Introduction
Colorectal cancer (CRC) is the third leading cause
of cancer-related mortality worldwide, with ~1.7 million newly
diagnosed CRC cases and 800,000 deaths annually (1,2). The most
common treatment for CRC is surgery, which has a high success rate
when patients are diagnosed early; however, the 5-year overall
survival rate remains unsatisfactory, as the majority of CRC
patients are diagnosed at an advanced stage. Consequently,
identifying novel biomarkers and therapeutic targets in CRC is
crucial.
MicroRNAs (miRNAs), which contain 22–24 nucleotides,
are small non-coding RNAs that affect a number of physiological
events (3), often by binding to the
3′-untranslated region (3′UTR) of target genes, thus inhibiting or
causing variations in mRNA transcripts (4–6). The
occurrence and progression of several tumor types have been found
to be associated with abnormal miRNA expression (7–11).
Reportedly, aberrant miRNA-203a-3p expression has been detected in
numerous cancers (12–18); however, its role and mechanism of
action in CRC remain elusive. Therefore, the present study was
conducted to investigate miRNA-203a-3p expression in CRC, and
elucidate the mechanism underlying the inhibition of apoptosis and
promotion of metastasis in CRC.
THBS2 affects interactions between cells and is a
potential tumor suppressor (19–21).
Accumulating evidence indicates that THBS2 is associated with CRC
(22,23). THBS2, secreted by stromal fibroblasts,
endothelial cells and immune cells, and belongs to the THBS family
of proteins, was identified in 1991 and its sequence was analyzed
in 1997 (24,25). Subsequently, it was found to be
associated with various cancer types (22,26–30). The
aim of the present study was to investigate the expression of THBS2
in CRC tissues, and determine its link with the overall survival
(OS) and disease-free survival (DFS) in patients with CRC. We
evaluated the associations among miRNA-203a-3p expression, THBS2
expression and CRC progression, in order to determine whether
miRNA-203a-3p and THBS2 may be used as biomarkers and therapeutic
targets in patients with CRC.
Materials and methods
Clinical sample collection
We collected 59 sets of cancer tissues and
corresponding adjacent non-tumor tissues that were resected from 26
female and 33 male CRC patients (aged 31–78 years) at the Zhejiang
Provincial People's Hospital (China) between October 2016 and May
2017. None of the patients received preoperative radiation or
chemotherapy. The study protocol was approved by the Ethics
Committee of Zhejiang Provincial People's Hospital. Written
informed consent was obtained from all patients prior to
participation in the study.
Cell culture
The human CRC cell lines HT29, HCT15, SW480 and
SW620, and the normal human colon cell line NCM460, were purchased
from the Cell Bank of Shanghai Institute of Cell Biology. All the
cells were maintained in RPMI-1640 or minimal essential media (MEM;
HyClone; GE Healthcare Life Sciences) containing 10% fetal bovine
serum (FBS) (Biowest SAS France), and cultured in a 5%
CO2 incubator at 37°C. Cells were passaged at 75%
confluence with 0.02% EDTA/0.25% trypsin.
Total RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
analysis
Total cell RNA was extracted from fresh specimens
and cells with TRIzol® (Invitrogen; Thermo Fisher
Scientific, Inc.) in accordance with the manufacturer's
instructions. RT was performed with SYBR Premix Ex Taq according to
manufacturer's protocols (Takara Bio, Inc.). RNU6B and GAPDH were
used as endogenous controls. miRNA-203a-3p was reverse-transcribed
using the following stem-loop RT primer:
5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGTTGAA-3′. qPCR was
performed using FastStart Essential DNA Green Master (Roche
Diagnostics) with miRNA-specific primers (forward,
5′-GUGAAAUGUUUAGGACCACUAG3′ and reverse,
5′-AGUGGUCCUAAACAUUUCACUU-3′; U6, forward,
5′-ATTGGAACGATACAGAGAAGATT-3′ and reverse,
5′-GGAACGCTTCACGAATTTG-3′). GAPDH, forward,
5′-ATCGTCCACCGCAAATGCTTCTA-3′ and reverse,
5′-AGCCATGCCAATCTCATCTTGTT-3′. THBS2, forward,
5′-CGTGGACAATGACCTTGTTG-3′ and reverse, 5′-GCCATCGTTGTCATCATCAG-3′.
The reaction ran on the ABI 7900HT Sequence Detection System
(Applied Biosystems; Thermo Fisher Scientific, Inc.) in the
presence of SYBR-Green dye (Toyobo Life Science). qPCR was
conducted as follows: 95°C for 10 min, 40 cycles at 95°C for 10
sec, 60°C for 30 sec and 72°C for 10 sec. Relative expression
levels were calculated using the 2−ΔΔCq method.
Transfection assay
Prior to transfection, cells were plated
(3.0×105 cells per well) and cultured at 37°C in 6-well
dishes (15×104 cells per well) for 20 h. miRNA-203a-3p
mimics (Guangzhou RiboBio Co., Ltd.) were transfected into SW480
cells and HT29 cells, which have a relatively low expression of
miRNA-203a-3p compared with the normal colonic cells NCM460 and
other CRC cell lines. The negative control group (Guangzhou RiboBio
Co., Ltd.) was set up in parallel. Transfection of each siRNA (50
nM) was conducted with Lipofectamine® 3000 (Thermo
Fisher Scientific, Inc.) according to the manufacturer's
instructions (miR-203a-3p mimic, forward,
5′-GUGAAAUGUUUAGGACCACUAG-3′ and reverse,
5′-AGUGGUCCUAAACAUUUCACUU-3′; scramble miRNA,
5′-CAGUACUUUUGUGUAGUACAA-3′). The cells were collected for the
following experiments after 48 h of transfection.
Transwell assay
The CRC cells were cultured for 24 h after
transfection. Migration and invasion assays were performed with a
Transwell assay kit (Corning, Inc.) and invasion chambers (Corning,
Inc.). Transfected cells (6×104/well for migration and
1×105/well for invasion assays) were plated in the upper
chamber, which contained FBS-free MEM; the lower chamber contained
MEM supplemented with 10% FBS. After 48 h, cells that had migrated
or invaded through the membrane were fixed with methanol and
stained with 0.1% crystal violet in 5% CO2 at 37°C for
15 min. The cells were photographed under a phase-contrast
microscope (Olympus Corporation).
Apoptosis assay
At 24 h after transfection with miRNA- 203a-3p
mimics or negative controls, the cells were washed with PBS and
fixed with 70% ethanol for >12 h at 4°C. Propidium iodide (PI)
staining solution (500 µl) was then added to the centrifuged cells
(845 × g, 3 min) at room temperature, followed by incubation for 30
min in the dark at room temperature. Cell apoptosis was analyzed by
FACSCalibur flow cytometry (BD Biosciences). The percent of
apoptotic cells was obtained from FACSCalibur flow cytometry (BD
Biosciences) which was used for further calculation. The Annexin
V/PI Apoptosis Detection Kit (Beijing Solarbio Science &
Technology, Co., Ltd.) was used to assess apoptosis.
Western blot analysis
At 48 h after transfection, the cells were
harvested, washed and lysed with lysis buffer (Nanjing KeyGen
Biotech Co., Ltd.). The proteins were were quantified using BCA kit
(Beyotime Institute of Biotechnology) and were separated with 12%
SDS-PAGE (20 µg/lane), and then transferred to microporous
membranes (EMD Millipore). The membranes were blocked with
Tris-buffered saline with 3% bovine serum albumin (Solarbio Science
& Technology Co., Ltd.) for 1 h at room temperature and
incubated with primary antibodies against THBS2 (1:500; PA5-80123;
Thermo Fisher Scientific, Inc.) and GAPDH (1:1,000; 10494-1-AP;
ProteinTech Group, Inc.) for 1 h at room temperature. The
corresponding HRP-conjugated secondary antibody was applied at a
1:2,000 dilution after the primary antibodies. The membranes were
evaluated using the Chemi Doc™ XRS+ imaging system (Bio-Rad
Laboratories, Inc., Hercules, CA, USA).
Luciferase reporter assay
The Renilla vector was established by Ruibo
Biotechnology Co., Ltd. HT29 cells were added to 96-well plates and
cultured for 24 h. Renilla was used for normalization. The
cells were transfected with THBS2-3′UTR-wild-type (WT) or
THBS2-3′UTR-mutant (mut) and miR-203a-3p or miR-control vectors
using Lipofectamine 3000. After 48 h, luciferase activity was
evaluated by Dual-Luciferase Reporter Assay reagent (Promega
Corporation).
Cell proliferation analysis
proliferative ability was detected with A Cell
Counting Kit-8 (CCK-8) on the manufacturer's instructions (Dojindo
Molecular Technologies, Inc., Kumamoto, Japan). At 24, 48 and 72 h
following transfection, 2×103 cells/well were seeded
into 96-well plates and 10 µl of CCK-8 solution was added to assess
cell viability. The optical density (OD) was measured using a
microplate reader (Molecular Devices LLC) at an absorbance of 450
nm.
Immunohistochemistry
The paraffin-embedded tissue specimens were cut into
5-µm sections. After deparaffinization, antigens were retrieved
with 0.01 M citrate buffer (pH 6.0) and treated with 3%
H2O2 for 10 min at room temperature. The
sections were incubated with primary antibody against THBS2 (1:500,
PA5-80123; Thermo Fisher Scientific, Inc.) overnight at 4°C and
then treated with corresponding HRP-conjugated secondary antibody
(1:2,000) for 1 h at room temperature. After dehydration, the
sections were each covered with a single slide. Images were
captured with the NanoZoomer Digital Pathology 2.0RS (Hamamatsu
Photonics K.K.) and analyzed with NDP.view, version 2.7.25
(Hamamatsu Photonics K.K.). Upright microscope was used in these
experiments and the magnification is 200 times.
Bioinformation analysis
We predict the target gene of miRNA with TargetScan
(version 5.0; http://genes.mit.edu/targetscan). The level of THBS2
mRNA in the adjacent normal colonic mucosal tissues and CRC tissues
and the Kaplan-Meier survival curve analysis of THBS2 in CRC
patients in The Cancer Genome Atlas (TCGA) were analyzed with GEPIA
(http://gepia.cancer-pku.cn/).
Statistical analysis
Statistical analysis was conducted using SPSS
software version 22.0 (IBM Corp.). Data were presented as the mean
± standard deviation of experiments repeated in triplicate.
Significance between groups was analyzed with a Student's t-test.
The correlation between miR-203a-3p and THBS2 expression was
examined using Pearson's correlation analysis. Survival analyses
were conducted using the Kaplan-Meier method and differences in
survival were examined using the log-rank test. P<0.05 was
considered to indicate a significant.
Results
miRNA-203a-3p expression in CRC
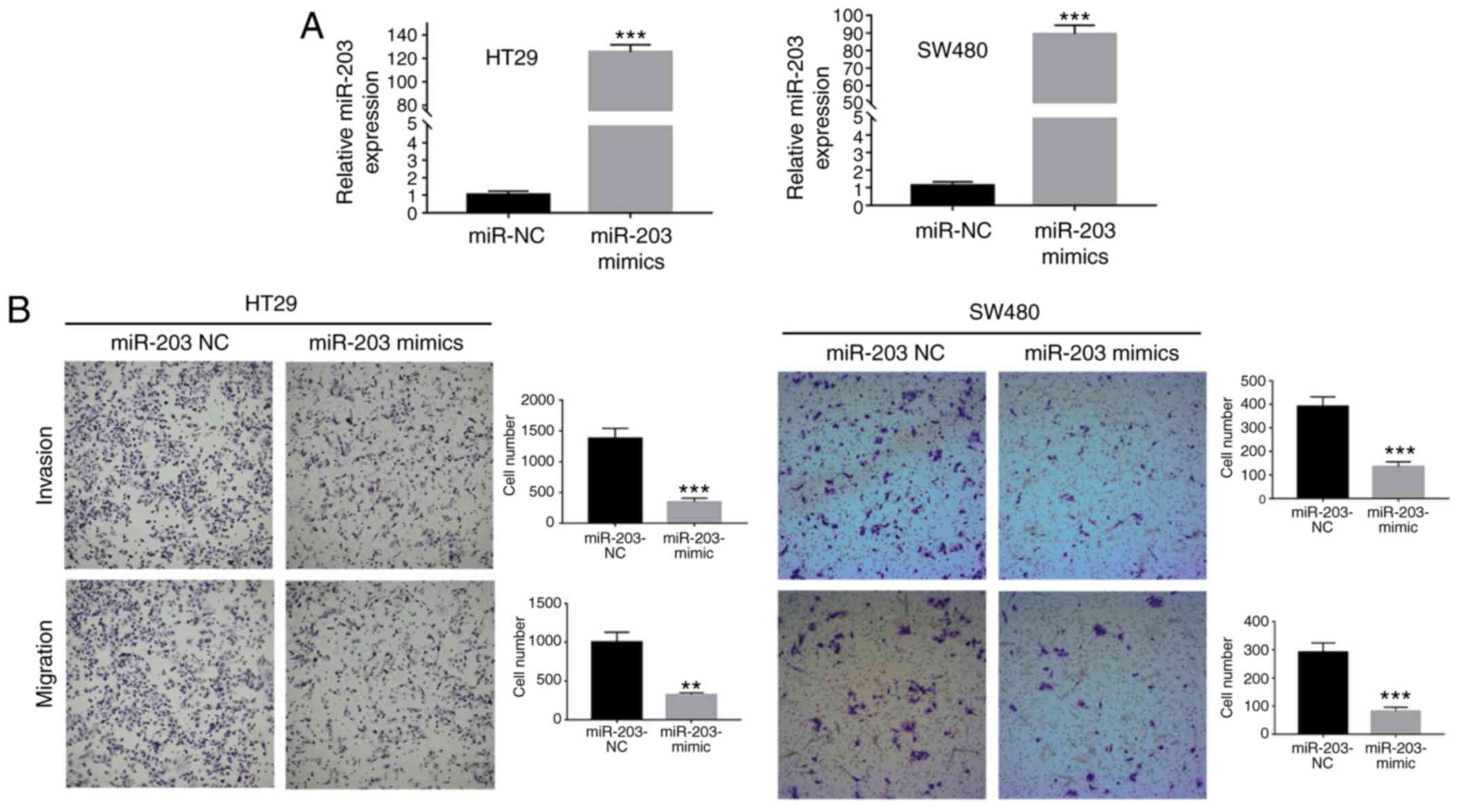
The expression of microRNA-203a-3p was found to be
significantly lower in the four CRC cell lines (SW480, SW620, HCT15
and HT29) compared with that in the NCM460 human colonic mucosal
epithelial cell line (Fig. 1A). Among
the CRC cell lines, HCT15 exhibited a relatively high level of
miRNA-203a-3p expression. The expression of miRNA-203a-3p in 59
paired CRC and adjacent normal colonic mucosal tissues was detected
by RT-qPCR, and was observed to be significantly downregulated in
CRC tissues compared with paired normal tissues (Fig. 1B and C).
miRNA-203-3p affects the invasion and
migration potentials of CRC cells
In order to certify the function of miRNA-203a-3p,
RT-qPCR was performed to identify the cell lines with lower
expression levels of miR-203a-3p. In these cell lines, mimics can
effectively activate gene expression. As a result, the HT29 and
SW480 cell lines, with lower expression of miRNA-203a-3p (Fig. 1), were employed for subsequent
analysis. The numbers of SW480 and HT29 cells that invaded and
migrated across the Transwell membrane were significantly lower for
those transfected with miRNA-203a-3p mimic compared with the
negative control group (Fig. 2).
These data indicate that miRNA-203a-3p affects the invasion and
migration potentials of CRC cells.
miRNA-203a-3p affects the apoptosis of
CRC cells
miRNA-203a-3p overexpression in HT29 cells was shown
to significantly reduce proliferation compared with the control
(Fig. 3A). To further investigate the
role of miRNA-203a-3p in CRC, HT29 cells were transfected with
miRNA-203a-3p mimics. Flow cytometry analysis indicated a
significant increase in the apoptotic rate of HT29 cells
transfected with miRNA-203a-3p mimics than the control (Fig. 3B).
THBS2 expression in CRC tissues
The protein expression of THBS2 in CRC tissues and
paired adjacent normal tissues was detected by immunohistochemical
staining, and was found to be notably upregulated in CRC tissues
compared with that in the adjacent tissues (Fig. 4A). Furthermore, the mRNA levels of
THBS2 in CRC tissues was significantly higher compared with that in
the adjacent normal colonic mucosal tissues in TCGA (Fig. 4B). Kaplan-Meier survival curve
analysis demonstrated that the OS and DFS of CRC patients with
higher THBS2 expression were significantly shorter compared with
those of patients with lower THBS2 levels in TCGA (log-rank test,
P<0.05; Fig. 4C).
miR-203a-3p suppresses tumor growth
and metastasis by epithelial-to-mesenchymal transition and
upregulates the expression of Bcl-2-associated X protein (BAX)
From western blot analysis, it was determined that
HT29 cells transfected with miR-203a-3p exhibited significantly
higher expression levels of E-cadherin, and BAX and lower
expression of N-cadherin and vimentin compared with the control
(Fig. 5).
miR-203a-3p target-gene luciferase
reporter assay
The TargetScan Release 5.0 database predicted that
THBS2 was a downstream target of miR-203a-3p, and revealed
miR-203a-3p target sites within the 3′UTR of THBS2 mRNA (Fig. 6A). A negative correlation was also
observed between miR-203a-3p and THBS2 expression in TCGA (Fig. 6B). To verify whether miR-203a-3p binds
directly to the 3′UTR region of THBS2, two groups of luciferase
reporter constructs were used. Subsequently, cells were
co-transfected with miR-203a-3p mimics and two luciferase reporter
constructs, one group in the presence of the wild-type (THBS2-wt)
3′UTR, and the other in the presence of the mutant (THBS2-mut)
3′UTR. Luciferase activity in the wt group was significantly lower
compared with that in the control group; however, the activity in
the mutant luciferase reporter group was markedly unaffected
(Fig. 6C). These data indicate that
the THBS2 3′UTR contains a specific miR-203a-3p target site. In
HT29 cells, overexpression of miR-203a-3p following transfection
with miR-203a-3p mimics caused downregulation of THBS2 expression
at the protein level (Fig. 6D). These
results indicated that THBS2 is a target gene of miR-203a-3p.
Discussion
Accumulating evidence has revealed associations
between miRNAs and a wide range of tumors (31,32) and,
more specifically, between aberrant miRNA-203a-3p expression and
tumorigenesis (13–15). miRNA-203a-3p serves different roles in
a variety of cancers, depending on the type of the tumor and the
targeted genes.
In the present study, we found that the expression
of miRNA-203a-3p in CRC tissues was significantly downregulated
compared with that in paired normal tissues, which indicates that
miRNA-203a-3p plays a tumor-suppressive role in human CRC.
miRNA-203a-3p overexpression was also found to suppress metastatic
ability in our study, further confirming that miRNA-203a-3p serves
an important role in CRC metastasis. It was also demonstrated that
miRNA-203a-3p affected CRC cell apoptosis.
To explore the function of miRNA-203a-3p in CRC and
to determine the underlying mechanism, the identification of
regulatory targets is crucial. Our western blot results indicated
that miRNA-203a-3p promotes tumor metastasis and growth.
Overexpression of miRNA-203a-3p expression led to downregulation of
N-cadherin, vimentin and the increased expression of E-cad and Bax,
which may cause tumor progression. By reviewing miRNA databases,
THBS2 was identified as a candidate downstream target of
miRNA-203a-3p. THBS2 is secreted by stromal fibroblasts,
endothelial cells and immune cells, and it belongs to the THBS
family of proteins (24). THBS2
affects interactions between cells and is a potent tumor suppressor
with a role in CRC (22,23). THBS2 was found to be significantly
upregulated in CRC tissues, and it was demonstrated that its
expression was negatively correlated with CRC prognosis.
To verify whether miRNA-203a-3p binds directly to
the 3′UTR region of THBS2, two groups of luciferase reporter
constructs were used. Subsequently, cells were co-transfected with
miRNA-203a-3p mimics and the two luciferase reporter constructs,
one group in the presence of THBS2-wt 3′UTR, and the other
in the presence of the THBS2-mut 3′UTR. Luciferase activity
in the wt group was found to be significantly lower compared with
that in the control group; however, activity in the mutant THBS2
luciferase reporter group was markedly unaffected. These data
indicate that the THBS2 3′UTR contains a specific
miRNA-203a-3p target site. Combining those results with our
findings that miRNA-203a-3p suppresses proliferation, apoptosis and
metastasis of CRC cells, it may be concluded that miRNA-203a-3p
exerts a suppressive effect on CRC cells through the suppression of
THBS2. In the future, we aim to employ an animal model to confirm
the results of our in vitro analysis.
In summary, the present study provided evidence that
miRNA-203a-3p is downregulated in CRC tissues and cell lines, and
affects CRC metastasis by targeting THBS2. These findings suggest
that miRNA-203a-3p may act as a tumor suppressor in CRC.
Acknowledgements
Not applicable.
Funding
The present study was financed by Grant-in-aid for
scientific research from the Zhejiang Provincial Natural Science
Foundation for the Youth of China (grant no. LQ18H160023).
Availability of materials and data
The datasets generated and analyzed in the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
SZ made substantial contributions to the study. ZQ,
LG, YM and YH performed the experiments and analyzed the data. ZQ
and LG wrote the manuscript. SZ and ZQ helped to revise the
manuscript. All the authors have read and approved the final
version of this manuscript for publication.
Ethics approval and consent to
participate
All the patients who provided the CRC tissue or
other tissue had signed informed consent forms prior to surgery.
The use of tissues in the experiment was approved by the Ethics
committee of Zhejiang Provincial People's Hospital.
Patient consent for publication
Not applicable.
Competing interests
All authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Fidler MM, Gupta S, Soerjomataram I,
Ferlay J, Steliarova-Foucher E and Bray F: Cancer incidence and
mortality among young adults aged 20–39 years worldwide in 2012: A
population-based study. Lancet Oncol. 18:1579–1589. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ameres SL and Zamore PD: Diversifying
microRNA sequence and function. Nat Rev Mol Cell Biol. 14:475–488.
2013. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jafri MA, Al-Qahtani MH and Shay JW: Role
of miRNAs in human cancer metastasis: Implications for therapeutic
intervention. Semin Cancer Biol. 44:117–131. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Friedman RC, Farh KH, Burge CB and Bartel
DP: Most mammalian mRNAs are conserved targets of microRNAs. Genome
Res. 19:92–105. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liu X, Chen X, Zeng K, Xu M, He B, Pan Y,
Sun H, Pan B, Xu X, Xu T, et al: DNA-methylation-mediated silencing
of miR-486-5p promotes colorectal cancer proliferation and
migration through activation of PLAGL2/IGF2/β-catenin signal
pathways. Cell Death Dis. 2018. View Article : Google Scholar
|
|
7
|
Feng L, Jing L, Han J, Wang G and Liu Y,
Zhang X, Wang Y, Wang F, Ma H and Liu Y: MicroRNA 486-3p directly
targets BIK and regulates apoptosis and invasion in colorectal
cancer cells. Onco Targets Ther. 11:8791–8801. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Han C, Song Y and Lian C: MiR-769 inhibits
colorectal cancer cell proliferation and invasion by targeting
HEY1. Med Sci Monit. 24:9232–9239. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mizoguchi A, Takayama A, Arai T, Kawauchi
J and Sudo H: MicroRNA-8073: Tumor suppressor and potential
therapeutic treatment. PLoS One. 13:e02097502018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yang X, Lou Y, Wang M, Liu C, Liu Y and
Huang W: miR675 promotes colorectal cancer cell growth dependent on
tumor suppressor DMTF1. Mol Med Rep. 19:1481–1490. 2019.PubMed/NCBI
|
|
11
|
Mazeh H, Mizrahi I, Ilyayev N, Halle D,
Brucher B, Bilchik A, Protic M, Daumer M, Stojadinovic A, Itzhak A
and Nissan A: The diagnostic and prognostic role of microRNA in
colorectal cancer-a comprehensive review. J Cancer. 4:281–295.
2013. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
DeCastro AJ, Dunphy KA, Hutchinson J,
Balboni AL, Cherukuri P, Jerry DJ and DiRenzo J: MiR203 mediates
subversion of stem cell properties during mammary epithelial
differentiation via repression of ΔNP63alpha and promotes
mesenchymal-to-epithelial transition. Cell Death Dis. 4:e5142013.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hu D, Hu Y, Xu W, Yu H, Yang N, Ni S and
Fu R: miR203 inhibits the expression of collagen-related genes and
the proliferation of hepatic stellate cells through a
SMAD3-dependent mechanism. Mol Med Rep. 16:1248–1254. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liang M, Shi B, Liu J, He L, Yi G, Zhou L,
Yu G and Zhou X: Downregulation of miR203 induces overexpression of
PIK3CA and predicts poor prognosis of gastric cancer patients. Drug
Des Devel Ther. 9:3607–3616. 2015.PubMed/NCBI
|
|
15
|
Xu D, Wang Q, An Y and Xu L: MiR203
regulates the proliferation, apoptosis and cell cycle progression
of pancreatic cancer cells by targeting Survivin. Mol Med Rep.
8:379–384. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Deng B, Wang B, Fang J, Zhu X, Cao Z, Lin
Q, Zhou L and Sun X: MiRNA-203 suppresses cell proliferation,
migration and invasion in colorectal cancer via targeting of
EIF5A2. Sci Rep. 6:283012016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Xiao Z, Qu Z, Chen Z, Fang Z, Zhou K,
Huang Z, Guo X and Zhang Y: LncRNA HOTAIR is a prognostic biomarker
for the proliferation and chemoresistance of colorectal cancer via
MiR-203a-3p-mediated Wnt/ss-catenin signaling pathway. Cell Physiol
Biochem. 46:1275–1285. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liu S and Feng P: MiR-203 Determines poor
outcome and suppresses tumor growth by targeting TBK1 in
osteosarcoma. Cell Physiol Biochem. 37:1956–1966. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Burnside MN, Pyatt RE, Hughes A, Baker PB
and Pierson CR: Complex brain malformations associated with
chromosome 6q27 gain that includes THBS2, which encodes
thrombospondin 2, an astrocyte-derived protein of the extracellular
matrix. Pediatr Dev Pathol. 18:59–65. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhuo C, Li X, Zhuang H, Tian S, Cui H,
Jiang R, Liu C, Tao R and Lin X: Elevated THBS2, COL1A2, and SPP1
expression levels as predictors of gastric cancer prognosis. Cell
Physiol Biochem. 40:1316–1324. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sun R, Wu J, Chen Y, Lu M, Zhang S, Lu D
and Li Y: Down regulation of Thrombospondin2 predicts poor
prognosis in patients with gastric cancer. Mol Cancer. 13:2252014.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Fei W, Chen L, Chen J, Shi Q, Zhang L, Liu
S, Li L, Zheng L and Hu X: RBP4 and THBS2 are serum biomarkers for
diagnosis of colorectal cancer. Oncotarget. 8:92254–92264. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang X, Zhang L, Li W, Sun H, Zhang H and
Lai M: THBS2 is a potential prognostic biomarker in colorectal
cancer. Sci Rep. 6:333662016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Adolph KW, Liska DJ and Bornstein P:
Analysis of the promoter and transcription start sites of the human
thrombospondin 2 gene (THBS2). Gene. 193:5–11. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bornstein P, O'Rourke K, Wikstrom K, Wolf
FW, Katz R, Li P and Dixit VM: A second, expressed thrombospondin
gene (Thbs2) exists in the mouse genome. J Biol Chem.
266:12821–12824. 1991.PubMed/NCBI
|
|
26
|
Ao R, Guan L, Wang Y and Wang JN:
Silencing of COL1A2, COL6A3, and THBS2 inhibits gastric cancer cell
proliferation, migration, and invasion while promoting apoptosis
through the PI3k-Akt signaling pathway. J Cell Biochem.
119:4420–4434. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chang IW, Li CF, Lin VC, He HL, Liang PI,
Wu WJ, Li CC and Huang CN: Prognostic impact of thrombospodin-2
(THBS2) overexpression on patients with urothelial carcinomas of
upper urinary tracts and bladders. J Cancer. 7:1541–1549. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lin X, Hu D, Chen G, Shi Y, Zhang H, Wang
X, Guo X, Lu L, Black D, Zheng XW and Luo X: Associations of THBS2
and THBS4 polymorphisms to gastric cancer in a Southeast Chinese
population. Cancer Genet. 209:215–222. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tsai EA, Gilbert MA, Grochowski CM,
Underkoffler LA, Meng H, Zhang X, Wang MM, Shitaye H, Hankenson KD,
Piccoli D, et al: THBS2 is a candidate modifier of liver disease
severity in alagille syndrome. Cell Mol Gastroenterol Hepatol.
2:663–675. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wei WF, Zhou CF, Wu XG, He LN, Wu LF, Chen
XJ, Yan RM, Zhong M, Yu YH, Liang L and Wang W: MicroRNA-221-3p, a
TWIST2 target, promotes cervical cancer metastasis by directly
targeting THBS2. Cell Death Dis. 8:32202017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liu S, Dong H, Dai H, Liu D and Wang Z:
MicroRNA-216b regulated proliferation and invasion of non-small
cell lung cancer by targeting SOX9. Oncol Lett. 15:10077–10083.
2018.PubMed/NCBI:
|
|
32
|
Zhang Y, Su Y, Zhao Y, Lv G and Luo Y:
MicroRNA720 inhibits pancreatic cancer cell proliferation and
invasion by directly targeting cyclin D1. Mol Med Rep.
16:9256–9262. 2017. View Article : Google Scholar : PubMed/NCBI
|