Introduction
Malignant gliomas account for ~70% of all primary
brain tumors diagnosed in adults (1).
As the most common form of malignant glioma, glioblastoma
multiforme (GBM) is the most prevalent primary brain tumor and has
one of the worst prognoses of all different types of cancer
(2). Several genetic and epigenetic
factors have been hypothesized to serve critical roles in the
etiology and prognosis of GBM; however, they remain poorly defined
and insufficiently characterized (3).
The identification of novel markers may be beneficial for the
diagnosis and prognosis of patients with GBM.
MicroRNAs (miRs) are non-coding RNA sequences which
are frequently complementary to the 3′ untraslated region (UTR),
and may completely or incompletely match the sequence of the target
gene mRNA sequence. miRs degrade or inhibit translation of mRNAs
and thus regulate protein expression at the post-transcriptional
level (4). Bioinformatics prediction
demonstrated that miRs act on an extensive range of target genes
and may regulate the expression of >30% of human genes (4,5). miRs act
on a wide range of target genes and participate in the fine
regulation of multiple biological processes, including immunity,
metabolism, differentiation, proliferation and carcinogenesis
(6). Recent studies have identified
miRNAs which act as tumor suppressor and oncogenes that serve
important roles in tumorigenesis (7).
Aberrant expression of miR-378 has been observed in various types
of cancer (8–11), which suggests its potential role in
human carcinogenic events (12). Li
et al (13) reported that
miR-378 may serve as a tumor suppressor and served an important
role in inhibiting glioma tumor migration and invasion. However,
the potential roles and targets of miR-378a-3p involved in GBM
remain known.
The aim of the present study was to examine the
potential roles of miR-378a-3p/tetraspanin 17 (TSPAN17) in the
pathological progression of GBM. In the present study, expression
of miR-378a-3p and TSPAN17 was observed in GBM tumors in patients
with GBM and the human GBM cell lines, U87MG and MT-330. Luciferase
reporter assay was used to determine and confirm the potential
targets of miR-378a-3p and TSPAN17. The mRNA and protein expression
levels of TSPAN17 were measured in the GBM cells following
transfection with miR-378a-3p mimic or inhibitor. Proliferation,
migration and invasion were also assessed following
transfection.
Materials and methods
Ethics approval
All procedures performed in the present study
involving human participants were conducted in accordance with the
1964 Declaration of Helsinki and its later amendments or comparable
ethical standards. The present study was approved by The Medical
Ethics Committee of Kunming Medical University (Kunming, China).
GBM tumor specimens were obtained from patients at The First
People's Hospital of Yunnan Province (Yunnan, China).
Preparation of tumor tissues
A total of 53 patients provided written consent to
participate in the present study. The patients were registered at
the clinic of The First People's Hospital of Yunnan Province
between January 2010 and August 2011. All the patients recruited
were undergoing surgery in the hospital for the first time and had
not received anti-tumor treatment prior to surgery. The specimens
were histopathologically verified as primary GBM by three senior
pathologists. The primary carcinoma tissues and the matched
adjacent normal tissues (≥3 cm away from the tumor) were collected
and frozen at −80°C immediately for analysis of gene or protein
expression. Genetic features, including isocitrate dehydrogenase
(IDH), O-6-methylguanine-DNA methyltransferase (MGMT), 1p 19q,
telomerase reverse transcriptase (TERT) ATP-dependent helicase
(ATRX) status, were used as genetic markers of favorable prognosis
of GBM, and were used as indicators to distinguish between the
subclasses of glioma, and predict outcomes in glioma with high
grades (14,15). To determine the expression of these
markers, a DNeasy Blood & Tissue kit (Qiagen China Co., Ltd.)
was used to extract the DNA from tissue samples. The methylation
status of the MGMT promoter was analyzed using a customized
pyrosequencing assay, as previously described (16). A SALSA MLPA probemix (cat. no.
P088-C1; MRC-Holland BV) was used for analysis of copy number of
1p/19q as described previously (17).
The presence of hotspot mutations in the IDH gene (R132 and R172),
as well as the two mutation hotspots in the TERT promoter (C228T
and C250T), were assessed primarily using pyrosequencing and partly
using Sanger sequencing, as described previously (18,19).
Cell culture and treatment
The human normal skin fibroblast HF cell line, and
the GBM cell lines U87MG and MT-330, were purchased from The Cell
Bank of Type Culture Collection of the Chinese Academy of Sciences.
As it has been reported that the U87MG cell line has been
misidentified or contaminated and this cell line is considered as a
glioblastoma with unknown origin, the U87MG cell line was
authenticated using STR profile detection by The Cell Bank of Type
Culture Collection of the Chinese Academy of Sciences. The results
showed that between the test sample and American Type Culture
Collection standard data, the match ratio was 94.4%; thus, the cell
line was identified as a U87MG cell line. The cells were cultured
in Dulbecco's Modified Eagles medium (Thermo Fisher Scientific,
Inc.) at 37°C in a humidified atmosphere containing 5%
CO2. The medium was supplemented with 10% fetal bovine
serum (Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and 100
µg/ml streptomycin.
After culturing for five passages, the U87MG and
MT-330 cells were transiently transfected with either miR-378a-3p
(Esembl ID: MIMAT0000732, hsa-miR-378a-3p) mimic (1.5 µl, 2.5 µM)
/inhibitor (3 µl, 2.5 µM) or the matched negative control (NC, 2.5
µM) with Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). The miR-378a-3p mimic, miR-378a-3p antagomir and
the matched miR NC (mimic-NC or anta-NC) were purchased from
Invitrogen (Thermo Fisher Scientific, Inc.). The sequences of the
miRNAs used are described as: miR-378a-3p mimic:
CUGGACUUGGAGUCAGAAGG; mimic-NC: AGUGCAUGUUAUGCCUACG; miR-378a-3p
antagomir: AGUUCAGGUUCUGACUCCU; anta-NC: UGGUCCGUGUAGGCCUACUA.
According to the manufacturer's protocols, a final concentration of
2×105 cells were seeded into each well of a 6-well plate
and transfected for 48 h after which the cells were collected for
further analysis.
Reverse-transcription-quantitative
(RT-q)PCR
The expression of miR-378a-3p and TSPAN17 in human
GBM cells or specimens from patients with GBM were determined using
RT-qPCR. Total RNA was isolated using TRIzol® and a
miRNeasy kit (Invitrogen; Thermo Fisher Scientific, Inc.) according
to the manufacturer's protocol. RT-qPCR was performed using an
All-in-One™ miRNA quantitative RT-PCR detection kit according to
the manufacturer's protocol (GeneCopoeia, Inc.).
The miRNA primers for hsa-miR-378a-3p and U6 were
purchased from Applied Biosystems (Thermo Fisher Scientific, Inc.).
The thermocycling conditions were: 95°C For 1 min; followed by 45
cycles of 95°C for 10 sec, 59°C for 10 sec and 72°C for 30 sec. The
expression of miRNAs was normalized to U6. The sequences of primers
are described as: U6, forward: 5′-CTCGCTTCGGCAGCACA-3′, reverse:
5′-AACGCTTCACGAATTTGCGT-3′; hsa-miR-378a-3p, forward:
5′-ACUGGACUUGGAGUCAGAAGGC-3′, reverse:
5′-GCTGTCAACGATACGCTACGTAACG-3′.
For the detection of TSPAN17 mRNA expression,
Moloney Murine Leukemia Virus Reverse Transcriptase (Takara Bio,
Inc.) and a 2X SYBR Green I mix (Takara Bio, Inc.) were used. The
thermocycling conditions were: 95°C For 3 min; followed by 39
cycles of 95°C for 10 sec, 57°C for 15 sec and 72°C for 30 sec;
followed by 95°C for 10 sec, 65°C for 5 sec and a final 95°C for 10
sec. GAPDH was used as the control. The primers were synthesized by
Invitrogen (Thermo Fisher Scientific, Inc.) and the sequences of
the primers were: GAPDH forward, 5′-CGAGATCCCTCCAAAATCAA-3′ and
reverse, 5′-TTCACACCCATGACGAACAT-3′; and TSPAN17 forward
5′-caccagcatttccaggaacc-3′ and reverse, 5′-CGCCTCCAACTACCACAAAC-3′.
The relative expression levels of the genes were determined using
the 2−ΔΔCq method (20).
Bioinformatics prediction
TargetScan (http://www.targetscan.org/vert_72/), miRanda
(http://miranda.org.uk) and PicTar (https://pictar.mdc-berlin.de), were used to predict
the candidate target genes of miR-378a-3p. According to the results
predicted by all three databases and the clinical data, TSPAN17
gene was selected for further analysis as a potential target
involved in GBM.
Luciferase reporter assays
The target of miR-378a-3p, TSPAN17, was validated
using a luciferase reporter assay in 293T cells (The Cell Bank of
Type Culture Collection of the Chinese Academy of Sciences). The
wild-type (wt) or the mutant (mut) of two potential seed regions of
human TSPAN17 mRNA 3′UTR, which included a potential target
position for miR-378a-3p, were cloned and inserted into pLUC vector
(Invitrogen; Thermo Fisher Scientific, Inc.) to generate the
TSPAN17 pLUC-UTR-wt or TSPAN17 pLUC-UTR-mut vector. Briefly, 293T
cells were plated into 96-well plates at 70% confluence for 24 h
prior to transfection. A total of 25 ng of pLUC-UTR-wt or
pLUC-UTR-mutTSPAN17 was co-transfected with 50 nM miR-378a-3p
mimic/anta-NC control), or 10 ng Renilla luciferase into
293T cells using Lipofectamine® 2000. Firefly luciferase
activity was normalized to Renilla, and the value of firefly
luciferase activity/Renilla luciferase activity was analyzed
using an iMark Fluorometer Microplate Reader (Bio-Rad,
Laboratories, Inc.). A total of five independent experiments were
performed in triplicate.
Knockdown of TSPAN17
Small interfering (si)RNA sequences targeting
TSPAN17 and the NC (si-NC) were designed and purchased from
Invitrogen (Thermo Fisher Scientific, Inc.). Lentivirus was
packaged in 293T cells using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) for TSPAN17 gene
knockdown. The GBM cells lines, U87MG and MT-330, as well as the
normal HF cell line, were infected with 1×106
recombinant lentivirus-transducing units. The efficiency of siRNA
knockdown was determined using RT-qPCR and western blotting.
Following transfection with miR-378a-3p antagomir, the U87MG and
MT-330 cells were infected with 1×106 recombinant
lentivirus-transducing TSPAN17 siRNA units. A total of 24 h after
transfection, the GBM cells were collected for further analysis.
The effects of miR-378a-3p antagomir and TSPAN17 siRNA co-treated
in GBM cells were also determined.
Western blotting
The protein expression levels of TSPAN17 were
evaluated in the tumor tissues and different cell lines as
described previously (21). Lysis
buffer supplemented with protease inhibitors (cat. no. P1006,
Beyotime Institute of Biotechnology) was used to extract protein
from cells and tissues. A bicinchoninic acid assay (Beyotime
Institute of Biotechnology) was used to quantify the concentration
of the protein samples, and 35 mg of protein extract from either
the cells or tissues were used for analysis. The proteins were
resolved using 15% SDS-PAGE and transferred to nitrocellulose
membranes. The membranes were incubated with anti-TSPAN17 (cat. no.
PA5-69207; 1:1,000; Invitrogen; Thermo Fisher Scientific, Inc.) and
anti-GAPDH (cat. no. TAB1001; 1:800; Invitrogen; Thermo Fisher
Scientific, Inc.) antibodies at 4°C for 12 h. The membranes were
washed and incubated with the horseradish peroxidase-conjugated
anti-rabbit antibody (1:2,000; cat. no. PI-1000; Vector
Laboratories, Inc.) at 37°C for 1 h. The signals were visualized
using enhanced chemiluminescence-based detection (BeyoECL Plus,
cat. no. P0018M; Beyotime Institute of Biotechnology). The quantity
of the target proteins were analyzed for each group. The blots were
quantified by densitometric analysis using a ChemiDoc™ RS+ imaging
system and Image Lab™ software v5.2.1 (Bio-Rad, Laboratories,
Inc.).
MTT assay
Briefly, 1×105 cells were plated into
96-well culture plates in 200 µl culture medium per well. After 48
h of culture with the aforementioned treatments, 20 µl 5 mg/ml MTT
was added to each well and incubated at 37°C for 4 h. The medium
was gently aspirated and 150 µl of DMSO was added to each well to
solubilize the formazan crystals. The absorbance of each sample was
immediately measured using a microplate reader (Multiskan Mk3;
Thermo Fisher Scientific, Inc.) at 570 nm.
Apoptosis assay
An Annexin V-fluorescein isothiocyanate-flow
cytometry assay kit (Beijing 4A Biotech Co.) was used to detect the
apoptotic rate in the U87MG and MT-330 cells following transfection
with the vectors, as described previously (22,23). The
kit was used according to the manufacturer's protocols. For flow
cytometry, a FACScan flow cytometer (BD Immunocytometry Systems)
was used and the data were analyzed using Cellquest software 10.6.4
version (BD Immunocytometry Systems). The apoptotic rate is
indicated as the percentages of cells staining with both Annexin
V+/PI− (early apoptosis) and
AnnexinV+/PI+ (late apoptosis). The
calculated formula: apoptotic rate = the percentages of (Q1-UR
quadrant + Q1-LR quadrant).
Migration and invasion assays
Cells (1×105) in serum-free medium were
placed into the upper chamber of a Transwell insert (BD Bioscience)
with or without Matrigel®. After 48 h of incubation,
cells remaining in the upper chamber or on the upper membrane were
carefully removed. Cells adhering to the lower membrane were
stained with 0.1% crystal violet for 20 min and fixed with 20%
methanol at room temperature for 30 sec, imaged, and counted using
an IBX3 inverted microscope (Olympus Corporation) (24). Under ×200 magnification, five random
fields of view were analyzed.
Statistical analysis
Data are presented as the mean ± standard deviation.
SPSS version 21.0 (IBM Corp.) was used for analysis. Based on the
median expression levels of miR-378a-3p or TSPAN17, the enrolled
patients were divided into two cohorts, high and low expression
(25). 5-year overall survival curves
of GBM patients with different expressional levels of miR-378a-3p
or TSPAN17 were analyzed by the Kaplan-Meier method and compared
using the log-rank test. A χ2 test was used to compare
the test the significant differences in observed variables. Pearson
correlation analysis was performed to analysis the correlation
between miR-378a-3p expression and TSPAN17 level in patients with
GBM. Multivariate logistical regression analysis was performed to
evaluate the association between TSPAN17 gene expression levels
with clinicopathologic characteristics of GBM. A paired Student's
t-test was used to compare the expression levels of TSPAN17 and
miR-378a-3p between GBM and the matched adjacent normal groups. The
statistical significance of RNA expression, apoptotic rates, cell
activity, migration and invasion among cells in the different
treatment groups were analyzed using a one-way ANOVA. The Least
Significant Difference post-hoc test was used to obtain individual
P-values following ANOVA. P<0.05 was considered to indicate a
statistically significant difference unless otherwise
indicated.
Results
Decreased expression of miR-378a-3p
and increased expression of Tspan17 are associated with GBM
The expression levels of miR-378a-3p and its
potential target gene TSPAN17 were detected in both the tumor
tissues of patients with GBM and GBM cell lines using RT-qPCR. The
results showed that the expression of miR-378a-3p in GBM tissues
was significantly lower compared with the adjacent non-neoplastic
tissues (P=0.00021; Fig. 1A).
Similarly, the expression of miR-378a-3p was significantly
decreased in U87MG and MT-330 cells compared with HF cells
(P=0.00017, P=0.00028; Fig. 1B).
Conversely, the mRNA expression levels of TSPAN17
were significantly increased in the tumor tissues compared with the
control (P=0.00008; Fig. 1C), which
was also observed in the GBM cells compared with the HF cells
(P=0.00007, P=0.00011; Fig. 1D).
Correlation analysis showed that decreased
miR-378a-3p expression was inversely correlated with TSPAN17
expression in patients with GBM (r=−0.901, P<0.05; Fig. 1E).
The 5-year overall survival (OS) analysis revealed
that patients with GBM with high expression of miR-378a-3p (cut-off
level 0.49; >0.49 high expression) demonstrated a higher rate OS
compared with patients with low expression (≤0.49, low expression)
(χ2=13.53, P=0.00048; Fig.
1F). The OS rate of patients with low levels of TSPAN17
(cut-off level 3.78; ≤3.78, low expression) in tumors was higher
compared with high levels of TSPAN17 (>3.78, high expression)
(χ2=17.93, P=0.00032; Fig.
1G).
The association between expression of TSPAN17 and
the clinicopathological characteristics of patients were determined
using the GBM tissues. The results showed that high expression of
TSPAN17 was significantly associated with age (50–60 years old;
rs=0.313, P=0.019), tumor size (rs=0.632,
P=0.00009), World Health Organization (WHO) grade
(rs=0.554, P=0.0013), IDH status (rs=0.701,
P<0.001), MGMT status (rs=0.254, P=0.045) and ATRX
status (rs=0.821, P<0.001; Table I). The TERT promoter mutations,
coinciding with IDH1/2 mutations and 1p/19q codeletion were the
most common genotypes detected in the oligodendrogliomas, while a
combination genotype of IDH-wt and TERT promoter mutation were
always present in patients with GBM (14). ATRX mutations, a molecular marker for
astrocytic tumors, combined with p53 mutations can predict the
prognosis of patients with glioma (26). The results of the present study
indicated that the high expression of TSPAN17 was significantly
associated with a poor survival rate, larger tumor and higher WHO
grade of glioma, as well as the aforementioned genetic markers.
Combined with the clinicopathologic features and common genotypes
present, high expression of TSPAN17 in carcinomas may be used as a
molecular indicator of poor prognosis. The multivariate analysis of
miR-378a-3p and TSPAN17 levels in patients with GBM were also
analyzed (Table II). The data
suggested that decreased miR-378a-3p and increased TSPAN17 levels
were associated with GBM.
 | Table I.TSPAN17 levels are associated with
clinicopathologic and genetic features of patients with
glioblastoma multiforme. |
Table I.
TSPAN17 levels are associated with
clinicopathologic and genetic features of patients with
glioblastoma multiforme.
|
| TSPAN17 levels
(mRNA) |
|
|
|---|
|
|
|
|
|
|---|
| Clinicopathological
characteristics | Low level n=12 | High level
n=41 | rs | P-value |
|---|
| Ages
(years)b |
|
| 0.313 |
0.019a |
|
40–49 | 4 | 7 |
|
|
|
50–60 | 5 | 25 |
|
|
|
>60 | 3 | 9 |
|
|
| Sex |
|
| 0.114 | 0.31 |
|
Male | 7 | 20 |
|
|
|
Female | 5 | 21 |
|
|
| Tumor size
(cm) |
|
| 0.632 |
0.0009a |
| <5
cm | 7 | 9 |
|
|
| ≥5
cm | 5 | 32 |
|
|
| WHO grade |
|
| 0.554 |
0.0013a |
|
I–II | 6 | 7 |
|
|
|
III | 5 | 10 |
|
|
| IV | 1 | 24 |
|
|
| IDH
status |
|
| 0.701 |
0.001a |
|
Wild-type | 10 | 33 |
|
|
|
Mutant | 2 | 8 |
|
|
| MGMT
status |
|
| 0.254 |
0.045a |
|
Methylated | 7 | 18 |
|
|
|
Unmethylated | 5 | 23 |
|
|
| 1p 19q
status |
|
| 0.198 | 0.122 |
|
Non-codeletion | 4 | 20 |
|
|
|
Codeletion | 8 | 21 |
|
|
| TERT |
|
| 0.107 | 0.33 |
|
Wild-type | 6 | 19 |
|
|
|
Mutant | 6 | 22 |
|
|
| ATRX |
|
| 0.821 |
<0.001a |
|
Wild-type | 5 | 3 |
|
|
|
Mutant | 7 | 38 |
|
|
 | Table II.Multivariate analysis of miR-378a-3p
and Tspan17 levels in patients with glioblastoma multiforme. |
Table II.
Multivariate analysis of miR-378a-3p
and Tspan17 levels in patients with glioblastoma multiforme.
|
| miR-378a-3p
expression |
| TSPAN17
expression |
|
|---|
|
|
|
|
|
|
|---|
|
| Low level (≤median)
n (%) | High level
(>median) n (%) | Adjusted OR (95%
CI) | Low level (≤median)
n (%) | High level n
(%) | Adjusted OR (95%
CI) |
|---|
| WHO grade |
|
I–II | 2
(3.7) | 5 (9.4) | 2.24
(1.54–4.02) | 6 (11.3) | 7
(13.2) | 1.58
(0.98–4.21) |
|
III | 23 (43.4) | 2 (3.7) | 16.19
(12.36–28.21) | 5 (9.4) | 10 (18.8) | 14.11
(9.45–29.2) |
| IV | 21 (39.6) | 0 (0) | 21.15
(12.31–31.58) | 1 (1.8) | 24 (45.2) | 10.34
(5.01–23.95) |
| Tumor size
(cm) |
|
<5 | 12 (22.6) | 6 (11.3) | 1.82
(1.33–5.01) | 7 (13.2) | 22 (41.5) | 1.23
(0.73–6.49) |
| ≥5 | 34 (64.1) | 1 (1.8) | 42.03
(28.93–57.38) | 5 (9.4) | 19 (35.8) | 1.49
(0.87–3.97) |
Validating TSPAN17 as a target gene of
miR-378a-3p
The expression levels of miR-378a-3p were measured
in cells transfected with miR-378a-3p mimics or inhibitor. The
results demonstrated that transfection with miR-378a-3p mimics
significantly upregulated the expression levels of miR-378a-3p in
U87MG cells (mimic, 2.08±0.15; mimic-NC, 0.53±0.07; P<0.05) and
MT-330 cells (mimic, 1.98±0.21; mimic-NC, 0.61±0.08; P<0.05)
compared with the control. Following miR-378a-3p antagomir
transfection, miR-378a-3p expression was significantly decreased in
U87MG (antagomir, 0.12±0.01; anta-NC, 0.56±0.02; P<0.05) and
MT-330 cells compared with the control (antagomir, 0.22±0.01;
anta-NC, 0.59±0.01; P<0.05) (Fig.
2).
It was determined that miR-378a-3p has two conserved
putative binding sites for TSPAN17 mRNA on the 3′UTR at positions
179–186 and 891–897 (Fig. 3A).
Transfection of TSPAN17 pLUC-UTR-wt in combination with miR-378a-3p
mimic significantly reduced the luciferase activity compared with
the corresponding control; this reduction of luciferase activity
was not observed in cells transfected with TSPAN17 pLUC-UTR-mut
vector. Furthermore, mutation of the putative miR-378a-3p binding
sites abrogated the suppressive effects of miR-378a-3p mimic on the
luciferase activity in 293T cells (Fig.
3B).
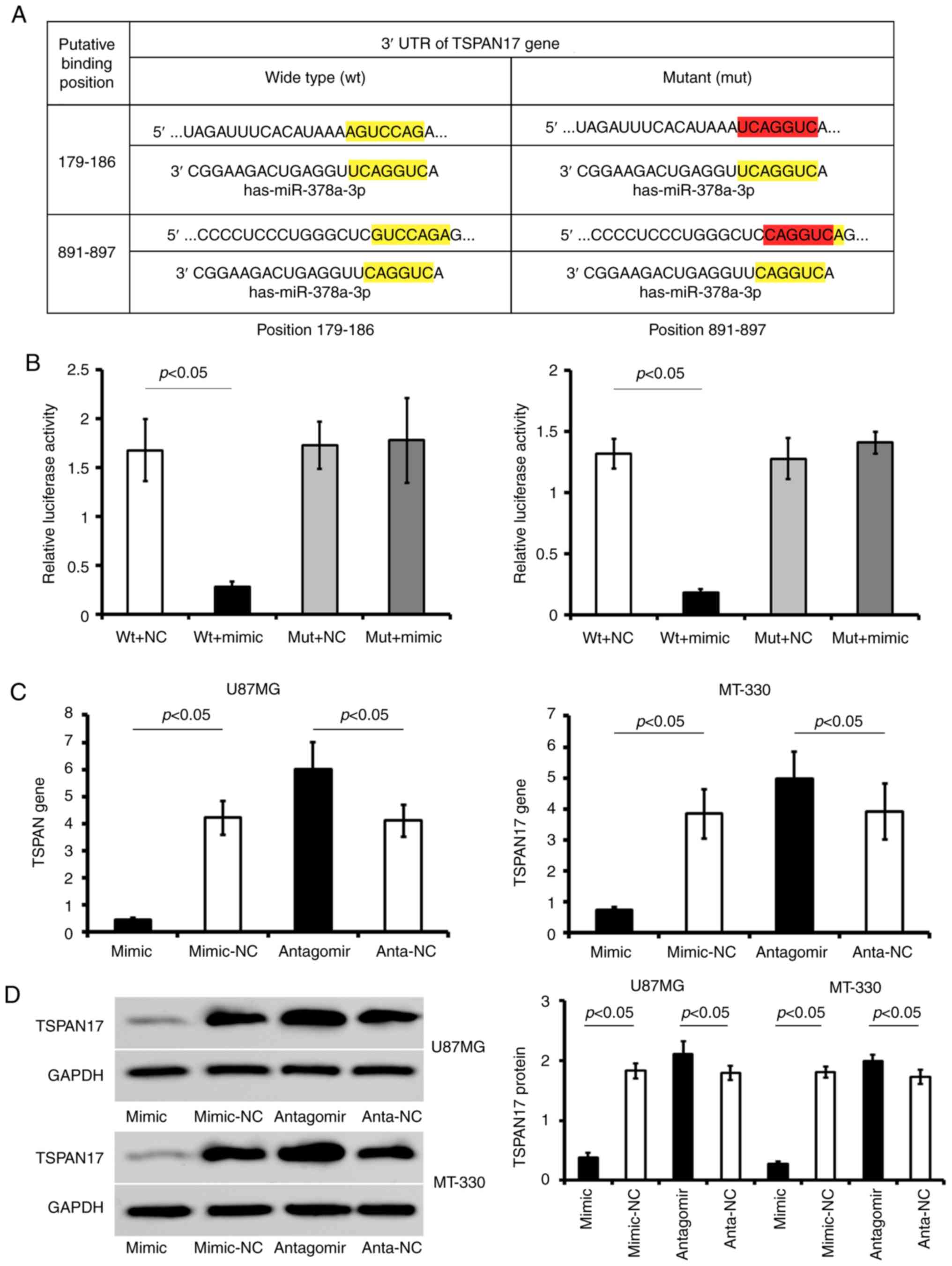 | Figure 3.TSPAN17 is a potential target gene
for miR-378a-3p. (A) Putative binding sites directly regulated by
miR-378a-3p and the pattern of mutating the two binding sites
(179–186 and 891–897) within TSPAN17 3′UTR. (B) Detection of the
relative luciferase activity of miR-378a-3p mimic co-transfected
with TSPAN17 3′UTR-wt or -mut by dual-luciferase reporter assay.
(C) relative expression of TSPAN17 gene in U87MG and MT-330 cells
treated with miR-378a-3p mimic or antagomir. (D) Representative
blots for TSPAN17 and GAPDH, as well as the relative expression of
TSPAN17 protein in U87MG and MT-330 cells treated with miR-378a-3p
mimic or antagomir. Data are represented as mean ± standard
deviation. All the data are representative of five independent
experiments (n=5). miR, microRNA; mimic, miR-378a-3p mimic;
mimic-NC, miR-378a-3p mimic negative control; antagomir,
miR-378a-3p antagomir; anta-NC, miR-378a-3p antagomir negative
control; mut, mutant; TSPAN17, tetraspanin 17; UTR, untranslated
region; wt, wild-type. |
Transfection with miR-378a-3p mimic significantly
reduced the expression of TSPAN17 in the GBM cell lines compared
with miR-NC (P<0.05). On the contrary, transfection with
miR-378a-3p antagomir increased the expression of TSPAN17 mRNA
levels in GBM cells compared with anta-NC transfected cells
(P<0.05; Fig. 3C and D).
Therefore, the results suggested that miR-378a-3p directly binds to
TSPAN17 3′ UTR and inhibited its expression.
TSPAN17 knockdown
The U87MG and MT-330 cells, as well as the control
HF cells were stably transfected with TSPAN17-siRNA or si-NC
lentiviral vector. The data showed that siRNA significantly reduced
mRNA and protein expression levels of TSPAN17 (normalized by si-NC
control) in U87MG, MT-330 and HF cells (Fig. 4). The mRNA levels of TSPAN17 were
significantly reduced following TSPAN17-siRNA transfection in U87MG
(0.11±0.001 vs. si-NC; P<0.05), MT-330 (0.13±0.006 vs. si-NC;
P<0.05) and HF (0.08±0.001 vs. si-NC; P<0.05) cells. In
accordance with the RT-qPCR results, TSPAN17 protein levels were
significantly lower in TSPAN17 siRNA- lentiviral transduced cells
compared with the si-NC control of U87MG (0.08±0.009), MT-330
(0.07±0.005) and HF (0.2±0.003) cells (vs. si-NC, P<0.05;
Fig. 4A-C).
Additionally, the effects of miR-378a-3p antagomir
and TSPAN17 siRNA co-treated were confirmed by RT-qPCR and western
blotting (Fig. 2B and C). Following
co-treatment with miR-378a-3p antagomir and TSPAN17 siRNA, the
levels of TSPAN17 mRNA were significantly increased in U87MG (anta
+ si, 2.89±0.14 vs. antagomir, 5.93±0.24, P<0.05; vs.
si-TSPAN17, 0.4±0.02, P<0.05; vs. anta-NC, 4.02±0.16, P<0.05;
vs. si-NC, 4.1±0.22, P<0.05) and MT-330 cells (anta + si,
2.51±0.21 vs. antagomir, 5.1±0.19, P<0.05; vs. si-TSPAN17,
0.38±0.06, P<0.05; vs. anta-NC, 3.94±0.11, P<0.05; vs si-NC,
3.89±0.09, P<0.05) (Fig. 2B). The
results of western blotting were consistent with that of the
RT-qPCR. TSPAN17 protein levels in the con-transfected U87MG (anta
+ si, 2.1±0.16 vs. antagomir, 2.8±0.24, P<0.05; vs si-TSPAN17,
0.28±0.03; P<0.05; vs anta-NC, 1.56±0.23. P<0.05; vs si-NC,
1.51±0.09, P<0.05) and MT-330 (anta+si, 1.9±0.14) (vs antagomir,
2.9±0.17; P<0.05; vs si-TSPAN17, 0.23±0.05; P<0.05; vs
anta-NC, 1.37±0.06. P<0.05; vs si-NC, 1.43±0.04, P<0.05)
cells were restored in part (Fig.
2C). However, the expressions of TSPAN17 didn't return to the
level of negative control (P<0.05, Fig. 2B and C).
Effects of miR-378a-3p/TSPAN17 on
proliferation and apoptosis
Transfection with miR-378a-3p mimic or TSPAN17 siRNA
significantly decreased cell proliferation and increased the
apoptotic rate in U87MG (P<0.05; Fig.
5) and MT-330 cells, compared with the corresponding controls
(P<0.05; Fig. 6). Transfection
with miR-378a-3p antagomir resulted in a significant increase in
cell proliferation and reduced apoptosis in both U87MG (P<0.05;
Fig. 5) and MT-330 cells (P<0.05;
Fig. 6) compared with the
corresponding controls. Co-treatment with miR-378a-3p antagomir and
TSPAN17 siRNA partially attenuated these effects induced by
miR-378a-3p antagomir.
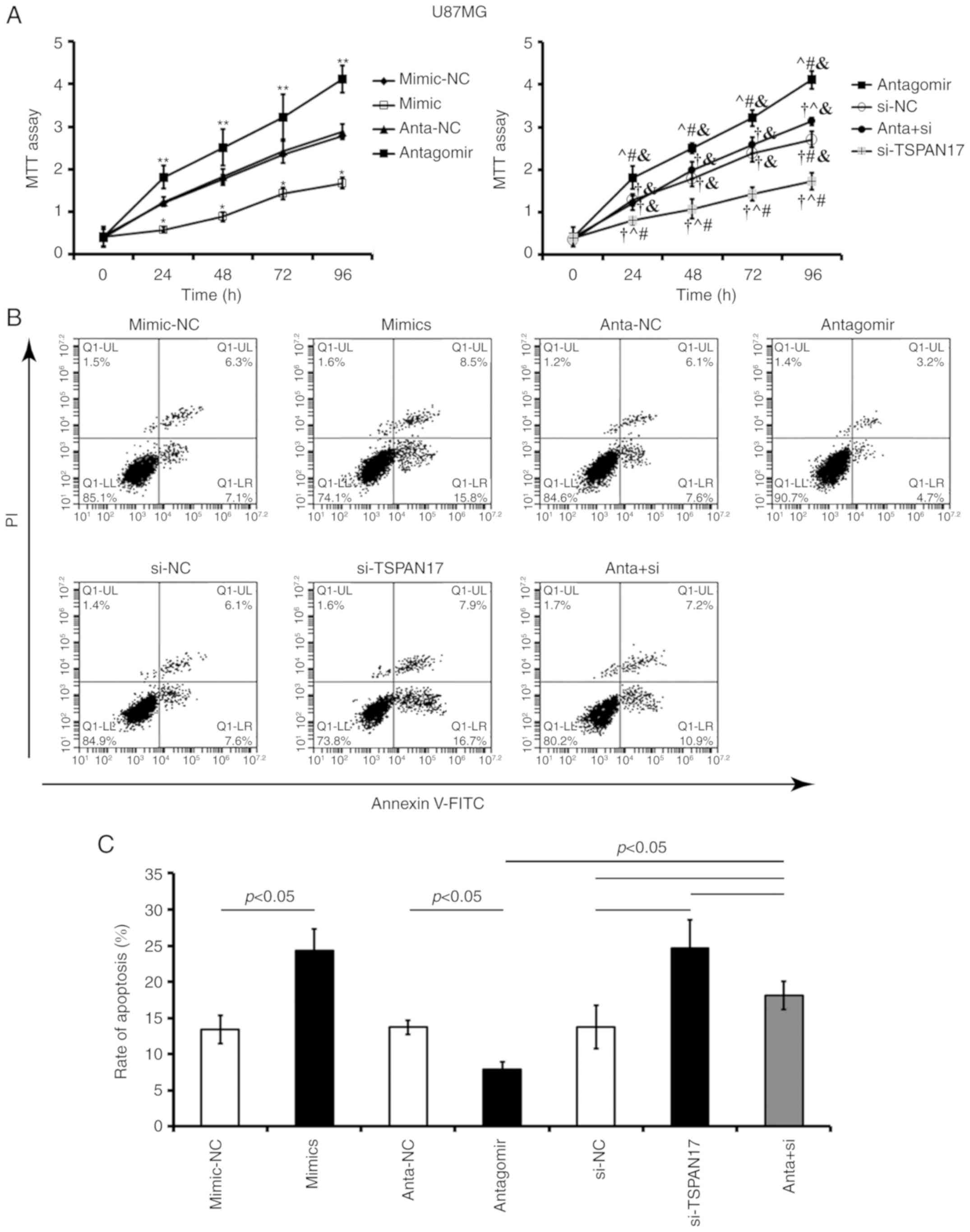 | Figure 5.Effects of miR-378a-3p on the
proliferation and apoptosis of U87MG cells by targeting TSPAN17.
(A) Cell proliferation was analyzed by an MTT assay after
transfection; *P<0.05 vs. the mimic-NC control; **P<0.05 vs.
anta-NC; †P<0.05 vs. antagomir; ^P<0.05 vs. si-NC;
#P<0.05 vs. anta + si; &P<0.05 vs.
si-TSPAN17. (B and C) Apoptosis rates were evaluated by flow
cytometry after transfection. Data are presented as mean ± standard
deviation. All the data are representative of five independent
experiments (n=5). FITC, fluorescein isothiocyanate; PI, propidium
iodide; miR, microRNA; mimic-NC, miR-378a-3p mimic negative
control; mimic, miR-378a-3p mimic; anta-NC, antagomir negative
control; anta, antagomir, miR-378a-3p antagomir; si-NC, small
interfering RNA negative control; TSPAN17, tetraspanin 17;
si-TSPAN17, TSPAN17 small interfering RNA; anta + si,
miR-378a-3p antagomir + TSPAN17 small interfering RNA. |
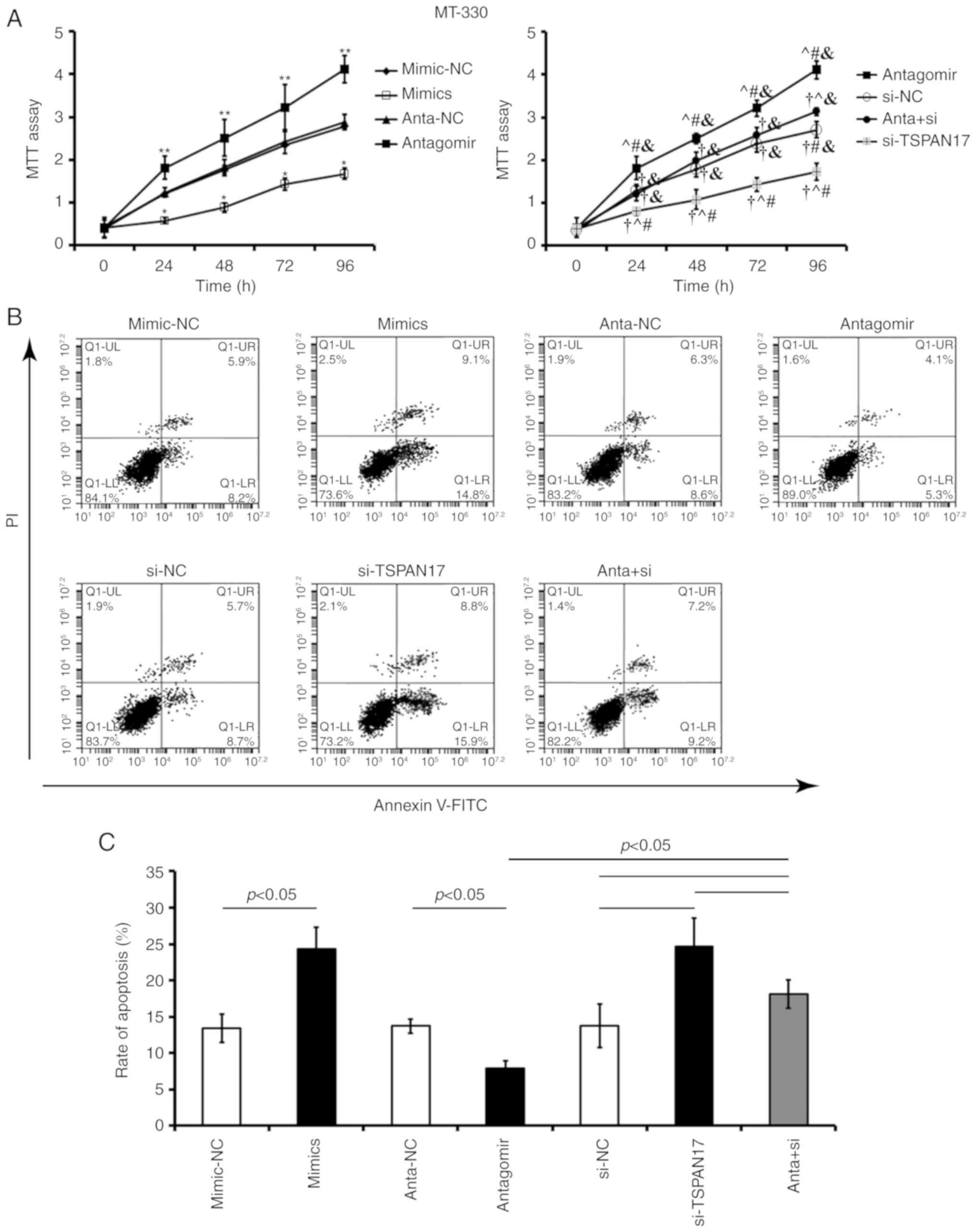 | Figure 6.Effects of miR-378a-3p on the
proliferation and apoptosis of MT-330 cells by targeting TSPAN17.
(A) Cell proliferation was detected by an MTT assay after treatment
various reagents. *P<0.05 vs. the mimic-NC control; **P<0.05
vs. anta-NC; †P<0.05 vs. antagomir;
^P<0.05 vs. si-NC; #P<0.05 vs anta +
si; &P<0.05 vs. si-TSPAN17. (B and C) Apoptosis
rates were evaluated by flow cytometry after transfection. Data are
represented as mean ± standard deviation. All the data are
representative of five independent experiments (n=5). FITC,
fluorescein isothiocyanate; PI, propidium iodide; miR, microRNA;
mimic-NC, miR-378a-3p mimic negative control; mimic, miR-378a-3p
mimic; anta-NC, antagomir negative control; antagomir, miR-378a-3p
antagomir; si-NC, small interfering RNA negative control; TSPAN17,
tetraspanin 17; si-TSPAN17, small interfering RNA; anta +
si, miR-378a-3p antagomir + TSPAN17 small interfering RNA. |
Migration and invasion
The migration assay revealed that there were
significantly fewer cells stained with crystal violet in the
miR-378a-3p mimic-transfected cells, compared with the matched NC
transfected cells. Additionally, transfection with TSPAN17 siRNA
significantly decreased migration and invasion in both U87MG
(Fig. 7) and MT-330 (Fig. 8) cells compared with the respective NC
cells (P<0.05). Furthermore, co-treatment with miR-378a-3p
antagomir and TSPAN17 siRNA attenuated the effects on migration and
invasion induced by miR-378a-3p antagomir transfection (Figs. 7 and 8).
The results showed that overexpression of miR-378a-3p significantly
inhibited migration and invasion in GBM cells by targeting
TSPAN17.
Discussion
The present study showed that miR-378a-3p expression
was downregulated in GBM tissues and cell lines, whereas TSPAN17
expression was upregulated. High levels of miR-378a-3p and low
levels of TSPAN17 expression were linked to better survival in
patients with GBM. Additionally, high expression levels of TSPAN17
were associated with poor prognosis in patients with GBM aged 50–60
years, tumor size and WHO grade. TSPAN17 was identified and
confirmed as a direct target of miR-378a-3p using a luciferase
reporter assay in human glioma cell lines. Overexpression of
miR-378a-3p in both U87MG and MT-330 cells decreased TSPAN17
expression, induced apoptosis, and suppressed proliferation,
migration and invasion. Functional inhibition of miR-378a-3p using
the antagomir in GBM cells increased TSPAN17 expression levels,
reduced apoptosis rates, and promoted proliferation, migration and
invasion. Co-treated with the miR-378a-3p antagomir and
TSPAN17-siRNA attenuated the effects induced by antagomir
transfection alone. These data suggested that miR-378a-3p acted as
a suppressor of GBM by targeting TSPAN17 mRNA.
miR-378a is a small noncoding RNA molecule that can
regulate gene expression at the post-transcriptional level.
miR-378a-3p, one of two mature strands of miR-378a, originates from
the first intron of the peroxisome proliferator-activated
receptor-g, coactivator 1β gene encoding PGC-1β (27–29).
Reports demonstrated that miR-378a-3p regulated skeletal muscle
growth and promoted the differentiation of myoblasts through the
post-transcriptional downregulation of histone acetylation enzyme 4
(30), and was involved in myotonic
dystrophy type-2 (31) and
adipogenesis by targeting mitogen-activated protein kinase 1
(32). miR-378a-3p has multiple roles
in various pathological diseases by post-transcriptionally
modifying its potential targets, including in tuberous sclerosis
(33), liver fibrosis (34), breast cancer (35) and colorectal cancer (36). The results of the present study showed
that: i) The downregulated expression of miR-378a-3p in carcinoma
of patients with GBM and in GBM cells; and ii) high miR-378a-3p
levels were associated with better survival in patients with GBM.
Similar observations have been demonstrated in other studies, which
support the hypothesis that miR-378a-3p may function as a tumor
suppressor in GBM (37,38).
Subsequently, it was identified and confirmed that
TSPAN17 was a direct target of miR-378a-3p, which was associated
with a poor prognosis in patients with GBM aged 50–60 years.
Overexpression of miR-378a-3p in either the U87MG or MT-330 cells
inhibited TSPAN17 expression, induced apoptosis, and suppressed
proliferation, migration and invasion. Functional inhibition of
miR-378a-3p using the antagomir in GBM cells increased the
expression levels of TSPAN17, reduced apoptosis rates, and promoted
migration and invasion. These data suggest that miR-378a-3p may
function as a tumor suppressor in GBM by targeting TSPAN17.
Tetraspanins are a heterogeneous group of
four-transmembrane proteins that segregate into
tetraspanin-enriched micro domains (TEMs), which interact laterally
with each other and different partners, such as integrins,
immunoglobulin-domain-containing proteins, growth factors and
cytokine receptors (39). TEMs of
various types are reportedly involved in the regulation of cell
growth, migration and invasion of several tumor cell types, both as
suppressors and supporting structures (40). Other members of the tetraspanin
family, such as Tspan9, Tspan1 and Tspan5, were demonstrated to
serve possible roles in pathophysiological processes and cancer
biology, which highlight their contribution to tumorigenesis
(41–44). Using co-immunoprecipitation, it has
been demonstrated that A disintegrin and metalloprotease 10
(ADAM10) interacted with TSPAN17, which defined TspanC8
tetraspanins as essential regulators of ADAM10 maturation and
trafficking to the cell surface (45). However, the role of TSPAN17 in gliomas
has not been demonstrated previously, to the best of our knowledge.
The present study illustrated the novel roles of miR-378a-3p in
regulating TSPAN17 expression and thus its involvement in GBM.
Considering that upregulated TSPAN17 expression in
GBM tissues and cell lines was associated with the poor prognosis
of patients and malignant features of GBM cells, we proposed that
TSPAN17 upregulation in normal cell lines might greatly increase
the risk of glioma oncogenesis and development. However, further
in vivo evaluation is necessary to validate the results
obtained from this study. For example, a glioma xenograft nude
mouse model could be developed by intradermal injection with
TSPAN17 overexpressed/downregulated HF cells or glioma cells in the
future. After seeding the tumor cells, tumor growth, and mouse
survival may be measured to further confirm the potential effects
of TSPAN17 in tumorigenesis of GBM. Additionally, as of the
relatively small sample size (n=53) employed in this study,
external validation is required in further investigation. We should
use public datasets, taking The Cancer Genome Atlas GBM as an
example, to examine any consistencies or variations from the
results of our study.
In conclusion, in the present study, it was
demonstrated that: i) Upregulation of TSPAN17 was associated with a
poor prognosis in patients with GBM aged 50–60 years; ii) TSPAN17
was identified and confirmed as a direct target of miR-378a-3p by a
luciferase report assay; and iii) overexpression of miR-378a-3p in
both U87MG and MT-330 cells inhibited TSPAN17 expression, induced
apoptosis and suppressed proliferation, migration and invasion, and
functional inhibition of miR-378a-3p by antagomir reversed these
effects. TSPAN17-siRNA transfection also reversed the effects
induced by miR-378a-3p antagomir transfection in GBM cells. The
results highlight the potential role of miR-378a-3p as a suppressor
of GBM by targeting TSPAN17.
Acknowledgements
We thank Doctor Jing Wu for his valuable work in
tumor specimen collection.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant nos. 81660613, 81160401,
81260493 and 30660212); the Natural Science Foundation of Yunnan
Province (grant nos. 2008CC009, 2013FB103 and 2015FA021); the Joint
Special Funds for the Department of Science and Technology of
Yunnan Province-Kunming Medical University (grant no. K13201234);
and the Science Foundation of Yun-nan Provincial Education Bureau
(grant no. 2015Y154 and 2017zDX164).
Availability of data and materials
The datasets used and analyzed in the present study
are available from the corresponding author on reasonable
request.
Authors' contributions
XBG, ZQS, and XCZ designed this study and wrote the
manuscript. LMM, XBG, ZQS, and XCZ conducted the statistical
analysis. PC and XCZ performed tumor tissues preparation and cell
culture. XBG, ZQS, PC and LMM performed cell experiments, RT-qPCR,
the luciferase reporter assay and western blotting. XBG, XCZ and
ZQS performed the MTT assay and flow cytometry analysis. PC and LMM
performed the migration and invasion assays. All authors have read
and approved the final manuscript.
Ethics approval and consent to
participate
All procedures performed in studies involving human
participants were in accordance with the 1964 Helsinki declaration
and its later amendments or comparable ethical standards. This
study was approved by The Medical Ethics Committee of Kunming
Medical University (Kunming, China). GBM tumor specimens were
obtained from patients at The First People's Hospital of Yunnan
Province (Yunnan, China) who provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no acompeting
interests.
References
|
1
|
Fisher JL, Schwartzbaum JA, Wrensch M and
Wiemels JL: Epidemiology of brain tumors. Neurol Clin. 25:867–890.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bondy ML, Scheurer ME, Malmer B,
Barnholtz-Sloan JS, Davis FG, Il'yasova D, Kruchko C, McCarthy BJ,
Rajaraman P, Schwartzbaum JA, et al: Brain tumor epidemiology:
Consensus from the brain tumor epidemiology consortium. Cancer. 113
(Suppl 7):S1953–S1968. 2008. View Article : Google Scholar
|
|
3
|
Feng J, Kim ST, Liu W, Kim JW, Zhang Z,
Zhu Y, Berens M, Sun J and Xu J: An integrated analysis of germline
and somatic, genetic and epigenetic alterations at 9p21.3 in
glioblastoma. Cancer. 118:232–240. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Caserta S, Kern F, Cohen J, Drage S,
Newbury SF and Llewelyn MJ: Circulating plasma microRNAs can
differentiate human sepsis and systemic inflammatory response
syndrome (SIRS). Sci Rep. 6:280062016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
McClure C, McPeak MB, Youssef D, Yao ZQ,
McCall CE and El Gazzar M: Stat3 and C/EBPβ synergize to induce
miR-21 and miR-181b expression during sepsis. Immunol Cell Biol.
95:42–55. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Rolle K: miRNA Multiplayers in glioma:
From bench to bedside. Acta Biochim Pol. 62:353–365. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Browne G, Dragon JA, Hong D, Messier TL,
Gordon JA, Farina NH, Boyd JR, VanOudenhove JJ, Perez AW, Zaidi SK,
et al: MicroRNA-378-mediated suppression of Runx1 alleviates the
aggressive phenotype of triple-negative MDA-MB-231 human breast
cancer cells. Tumour Biol. 37:8825–8839. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mirzaei H, Khataminfar S, Mohammadparast
S, Sales SS, Maftouh M, Mohammadi M, Simonian M, Parizadeh SM,
Hassanian SM and Avan A: Circulating microRNAs as potential
diagnostic biomarkers and therapeutic targets in gastric cancer:
Current status and future perspectives. Curr Med Chem.
23:4135–4150. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Peng J, Xie Z, Cheng L, Zhang Y, Chen J,
Yu H, Li Z and Kang H: Paired design study by real-time PCR:
miR-378* and miR-145 are potent early diagnostic biomarkers of
human colorectal cancer. BMC Cancer. 15:1582015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Scapoli L, Palmieri A, Lo Muzio L,
Pezzetti F, Rubini C, Girardi A, Farinella F, Mazzotta M and
Carinci F: MicroRNA expression profiling of oral carcinoma
identifies new markers of tumor progression. Int J Immunopathol
Pharmacol. 23:1229–1234. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhu XW, Wen XM, Zhang YY, Yang L, Guo H,
Yang J, Zhang M, Yin JY, Ma JC, Lin J, et al: The 5′flanking region
of miR-378 is hypomethylated in acute myeloid leukemia. Int J Clin
Exp Pathol. 8:4321–4331. 2015.PubMed/NCBI
|
|
13
|
Li B, Wang Y, Li S, He H, Sun F, Wang C,
Lu Y, Wang X and Tao B: Decreased expression of miR-378 correlates
with tumor invasiveness and poor prognosis of patients with glioma.
Int J Clin Exp Pathol. 8:7016–7021. 2015.PubMed/NCBI
|
|
14
|
Arita H, Yamasaki K, Matsushita Y,
Nakamura T, Shimokawa A, Takami H, Tanaka S, Mukasa A, Shirahata M,
Shimizu S, et al: A combination of TERT promoter mutation and MGMT
methylation status predicts clinically relevant subgroups of newly
diagnosed glioblastomas. Acta Neuropathol Commun. 4:792016.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Turkalp Z, Karamchandani J and Das S: IDH
mutation in glioma: New insights and promises for the future. JAMA
Neurol. 71:1319–1325. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mulholland S, Pearson DM, Hamoudi RA,
Malley DS, Smith CM, Weaver JM, Jones DT, Kocialkowski S, Bäcklund
LM, Collins VP and Ichimura K: MGMT CpG island is invariably
methylated in adult astrocytic and oligodendroglial tumors with
IDH1 or IDH2 mutations. Int J Cancer. 131:1104–1113. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Okita Y, Narita Y, Miyakita Y, Ohno M,
Matsushita Y, Fukushima S, Sumi M, Ichimura K, Kayama T and Shibui
S: IDH1/2 mutation is a prognostic marker for survival and predicts
response to chemotherapy for grade II gliomas concomitantly treated
with radiation therapy. Int J Oncol. 41:1325–1336. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Arita H, Narita Y, Matsushita Y, Fukushima
S, Yoshida A, Takami H, Miyakita Y, Ohno M, Shibui S and Ichimura
K: Development of a robust and sensitive pyrosequencing assay for
the detection of IDH1/2 mutations in gliomas. Brain Tumor Pathol.
32:22–30. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Arita H, Narita Y, Fukushima S, Tateishi
K, Matsushita Y, Yoshida A, Miyakita Y, Ohno M, Collins VP,
Kawahara N, et al: Upregulating mutations in the TERT promoter
commonly occur in adult malignant gliomas and are strongly
associated with total 1p19q loss. Acta Neuropathol. 126:267–276.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Skup M, Dwornik A, Macias M, Sulejczak D,
Wiater M and Czarkowska-Bauch J: Long-term locomotor training
upregulates TrkBFL receptor-like proteins, brain-derived
neurotrophic factor, and neurotrophin 4 with different topographies
of expression in oligodendroglia and neurons in the spinal cord.
Exp Neurol. 176:289–307. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Minemura H, Takagi K, Miki Y, Shibahara Y,
Nakagawa S, Ebata A, Watanabe M, Ishida T, Sasano H and Suzuki T:
Abnormal expression of miR-1 in breast carcinoma as a potent
prognostic factor. Cancer Sci. 106:1642–1650. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hu T, Li YS, Chen B, Chang YF, Liu GC,
Hong Y, Chen HL and Xiyang YB: Elevated glucose-6-phosphate
dehydrogenase expression in the cervical cancer cases is associated
with the cancerigenic event of high-risk human papillomaviruses.
Exp Biol Med (Maywood). 240:1287–1297. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liang L, Wong CM, Ying Q, Fan DN, Huang S,
Ding J, Yao J, Yan M, Li J, Yao M, et al: MicroRNA-125b suppressed
human liver cancer cell proliferation and metastasis by directly
targeting oncogene LIN28B2. Hepatology. 52:1731–1740. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
He Z, Xia Y, Liu B, Qi X, Li Z, Wang J,
Chen L and Chen Y: Down-regulation of miR-452 is associated with
poor prognosis in the non-small-cell lung cancer. J Thorac Dis.
8:894–900. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li Z, Piao YS, Zhang LY, Wang LM, Wang DD,
Fu YJ, Cai YN and Lu DH: Application of ATRX in diagnosis and
prognostic evaluation of glioma. Zhonghua Bing Li Xue Za Zhi.
46:690–694. 2017.(In Chinese). PubMed/NCBI
|
|
27
|
Krist B, Florczyk U,
Pietraszek-Gremplewicz K, Józkowicz A and Dulak J: The role of
miR-378a in metabolism, angiogenesis, and muscle biology. Int J
Endocrinol. 2015:2817562015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Eichner LJ, Perry MC, Dufour CR, Bertos N,
Park M, St-Pierre J and Giguère V: miR-378(∗) mediates
metabolic shift in breast cancer cells via the PGC-1β/ERRγ
transcriptional pathway. Cell Metab. 12:352–361. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kasashima K, Nakamura Y and Kozu T:
Altered expression profiles of microRNAs during TPA-induced
differentiation of HL-60 cells. Biochem Biophys Res Commun.
322:403–410. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wei X, Li H, Zhang B, Li C, Dong D, Lan X,
Huang Y, Bai Y, Lin F, Zhao X and Chen H: miR-378a-3p promotes
differentiation and inhibits proliferation of myoblasts by
targeting HDAC4 in skeletal muscle development. RNA Biol.
13:1300–1309. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Greco S, Perfetti A, Fasanaro P, Cardani
R, Capogrossi MC, Meola G and Martelli F: Deregulated microRNAs in
myotonic dystrophy type 2. PLoS One. 7:e397322012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Huang N, Wang J, Xie W, Lyu Q, Wu J, He J,
Qiu W, Xu N and Zhang Y: miR-378a-3p enhances adipogenesis by
targeting mitogen-activated protein kinase 1. Biochem Biophys Res
Commun. 457:37–42. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Trelinska J, Fendler W, Dachowska I,
Kotulska K, Jozwiak S, Antosik K, Gnys P, Borowiec M and Mlynarski
W: Abnormal serum microRNA profiles in tuberous sclerosis are
normalized during treatment with everolimus: Possible clinical
implications. Orphanet J Rare Dis. 11:1292016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hyun J, Wang S, Kim J, Rao KM, Park SY,
Chung I, Ha CS, Kim SW, Yun YH and Jung Y: MicroRNA-378 limits
activation of hepatic stellate cells and liver fibrosis by
suppressing Gli3 expression. Nat Commun. 7:109932016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ikeda K, Horie-Inoue K, Ueno T, Suzuki T,
Sato W, Shigekawa T, Osaki A, Saeki T, Berezikov E, Mano H and
Inoue S: miR-378a-3p modulates tamoxifen sensitivity in breast
cancer MCF-7 cells through targeting GOLT1A. Sci Rep. 5:131702015.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kara M, Yumrutas O, Ozcan O, Celik OI,
Bozgeyik E, Bozgeyik I and Tasdemir S: Differential expressions of
cancer-associated genes and their regulatory miRNAs in colorectal
carcinoma. Gene. 567:81–86. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Megiorni F, Cialfi S, McDowell HP, Felsani
A, Camero S, Guffanti A, Pizer B, Clerico A, De Grazia A, Pizzuti
A, et al: Deep sequencing the microRNA profile in rhabdomyosarcoma
reveals down-regulation of miR-378 family members. BMC Cancer.
14:8802014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Li H, Dai S, Zhen T, Shi H, Zhang F, Yang
Y, Kang L, Liang Y and Han A: Clinical and biological significance
of miR-378a-3p and miR-378a-5p in colorectal cancer. Eur J Cancer.
50:1207–1221. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Masse I, Agaësse G and Berthier-Vergnes O:
Tetraspanins in cutaneous physiopathology. Med Sci (Paris).
32:267–273. 2016.(In French). View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Hölters S, Anacker J, Jansen L,
Beer-Grondke K, Dürst M and Rubio I: Tetraspanin 1 promotes
invasiveness of cervical cancer cells. Int J Oncol. 43:503–512.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
He P, Wang S, Zhang X, Gao Y, Niu W, Dong
N, Shi X, Geng Y, Ma Q, Li M, et al: Tspan5 is an independent
favourable prognostic factor and suppresses tumour growth in
gastriccancer. Oncotarget. 7:40160–40173. 2016.PubMed/NCBI
|
|
42
|
Yang YG, Sari IN, Zia MF, Lee SR, Song SJ
and Kwon HY: Tetraspanins: Spanning from solid tumors to
hematologic malignancies. Exp Hematol. 44:322–338. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wang XF, Gao GD, Liu J, Guo R, Lin YX, Chu
YL, Han FC, Zhang WH and Bai YJ: Identification of differentially
expressed genes induced by angiotensin II in rat cardiac
fibroblasts. Clin Exp Pharmacol Physiol. 33:41–46. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Hou FQ, Lei XF, Yao JL, Wang YJ and Zhang
W: Tetraspanin 1 is involved in survival, proliferation and
carcinogenesis of pancreatic cancer. Oncol Rep. 34:3068–3076. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Haining EJ, Yang J, Bailey RL, Khan K,
Collier R, Tsai S, Watson SP, Frampton J, Garcia P and Tomlinson
MG: The TspanC8 subgroup of tetraspanins interacts with A
disintegrin and metalloprotease 10 (ADAM10) and regulates its
maturation and cell surface expression. J Biol Chem.
287:39753–39765. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zhang YA, Zhou Y, Luo X, Song K, Ma X,
Sathe A, Girard L, Xiao G and Gazdar AF: SHOX2 is a potent
independent biomarker to predict survival of WHO grade II–III
diffuse gliomas. EBioMedicine. 13:80–89. 2016. View Article : Google Scholar : PubMed/NCBI
|






















