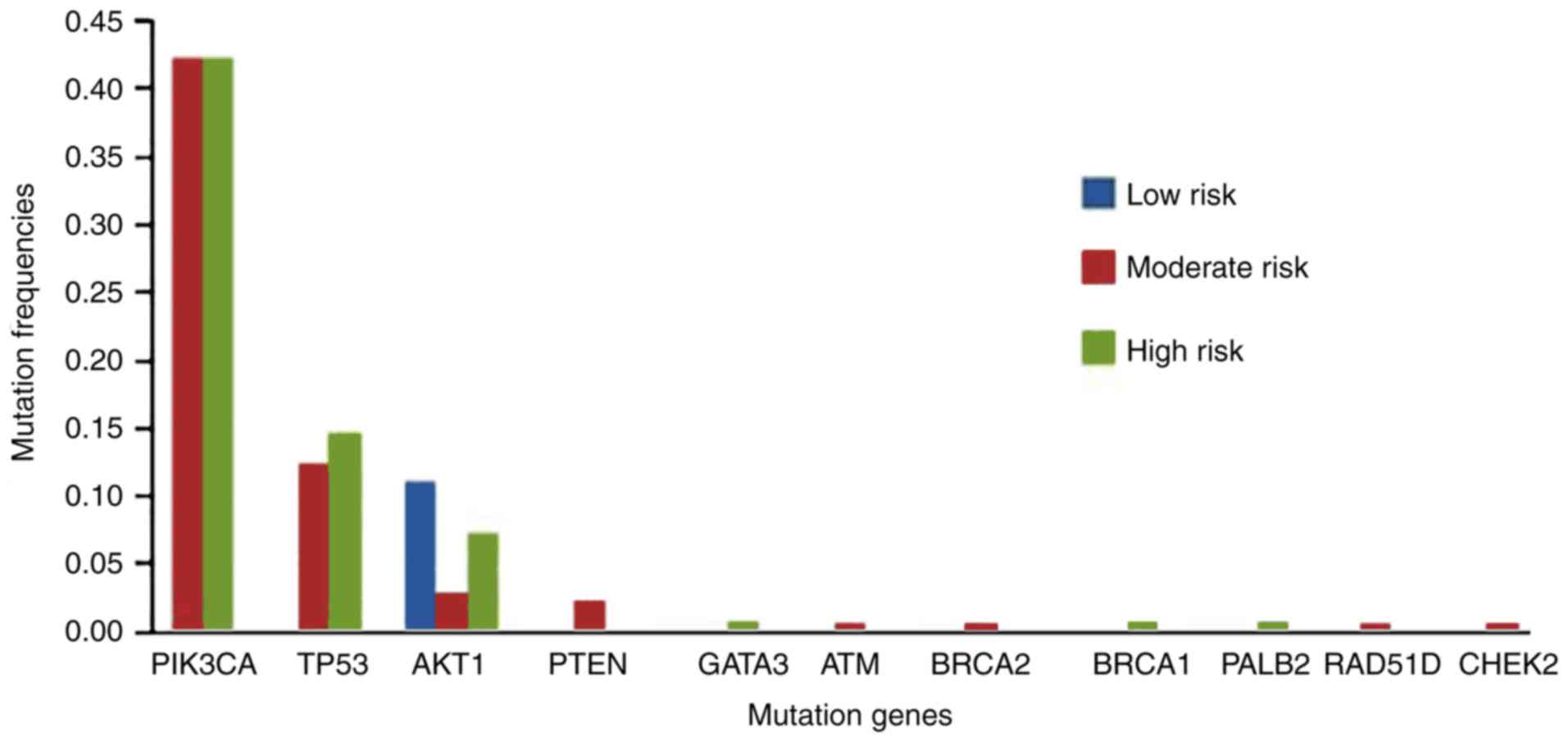 | Figure 1.Distribution of the mutation
detection rates in groups with differing risk of recurrence.
PIK3CA, phosphatidylinositol-4,5-bisphosphate 3-kinase
catalytic subunit α; TP53, tumor protein P53; AKT1,
AKT serine/threonine kinase 1; PTEN, phosphatase and tensin
homolog; GATA3, GATA binding protein 3; ATM, ATM
serine/threonine kinase; BRCA2, BRCA2 DNA repair associated;
BRCA1, BRCA1 DNA repair associated; PALB2, partner
and localizer of BRCA2; RAD51D, RAD51 paralog D;
CHEK2, checkpoint kinase 2. |
Gene mutation frequencies according to
molecular subtype
The Luminal A, Luminal B, HER2-positive, and
triple-negative subtype groups included 76, 120, 29, and 35,
respectively. As shown in Fig. 2, the
percentages of patients who carried gene mutations differed among
the molecular subtype groups. In the Luminal A subtype group
(Fig. 2A), the mutation rate of
PIK3CA was the highest at 55.3%, followed by those of
TP53 (10.5%) and AKT1 (6.6%). In the Luminal B
subtype group (Fig. 2B), the
PIK3CA mutation rate was the highest at 38.3%, followed by
those of TP53 (12.5%) and AKT1 (5.8%). In the
HER2-positive subtype group (Fig.
2C), the frequency of mutations in the PIK3CA gene was
41.4% and that in the TP53 gene was only 6.9%. In the
triple-negative subtype group (Fig.
2D), the PIK3CA and TP53 genes had the highest
mutation frequencies at 21.8% each. In the
HR+/HER2− group, 59.8% of the mutations were
found in 29% of the genes, while in the
HR−/HER2− group, 14.5% of the mutations were
found in only 12.9% of the genes, which was significantly
different. Furthermore, PIK3CA and TP53 mutations
showed significantly different frequencies between
HR+/HER2− and triple-negative tumors
(Table IV).
 | Figure 2.Gene mutation frequencies according
to four molecular subtypes of breast cancer: (A) Luminal A, (B)
Luminal B, (C) HER2-positive and (D) triple-negative.
PIK3CA, phosphatidylinositol-4,5-bisphosphate 3-kinase
catalytic subunit α; TP53, tumor protein P53; AKT1,
AKT serine/threonine kinase 1; PTEN, phosphatase and tensin
Homolog; GATA3, GATA binding protein 3; ATM, ATM
serine/threonine kinase; PALB2, partner and localizer of
BRCA2; RAD51D, RAD51 paralog D; CHEK2,
checkpoint kinase 2. |
 | Table IV.Comparison of the gene mutations in
PIK3CA, TP53, AKT1 and PTEN between the
HR+/Her2− and triple-negative groups. |
Table IV.
Comparison of the gene mutations in
PIK3CA, TP53, AKT1 and PTEN between the
HR+/Her2− and triple-negative groups.
| Gene |
HR+/Her2− group
(n=149) | Triple-negative
group (n=55) | P-value |
|---|
| PIK3CA | 73 | 12 | <0.01 |
| TP53 | 15 | 12 | <0.05 |
| AKT1 | 10 | 1 | >0.05 |
| PTEN | 4 | 0 |
|
| GATA3 | 1 | 0 |
|
| BRCA1 | 1 | 0 |
|
| BRCA2 | 1 | 0 |
|
| PALB2 | 1 | 0 |
|
| RAD51D | 0 | 1 |
|
|
CHEK2(1) | 1 | 0 |
|
| Total | 107 | 26 | <0.01 |
Gene mutations identified in 72 sets
of cancerous, paracancerous, and normal tissues
The gene mutation spectra differed among cancerous,
paracancerous, and normal tissues (Table
V). The somatic non-synonymous mutations and pathogenic
mutations detected in tissues from 70 breast cancer patients after
removal of two primary cancer patients are shown in Fig. 3. Compared with normal and
paracancerous tissues, tumor cells are disorderly arranged and more
nuclear pleomorphism and divisional nucleus were observed in the
breast cancer tissues (Fig. 3). A
total of 914 somatic nonsynonymous mutations were detected in these
70 patients, and the cancer tissues harbored the highest number of
gene mutations. The correlations between gene mutations and
molecular subtypes in the three tissue types were analyzed, and the
results showed that nonsynonymous and pathogenic mutations had no
significant differences among the different molecular subtypes
(Table VI). We also found that some
of the eight common PIK3CA gene mutations were repeatedly
detected in the same patient (Table
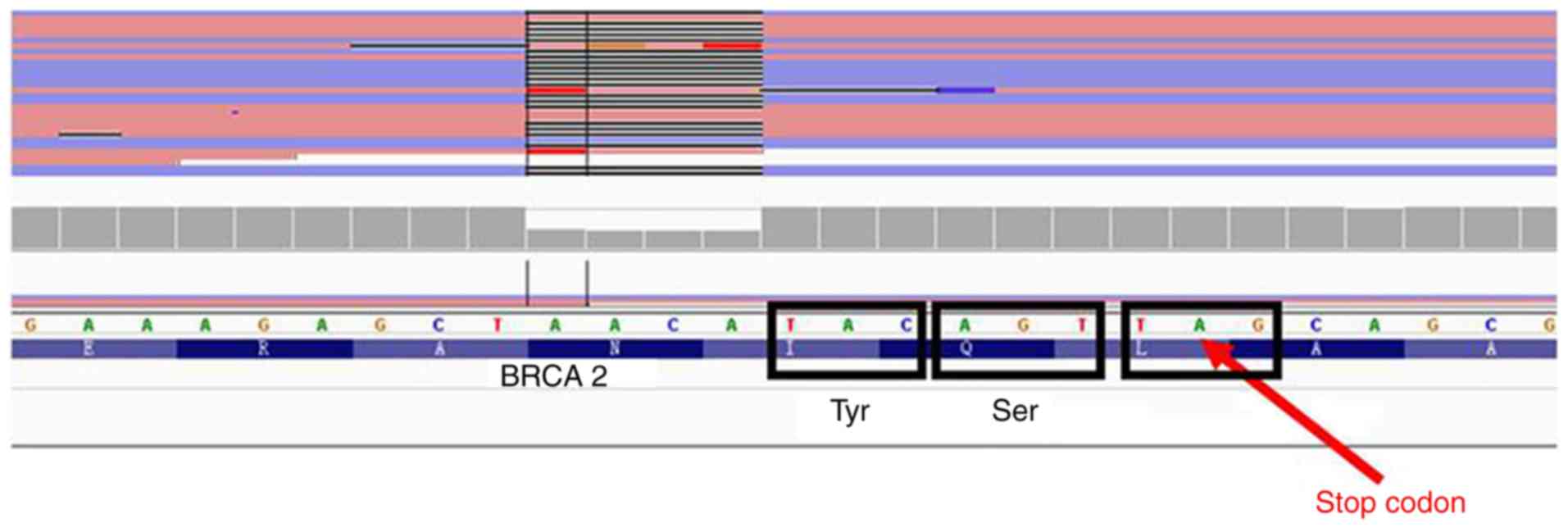
VII). A frameshift mutation in the BRCA2 gene was
identified in this patient, and this frameshift is likely to cause
truncation of BRCA2 (Fig. 4),
resulting in a protein lacking the last 374 amino acids. This
truncation is predicted to impair the binding of CDK and
RAD51, which have important functions during DNA repair
(Fig. 5).
 | Table V.Gene mutations identified in 72 sets
of cancerous, paracancerous, and normal tissues. |
Table V.
Gene mutations identified in 72 sets
of cancerous, paracancerous, and normal tissues.
| Gene | Cancerous
tissues | Paracancerous
tissues | Normal tissues |
|---|
| PIK3CA | 28 (20) | 8 (1) | 10 (3) |
| TP53 | 6 | 0 | 0 |
| AKT1 | 7 | 1 | 0 |
| GATA3 | 1 | 1 | 0 |
| ATM | 0 | 1 | 0 |
| PTEN | 0 | 0 | 3 |
| BRCA2 | 0 | 0 | 1 |
| Total | 42 | 11 | 14 |
 | Table VI.Correlation of mutation types with
breast cancer subtypes in 70 breast cancer patients from whom
cancerous, paracancerous and normal tissues were obtained. |
Table VI.
Correlation of mutation types with
breast cancer subtypes in 70 breast cancer patients from whom
cancerous, paracancerous and normal tissues were obtained.
|
|
| Molecular
subtype |
|
|
|---|
|
|
|
|
|
|
|---|
| Tissues | Mutation type | Luminal A
(n=21) | Luminal B
(n=30) | HER2+
(n=7) | Triple-negative
(n=12) | P-value |
|---|
| Cancerous
tissues | Nonsynonymous | 7 | 14 | 4 | 4 |
|
|
| Pathogenic | 14 | 16 | 3 | 8 | 0.584 |
| Paracancerous
tissues | Nonsynonymous | 18 | 24 | 6 | 11 |
|
|
| Pathogenic | 3 | 6 | 1 | 1 | 0.813 |
| Normal tissues | Nonsynonymous | 15 | 26 | 5 | 11 |
|
|
| Pathogenic | 6 | 4 | 2 | 1 | 0.358 |
 | Table VII.Analysis of PIK3CA gene
mutations detected in 72 sets of cancerous, paracancerous, and
normal tissues. |
Table VII.
Analysis of PIK3CA gene
mutations detected in 72 sets of cancerous, paracancerous, and
normal tissues.
| PIK3CA
mutation | Cancerous
tissues | Paracancerous
tissues | Normal tissues |
|---|
| Single
detection | 14 | 1 | 3 |
| Repeat
detection | 14 | 7 | 7 |
Correlations between mutations in the
PTEN/PI3K/AKT signaling pathway and the 5-year survival rate
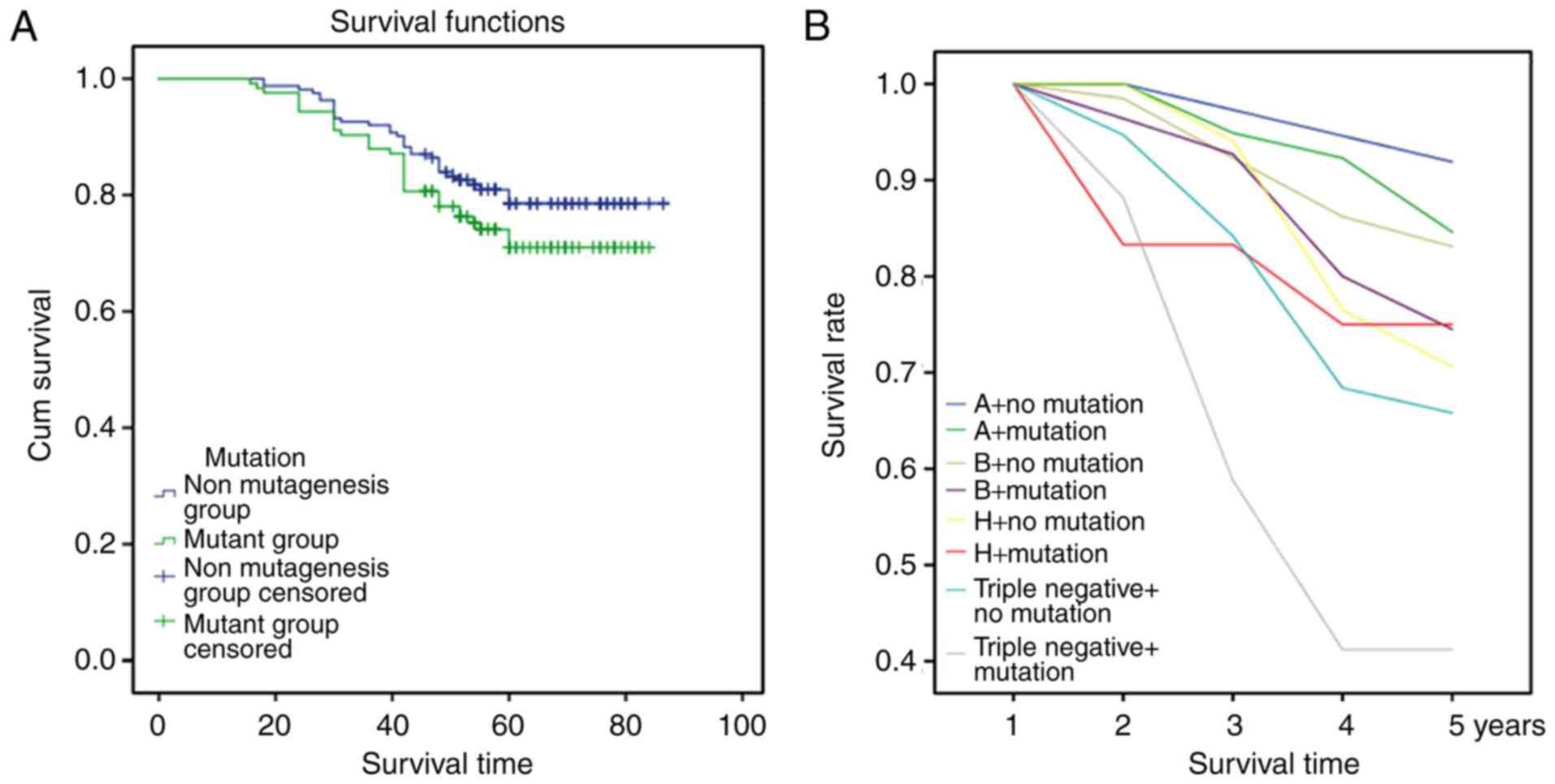
Cox multivariate analysis showed that patients with
gene mutations in the PTEN/PI3K/AKT signaling pathway had a poor
prognosis (P=0.044; Fig. 6A) We
further analyzed the correlations between gene mutations and
molecular subtypes of breast cancer, and the results showed that
patients in the triple-negative group with gene mutations of the
PTEN/PI3K/AKT signaling pathway had a poor prognosis (P=0.038;
Fig. 6B).
Discussion
Breast cancer development and progression are highly
variable, leading to genetic heterogeneity (22) and variable drug responses among tumor
cells of the same patient (23). In
the present study, the multigene mutation spectra of female breast
cancer patients in Northeast China were investigated to examine
correlations between mutations and clinicopathological
characteristics and identify genetic mutations potentially useful
for predicting the prognosis and survival of breast cancer
patients. The Ion Torrent sequencing platform was used to detect
gene mutations and based on comparisons with previous studies of
this platform, the results of our study are accurate and reliable
(24,25).
Our study showed that 54.2% of breast cancer
patients carried somatic nonsynonymous mutations, which is slightly
lower than the percentage reported by a previous study (62.1%)
(26). The mutation rate of PIK3CA
was higher than that of TP53 (39.2 vs. 12.9%), which is consistent
with previous results obtained in Chinese patients (35.2 vs. 15.2%)
(27) but different from the rates in
other countries (31.7 vs. 38.8%) (26). The mutation rate of AKT1 (4.9%)
was slightly lower in our study than previously reported in other
countries (6%), while the mutation rate of PTEN (1.4%) was
the same (1.4%) (26). The detected
PIK3CA gene mutation rate is among the highest in breast cancer
reaching 25–40%. In our study, the PIK3CA mutation detection
rate was 39.2%, similar to rates reported worldwide (28–30). The
frequency of the PIK3CA c.3140A>G (p.H1047R) mutation was
high in our cohort and similar to the rate previously reported
(28). Further analysis showed that
the gene mutation rates differed among the low, moderate, and high
recurrence risk groups. The gene mutation spectra for the moderate-
and high-risk groups were more complicated, and their mutation
frequencies were significantly higher than those for the low-risk
group. Our study demonstrated the practicality of using the
mutation spectrum for predicting the recurrence risk after surgery.
The spectra of gene mutations also varied among the different
molecular subtypes of breast cancer. Although the sample size for
the HER2-positive subtype was small (n=31) and mutations were
identified in only two genes, PIK3CA and TP53, the
mutations were detected at a frequency of 8.3% (15 of 179), which
is similar to the rate previously reported (26).
The gene mutation spectra differed between the
HR+/HER2− and HR/HER2− groups
(31). In the
HR+/HER2− group, clinical
immunohistochemistry results for p53 expression were positive for
all breast cancer, indicating that breast cancer patients without
p53 protein function due to TP53 gene mutation were
concentrated in the triple-negative subtype.
In our study, a total of 914 somatic nonsynonymous
mutations were detected in 70 patients, and the cancerous tissues
harbored the highest number of gene mutations. A total of 67
harmful/pathogenic mutations were detected in these tissues, and
cancerous tissues contained the highest number of mutated genes.
Overall, 96% of patients were found to have somatic nonsynonymous
mutations in all three tissue types, while only 7.7% of patients
were diagnosed with somatic pathogenic mutations in all 3 tissues,
and 34.2% of patients had pathogenic mutations in only cancerous
tissue. In all four patient groups, the mutation rates in cancerous
tissue were higher than those in paracancerous and normal tissues
after normalization.
Mutations in genes associated with the PTEN-PI3K/AKT
signaling pathway are closely related to breast cancer development
(32,33). Based on follow up of all enrolled
patients, the 5-year survival rate of the 286 patients was 77.3%,
which is similar to a previously reported rate (34). We also found that gene mutations
associated with the PTEN-PI3K/AKT signaling pathway were
independent factors affecting the survival rate as well as
predictors of poor prognosis for breast cancer patients, and these
findings differed from those of previous studies (35–37),
providing new evidence for evaluating the prognosis of patients
with breast cancer. In addition, we analyzed correlations of
specific mutations and the number of mutations with the 5-year
survival rate and found no significant differences.
Although our study primarily focused on somatic
nonsynonymous mutations, we found an interesting and unreported
genetic mutation in all four tissues of patient number 370. We
validated these mutations by sequencing and will continue to
explore this mutation status through the following steps: i)
Examine BRCA2 mRNA and protein levels in samples from
patient 370; ii) characterize the interaction between truncated
BRCA2 (if expressed) and RAD51 and CDK; iii)
transfect BRCA2−/− and BRCA2+/−
cell lines with truncated BRCA2; and iv) characterize the
DNA integrity in cells from patient 370. These studies are in
progress.
In summary, we analyzed the harmful/pathogenic gene
mutation spectra of breast cancer cases in Northeast China through
second-generation sequencing and provided new insights into these
spectra. The population coverage in our study was high, and the
results differed from those of studies conducted in Europe and the
United States (26,38,39);
Japan, South Korea and other Asian areas (40,41); and
southern China (42). In the context
of the global promotion of precision medicine, our study presents
valuable information for the clinical treatment of breast cancer
patients in Northeast China and other similar Asian areas.
Acknowledgements
The authors thank the Tissue Bank of the China-Japan
Union Hospital of Jilin University for their generous help.
Funding
This research was supported by the Science &
Technology Development Project of Jilin Province (grant no.
20160101033JC).
Availability of data and materials
We declared that materials described in the
manuscript, including all relevant raw data, will be freely
available to any scientist wishing to use them for non-commercial
purposes, without breaching participant confidentiality.
Authors' contributions
CF and NL conducted the experiments, collected the
data and analyzed the data. SF and ZY were major contributors in
interpreting the results and writing the manuscript. NY and KW
helped in designing the study, analyzing the data and revising the
manuscript. All authors read and approved the final manuscript and
agree to be accountable for all aspects of the research in ensuring
that the accuracy or integrity of any part of the work are
appropriately investigated and resolved.
Ethics approval and consent to
participate
All experiments were approved by the Ethics
Committee of the China-Japan Union Hospital of Jilin University,
and an informed consent form was signed by each enrolled
patient.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Arnold M, Karim-Kos HE, Coebergh JW,
Byrnes G, Antilla A, Ferlay J, Renehan AG, Forman D and
Soerjomataram I: Recent trends in incidence of five common cancers
in 26 European countries since 1988: Analysis of the European
Cancer Observatory. Eur J Cancer. 51:1164–1187. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Brenner DR, Brockton NT, Kotsopoulos J,
Cotterchio M, Boucher BA, Courneya KS, Knight JA, Olivotto IA, Quan
ML and Friedenreich CM: Breast cancer survival among young women: A
review of the role of modifiable lifestyle factors. Cancer Causes
Control. 27:459–472. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gray JM, Rasanayagam S, Engel C and Rizzo
J: State of the evidence 2017: An update on the connection between
breast cancer and the environment. Environ Health. 16:942017.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Makarem N, Chandran U, Bandera EV and
Parekh N: Dietary fat in breast cancer survival. Annu Rev Nutr.
33:319–348. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Brewer HR, Jones ME, Schoemaker MJ,
Ashworth A and Swerdlow AJ: Family history and risk of breast
cancer: An analysis accounting for family structure. Breast Cancer
Res Treat. 165:193–200. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang B, Shu XO, Delahanty RJ, Zeng C,
Michailidou K, Bolla MK, Wang Q, Dennis J, Wen W, Long J, et al:
Height and breast cancer risk: Evidence from prospective studies
and mendelian randomization. J Natl Cancer Inst. 107:djv2192015.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kwong A, Shin VY, Ho JC, Kang E, Nakamura
S, Teo SH, Lee AS, Sng JH, Ginsburg OM, Kurian AW, et al:
Comprehensive spectrum of BRCA1 and BRCA2 deleterious mutations in
breast cancer in Asian countries. J Med Genet. 53:15–23. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Stadler ZK, Schrader KA, Vijai J, Robson
ME and Offit K: Cancer genomics and inherited risk. J Clin Oncol.
32:687–698. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Smith DR, Quinlan AR, Peckham HE, Makowsky
K, Tao W, Woolf B, Shen L, Donahue WF, Tusneem N, Stromberg MP, et
al: Rapid whole-genome mutational profiling using next-generation
sequencing technologies. Genome Res. 18:1638–1642. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gilissen C, Hehir-Kwa JY and Thung DT:
Genome sequencing identifies major causes of severe intellectual
disability. Nature. 511:344–347. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kittaneh M, Montero AJ and Gluck S:
Molecular profiling for breast cancer: A comprehensive review.
Biomark Cancer. 5:61–70. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Goldhirsch A, Wood WC, Coates AS, Gelber
RD, Thürlimann B and Senn HJ; Panel members, : Strategies for
subtypes-dealing with the diversity of breast cancer: Highlights of
the St. Gallen International Expert Consensus on the primary
therapy of early breast cancer 2011. Ann Oncol. 22:1736–1747. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Marziali A and Akeson M: New DNA
sequencing methods. Annu Rev Biomed Eng. 3:195–223. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Xu X, Hou Y, Yin X, Bao L, Tang A, Song L,
Li F, Tsang S, Wu K, Wu H, et al: Single-cell exome sequencing
reveals single-nucleotide mutation characteristics of a kidney
tumor. Cell. 148:886–895. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hou Y, Song L, Zhu P, Zhang B, Tao Y, Xu
X, Li F, Wu K, Liang J, Shao D, et al: Single-cell exome sequencing
and monoclonal evolution of a JAK2-negative myeloproliferative
neoplasm. Cell. 148:873–885. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Clinical and laboratory standards
institute, . Nucleic acid sequencing methods in diagnostic
laboratory medicine. Approved guideline-Second Edition CLSI
document MM09. A2. (Wayne, PA). CLSI. 2014.
|
|
18
|
Sotiriou C and Pusztai L: Gene-expression
signatures in breast cancer. N Engl J Med. 360:790–800. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Roukos D and Ku CS: Clinical cancer genome
and precision medicine. Ann Surg Oncol. 19:3646–3650. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Damerla RR, Chatterjee B, Li Y, Francis
RJ, Fatakia SN and Lo CW: Ion Torrent sequencing for conducting
genome-wide scans for mutation mapping analysis. Mamm Genome.
25:154–156. 2014. View Article : Google Scholar
|
|
21
|
Meldrum C, Doyle MA and Tothill RW:
Next-generation sequencing for cancer diagnostics: A practical
perspective. Clin Biochem Rev. 32:177–195. 2011.PubMed/NCBI
|
|
22
|
Vogelstein B, Papadopoulos N, Velculescu
VE, Zhou S, Diaz LA Jr and Kinzler KW: Cancer genome landscapes.
Science. 339:1546–1558. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Almendro V and Fuster G: Heterogeneity of
breast cancer: Etiology and clinical relevance. Clin Transl Oncol.
13:767–773. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Fujita S, Masago K, Takeshita J, Okuda C,
Otsuka K, Hata A, Kaji R, Katakami N and Hirata Y: Validation of an
Ion Torrent sequencing platform for the detection of gene mutations
in biopsy specimens from patients with non-small-cell lung cancer.
PLoS ONE. 10:e01302192015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zanella I, Merola F, Biasiotto G, Archetti
S, Spinelli E and Di Lorenzo D: Evaluation of the Ion Torrent PGM
sequencing workflow for the routine rapid detection of BRCA1 and
BRCA2 germline mutations. Exp Mol Pathol. 102:314–320. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Roy-Chowdhuri S, de Melo Gagliato D,
Routbort MJ, Patel KP, Singh RR, Broaddus R, Lazar AJ, Sahin A,
Alvarez RH, Moulder S, et al: Multigene clinical mutational
profiling of breast carcinoma using next-generation sequencing. Am
J Clin Pathol. 144:713–721. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bai X, Zhang E, Ye H, Nandakumar V, Wang
Z, Chen L, Tang C, Li J, Li H, Zhang W, et al: PIK3CA and TP53 gene
mutations in human breast cancer tumors frequently detected by ion
torrent DNA sequencing. PLoS One. 9:e993062014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Klarenbeek S, van Miltenburg MH and
Jonkers J: Genetically engineered mouse models of PI3K signaling in
breast cancer. Mol Oncol. 7:146–164. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Samuels Y and Waldman T: Oncogenic
mutations of PIK3CA in human cancers. Curr Top Microbiol Immunol.
347:21–41. 2010.PubMed/NCBI
|
|
30
|
Bhat-Nakshatri P, Goswami CP, Badve S,
Magnani L, Lupien M and Nakshatri H: Molecular insights of pathways
resulting from two common PIK3CA mutations in breast cancer. Cancer
Res. 76:3989–4001. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Boyault S, Drouet Y, Navarro C, Bachelot
T, Lasset C, Treilleux I, Tabone E, Puisieux A and Wang Q:
Mutational characterization of individual breast tumors: TP53 and
PI3K pathway genes are frequently and distinctively mutated in
different subtypes. Breast Cancer Res Treat. 132:29–39. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tserga A, Chatziandreou I, Michalopoulos
NV, Patsouris E and Saetta AA: Mutation of genes of the PI3K/AKT
pathway in breast cancer supports their potential importance as
biomarker for breast cancer aggressiveness. Virchows Arch.
469:35–43. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lee MS, Jeong MH, Lee HW, Han HJ, Ko A,
Hewitt SM, Kim JH, Chun KH, Chung JY, Lee C, et al: PI3K/AKT
activation induces PTEN ubiquitination and destabilization
accelerating tumourigenesis. Nat Commun. 6:77692015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Fan L, Strasser-Weippl K, Li JJ, St Louis
J, Finkelstein DM, Yu KD, Chen WQ, Shao ZM and Goss PE: Breast
cancer in China. Lancet Oncol. 15:e279–e289. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Koundouros N and Poulogiannis G:
Phosphoinositide 3-kinase/Akt signaling and redox metabolism in
cancer. Front Oncol. 8:1602018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Miller TW, Rexer BN, Garrett JT and
Arteaga CL: Mutations in the phosphatidylinositol 3-kinase pathway:
Role in tumor progression and therapeutic implications in breast
cancer. Breast Cancer Res. 13:2242011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Papadavid E, Korkolopoulou P, Levidou G,
Saetta AA, Papadaki T, Siakantaris M, Nikolaou V, Oikonomidi A,
Chatziandreou I, Marinos L, et al: In situ assessment of PI3K and
PTEN alterations in mycosis fungoides: Correlation with
clinicopathological features. Exp Dermatol. 23:931–933. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Bozhanov SS, Angelova SG, Krasteva ME,
Markov TL, Christova SL, Gavrilov IG and Georgieva EI: Alterations
in p53, BRCA1, ATM, PIK3CA, and HER2 genes and their effect in
modifying clinicopathological characteristics and overall survival
of Bulgarian patients with breast cancer. J Cancer Res Clin Oncol.
136:1657–1669. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Pereira B, Chin SF, Rueda OM, Vollan HK,
Provenzano E, Bardwell HA, Pugh M, Jones L, Russell R, Sammut SJ,
et al: The somatic mutation profiles of 2,433 breast cancers
refines their genomic and transcriptomic landscapes. Nat Commun.
7:114792016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Kato T, Park JH, Kiyotani K, Ikeda Y,
Miyoshi Y and Nakamura Y: Integrated analysis of somatic mutations
and immune microenvironment of multiple regions in breast cancers.
Oncotarget. 8:62029–62038. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kim JY, Lee E, Park K, Park WY, Jung HH,
Ahn JS, Im YH and Park YH: Clinical implications of genomic
profiles in metastatic breast cancer with a focus on TP53 and
PIK3CA, the most frequently mutated genes. Oncotarget.
8:27997–28007. 2017.PubMed/NCBI
|
|
42
|
Yang X, Wu J, Lu J, Liu G, Di G, Chen C,
Hou Y, Sun M, Yang W, Xu X, et al: Identification of a
comprehensive spectrum of genetic factors for hereditary breast
cancer in a Chinese population by next-generation sequencing. PLoS
One. 10:e01255712015. View Article : Google Scholar : PubMed/NCBI
|