Introduction
Epithelial ovarian cancer (OC) is the most common
gynecological tumor with a high mortality rate worldwide (1). Epithelial OC is associated with a high
rate of metastasis at an early stage, and metastasis is the major
cause of the dismal prognosis (2);
however, the detailed mechanisms underlying the propensity of OC
for metastasis remain unclear. There is an urgent need to identify
methods for early diagnosis and effective treatment for OC.
MicroRNAs (miRNAs) are an endogenous class of
single-stranded small RNAs ~20–24 nucleotides in length, which play
a variety of important regulatory roles in cells (3). The first miRNA was identified in
Caenorhabditis elegans (4),
whereas hundreds of miRNAs have been identified to date and found
to be highly conserved in all types of organisms (5). In recent years, research on miRNAs has
markedly progressed; in particular, the mechanism of target gene
regulation, and numerous miRNAs and their target genes have been
identified and investigated. miRNAs are promising targets for the
diagnosis and treatment of diseases caused by multiple factors,
particularly cancer (6), and they
have also been studied in the context of OC (7). miR-26a has been reported to act as a
tumor suppressor in numerous human cancers, such as prostate cancer
(8–10), lung cancer (11–13),
thyroid cancer (14), breast cancer
(15–17), bladder cancer (18), hepatocellular carcinoma (19–21),
colorectal cancer (22,23), glioblastoma (24), melanoma (25) and esophageal squamous cell carcinoma
(26). However, there are few reports
of the role of transcription factor 12 (TCF12) in OC.
As a transcription factor, TCF12 belongs to the
basic helix-loop-helix (bHLH) E protein family, which can recognize
the E-box sequence CANNTG (27).
TCF12 has been proven to be a regulator of cell development and
differentiation (28), and has been
demonstrated to be highly expressed in several human cancers
(29–31). However, the role of TCF12 and the
association of miR-26a and TCF12 expression in OC have not been
extensively investigated to date.
The aim of the present study was to investigate the
expressions of miR-26a and TCF12 and their correlation in OC
patients. In addition, the regulatory effect of miR-26a on TCF12
expression was determined, and the proliferation, migration,
invasion and apoptosis of OC cells were also investigated.
Materials and methods
Clinical patient samples and cell
lines
A total of 27 cases of human fresh OC tissues and
paired normal ovarian tissues were collected from patients (mean
age 55.10±11.10 years, range 35–78 years) who were diagnosed with
primary OC and underwent initial surgery at the Department of
Obstetrics and Gynecology of the Affiliated Hospital of Nantong
University. Written informed consent was obtained from all the
patients and the study protocol was approved by the Ethics
Committee of the Affiliated Hospital of Nantong University.
The human OC cell lines SK-OV-3 and A2780, the
normal ovarian epithelial cell line IOSE80 and 293 cells were
cultured in Dulbecco's modified Eagle's medium (DMEM; Thermo Fisher
Scientific, Inc.) and supplemented with 10% fetal bovine serum
(FBS; Thermo Fisher Scientific, Inc.) at 37°C in a humidified
incubator with 5% CO2.
Oligos, plasmids and transfection
The human miR-26a mimic (sense:
5′-UUCAAGUAAUCCAGGAUAGGCU-3′, antisense:
5′-AGCCUAUCCUGGAUUACUUGAA-3′) was used to regulate the expression
of TCF12 in OC cells and a scrambled miR-26a mimic sequence used as
negative control (NC; sense: 5′-GGUCGUCUGAUAUACGAUACAA-3′;
antisense: 5′-UUGUAUCGUAUAUCAGACGACC-3′). The miR-26a binding site
of TCF12 was predicted using TargetScan online software (http://www.targetscan.org/). The 50 base pairs (bp)
cDNA of the 3′ untranslated region (UTR) sequence containing
miR-26a-binding sites from TCF12 mRNA was cloned into a
pGL3-control vector (Promega Corporation) downstream of the
luciferase gene, which was used as the wild-type TCF12 (wtTCF12)
plasmid. A point mutant miR-26a-binding site of TCF12 was
constructed and was used as the mutant TCF12 (muTCF12) plasmid.
Cell transfection of 50 pmol miRNA or 80 ng plasmid was performed
by Lipofectamine® 2000 transfection reagent (Thermo
Fisher Scientific, Inc.) according to the manufacturer's
instructions. miRNA, NC and DNA oligos were obtained from Biomics
Biotechnologies Co., Ltd.
Reverse transcription-quantitative PCR
(RT-qPCR) analysis
Small RNAs were isolated from tissues and cell lines
using the mirVana miRNA Isolation Kit (Thermo Fisher Scientific,
Inc.) according to the manufacturer's instructions, and the miR-26a
expression levels were detected by the stem-loop RT-qPCR method
(32). U6 small RNA was used as an
internal control. Total RNA was extracted from cells using
TRIzol® reagent (Thermo Fisher Scientific, Inc.) and
RT-qPCR reactions were carried out using the One-Step RT-qPCR kit
(Thermo Fisher Scientific, Inc.) according to the manufacturers'
instructions. The thermocycling conditions were as follows:
Pre-denaturation at 95°C for 10 min, followed by 40 cycles of
denaturation at 95°C for 20 sec, annealing at 58°C for 20 sec and
extension at 72°C for 30 sec. β-actin was used as an internal
control. miRNA and mRNA level were analyzed by the
2−ΔΔCq method (33). The
primer sequences were as follows: TCF12 forward:
5′-CTAATGAAGATGAGGATT-3′ and reverse: 5′-GATGAAGAATAAGGAGTT-3′;
β-actin forward: 5′-TTGCCGACAGGATGCAGAAGGA-3′ and reverse:
5′-AGGTGGACAGCGAGGCCAGGAT-3′; miR-26a forward:
5′-GCGTTGTCTGGAATGTAAGG-3′ and reverse:
5′-TGACGAGTTTAGAGCCGGATAG-3′; and U6 forward:
5′-AACGCTTCACGAATTTGCGT-3′ and reverse:
5′-CTCGCTTCGGCAGCACA-3′.
Dual luciferase reporter assay
The 293 cells were used to measure luciferase
activity. Briefly, 293 cells were plated into 24-well plates at
1×105 per well and cultured for 24 h. Subsequently, 80
ng wtTCF12 or muTCF12 plasmid and 50 nmol/l miR-26a mimic or NC
were co-transfected into 293 cells using Lipofectamine®
2000 transfection reagent (Thermo Fisher Scientific, Inc.) along
with pRL-TK plasmid (Promega Corporation). Cells were collected 48
h after transfection and firefly and Renilla luciferase activities
were measured using a dual luciferase reporter assay system
according to the manufacturer's instructions (Promega Corporation).
The results of firefly luciferase activity were normalized to the
Renilla luciferase activity.
Western blot analysis
Briefly, following treatment for 48 h as described
above, SK-OV-3 and A2780 cells were collected, and total protein
was extracted with RIPA lysis and extraction buffer (Thermo Fisher
Scientific, Inc.). Total proteins were quantified using the BCA
method, then separated by 10% SDS-PAGE with 50 µg per lane and
transferred onto a PVDF membrane (GE Healthcare). Subsequently,
after blocking with 5% non-fat dry milk for 2 h at room
temperature, the membrane was incubated with rabbit anti-human
TCF12 antibody (1:500 dilution, cat no. ab245540) or mouse
anti-human β-actin antibody (1:1,000 dilution, cat no. ab179467) at
4°C overnight. After washing with TBST with 0.1% Tween-20, the PVDF
membrane was incubated with a horseradish peroxidase-conjugated IgG
(1:2,000 dilution, cat no. ab205718) for 2 h at room temperature;
all antibodies were from Abcam, Inc. Finally, the specific proteins
were detected using Pierce ECL Western Blotting Substrate (Thermo
Fisher Scientific, Inc.), and ImageJ software 1.51 (National
Institutes of Health) was used for densitometry analysis.
Cell proliferation assay
Cell proliferation was detected using the MTT
method. In brief, 1.5×103 cells per well were grown in
96-well plates overnight, and then the cells were treated as
indicated above. At 24, 48 and 72 h after treatment, 10 µl MTT
solution (Promega Corporation) were added to each well. At 4 h
after incubation at 37°C protected from light, 150 µl DMSO were
added to each well and incubated at 37°C for 10 min. Finally, the
OD value of each well was measured using a microplate reader at 490
nm.
Transwell assay
Transwell assay was used to evaluate cell migration
and Matrigel-based Transwell assay was used to evaluate cell
invasion. The cells were plated onto 6-well plates and cultured for
24 h. After treatment for 48 h, as indicated above, cells were
collected and suspended in DMEM at a density of 1×106
cells/ml. Prior to treatment, Transwell chambers (8-µm pore size;
Corning, Inc.) were incubated with DMEM for 1 h at 37°C. For cell
invasion detection, Matrigel (BD Biosciences) was thawed at 4°C
overnight, and 100 µl (1:8 diluted in DMEM) were added into the
Transwell upper chamber and incubated at 37°C for 1 h. Cell
suspension (100 µl) was added into the upper chamber and 600 µl
DMEM supplemented with 10% FBS was added into the lower chamber.
Following incubation for 24 h at 37°C, the cells on upper surface
of the membrane were carefully removed using a cotton swab, and the
cells on the lower surface were fixed in 10% formaldehyde for 20
min at room temperature. After washing with PBS, the cells were
stained in 0.5% crystal violet solution for 10 min at room
temperature and washed with PBS. Finally, the stained cells on the
lower surface of upper chamber were observed under a light
microscope (magnification, ×100).
Cell apoptosis assay
Annexin V-FITC and propidium iodide (PI) double
staining followed by flow cytometry was used to evaluate cell
apoptosis. Briefly, after treatment for 48 h as described above,
1×105 SK-OV-3 or A2780 cells were collected and washed
with PBS, then transferred into centrifuge tubes and centrifuged at
800 × g for 5 min at 4°C. After washing with PBS, the cells were
re-suspended in 195 µl 1X Annexin V-FITC binding buffer and 5 µl
Annexin V-FITC (Sigma-Aldrich; Merck KGaA) and incubated for 10 min
at room temperature in the dark. Following centrifugation for 5 min
at 800 × g at 4°C, the cells were re-suspended in 190 µl 1X Annexin
V-FITC binding buffer followed by 10 µl PI (Sigma-Aldrich; Merck
KGaA). Finally, cell apoptosis was detected using a flow cytometer
(FACSCalibur; BD Biosciences) and the results were analyzed by BD
CellQuest software, version 5.1 (BD Biosciences).
Statistical analysis
All data are presented as mean ± standard deviation.
Statistical analyses were performed using SPSS 20.0 software (IBM
Corp.). Differences between two groups were tested by paired
Student's two-tailed t-test. One-way ANOVA followed by Dunnett's
post hoc test was used to compare multiple groups. The correlation
of miR-26a with TCF12 expression was analyzed by Spearman's
correlation analysis. P<0.05 was considered to indicate
statistically significant differences.
Results
Low miR-26a and high TCF12 expression
is observed in OC patients
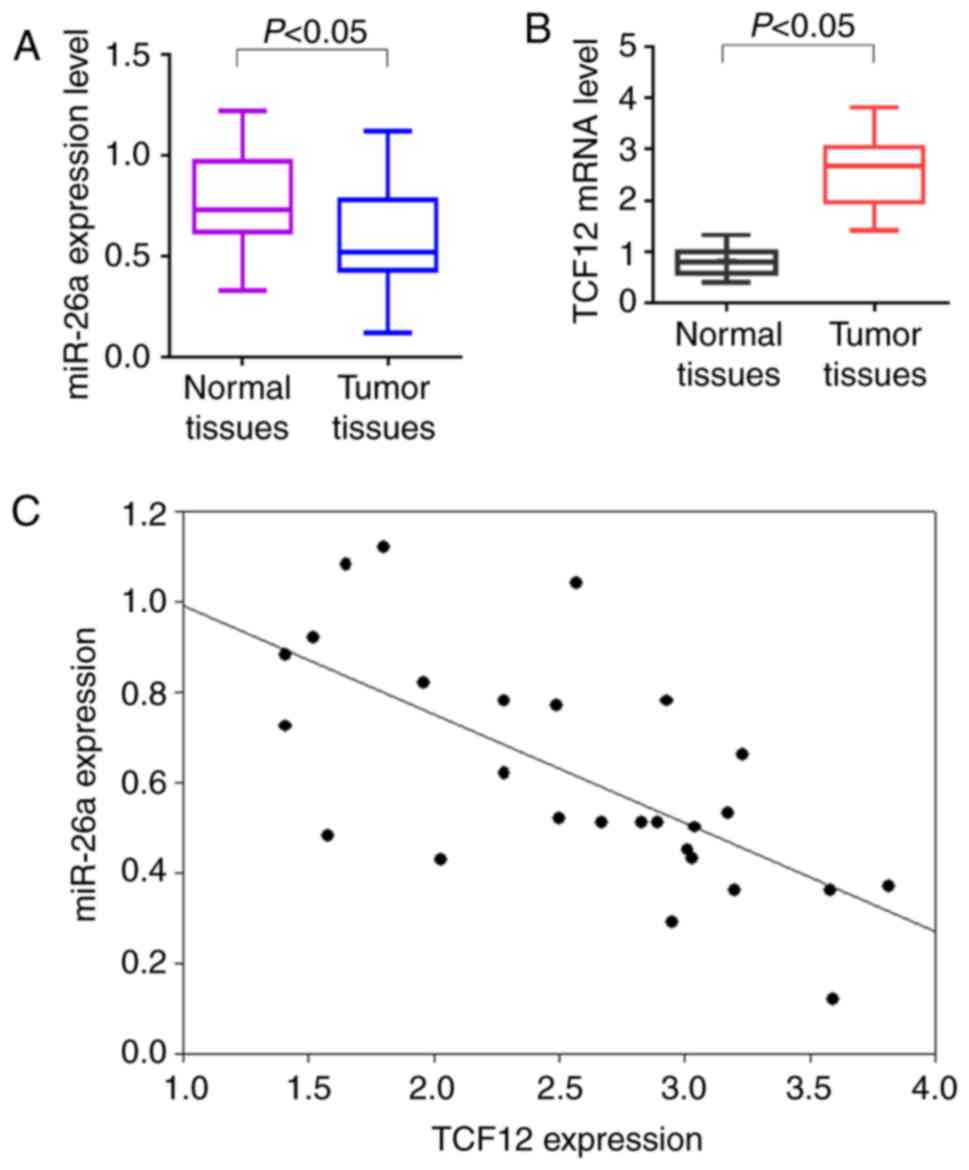
To investigate the expression of miR-26a and TCF12
in OC patients, 27 cases of fresh OC samples and paired normal
ovarian tissues were used for RT-qPCR analysis. The results
demonstrated that the miR-26a expression levels were low in OC
tissues, while the TCF12 expression levels were high, compared with
those in normal ovarian tissues (P<0.05; Fig. 1A and B). Spearman's correlation
analysis revealed that miR-26a was negatively correlated with TCF12
expression (r=−0.695, P<0.05; Fig.
1C).
miR-26a directly targets the 3′UTR
region of TCF12 mRNA
As predicted by software, TCF12 mRNA contains a
miR-26a-binding site at position 124 in the 3′UTR region. The
luciferase reporter plasmids wtTCF12 and muTCF12 were constructed
(Fig. 2A) and co-transfection of
wtTCF12 or muTCF12 plasmid and miR-26a mimic into 293 cells was
performed. Following transfection for 48 h, the luciferase activity
of wtTCF12 and miR-26a co-transfected cells was obviously decreased
compared with that in cells co-transfected with NC and wtTCF12 or
muTCF12 (P<0.05; Fig. 2B).
miR-26a inhibits the expression of
TCF12 in OC cells
Furthermore, the miR-26a expression levels were
found to be low in the OC cell lines SK-OV-3 and A2780, while the
TCF12 expression levels were high, compared with those in IOSE80
cells (P<0.05; Fig. 3A and B). To
further observe regulation of TCF12 expression by miR-26a in OC
cells, human miR-26a mimic was used to increase endogenous miR-26a
in OC cells, and the miR-26a levels in SK-OV-3 and A2780 cells were
significantly upregulated following miR-26a mimic transfection
(Fig. 3C). The protein expression of
TCF12 was significantly inhibited by miR-26a in both SK-OV-3 and
A2780 cells, compared with NC-treated or untreated cells
(P<0.05; Fig. 3D).
miR-26a inhibits the proliferation of
OC cells
MTT assay was used to investigate the inhibitory
effects of miR-26a on OC cell growth. TCF12 inhibition by miR-26a
clearly inhibited the growth of SK-OV-3 (Fig. 4A) and A2780 (Fig. 4B) cells at 48 and 72 h, compared with
NC-treated or untreated cells (P<0.05).
miR-26a inhibits the migration and
invasion of OC cells
Transwell and Matrigel-based Transwell assay was
used to evaluate the inhibitory effects of miR-26a on OC cell
migration and invasion, respectively. The results demonstrated that
miR-26a treatment significantly decreased SK-OV-3 and A2780 cell
migration and invasion, compared with NC-treated or untreated cells
(P<0.05; Fig. 5A and B).
miR-26a induces the apoptosis of OC
cells
Annexin-V-FITC/PI flow cytometry was used to
evaluate the effects of miR-26a on OC cell apoptosis. Treatment
with miR-26a for 48 h significantly increased apoptosis of both
SK-OV-3 and A2780 cells compared with cells treated with NC
(P<0.05; Fig. 5C).
Discussion
Lack of early diagnosis and effective treatment is a
major cause of death from OC among women with malignant
gynecological tumors (2).
Individualized cancer treatment based on molecular targeted therapy
is crucial for improving therapeutic efficacy, and molecules that
are heavily involved in the uncontrolled cell growth and
proliferation, inhibition of apoptosis and tumor metastasis may
represent promising targets for cancer therapy (34).
miRNAs are small endogenously expressed RNAs in
mammalian cells, which have been used in regulation of gene
expression at the post-transcriptional level in a sequence-specific
manner (35). miRNAs have been
demonstrated to play multiple functional roles in biological
processes, such as cell developmental control, regulation, and the
occurrence of various diseases, particularly cancer, due to their
high abundance, sequence conservation and tissue specificity
(5). Due to their different
expression patterns and functions, miRNAs involved in tumorigenesis
may be classified as oncogenes or tumor suppressors in different
human cancers (36); thus, miRNAs may
be used as novel targets or markers for detection, prognosis,
diagnosis and therapy (37). In
recent years, a number of studies have identified aberrant
expression of miRNAs in ovarian tumorigenesis leading to OC
(38). miR-26a was identified as a
tumor suppressor in a number of human cancers. miR-26a was shown to
exert an antiproliferative effect on prostate cancer cells by
decreasing survival and migration and inducing cell cycle arrest
and apoptosis (9). In gastric cancer,
miR-26a promoted cell proliferation, migration and invasion, and
phosphatase and tensin homolog was identified as a direct target of
miR-26a (10). miR-26a was
demonstrated to enhance the metastatic potential of lung cancer
cells by targeting glycogen synthase kinase (GSK)3β of the
β-catenin pathway; miR-26a expression was found to be negatively
correlated with GSK3β expression in patients with lung cancer, and
overexpression of GSK3β reversed the enhancing effect of miR-26a on
lung cancer cell migration and invasion. In addition, integrin β8
(13), lin-28 homolog B (39) and matrix metalloproteinase-2 (11) have also been identified as targets in
lung cancer. miR-26a has been proven to be associated with
metastasis, survival and apoptosis in breast cancer, acts as a
tumor suppressor and serves as a prognostic marker (15,16). The
proliferation, migration and invasion of hepatocellular carcinoma
(HCC) cells can also be inhibited by miR-26a by targeting diverse
genes and pathways, such as the epithelial-to-mesenchymal
transition (40), interleukin-6-Stat3
(13) and PI3K/Akt (23) pathways, and low miR-26a levels were
found to be closely associated with metastasis and progression of
HCC (19–21). In bladder cancer (18), colorectal cancer (23) and melanoma (25), miR-26a was found to play the same
functional roles. In recent years, the expression of miR-26a has
been reported to be low in OC tissues and cells, further
investigation demonstrated that the proliferation of OC cells was
inhibited while their apoptosis was induced by miR-26a, suggesting
that miR-26a downregulation may be predictive of poor prognosis in
epithelial OC (41,42). Moreover, TCF12 was found to be highly
expressed in OC patients in our recent study (43), and its overexpression was correlated
with histological grade and metastasis of OC, whereas TCF12
downregulation inhibited the growth, migration and invasion of OC
cells and promoted apoptosis. In the present study, the expression
of miR-26a was also found to be low in OC patients and cell lines,
whereas the expression of TCF12 was high, and low miR-26a
expression was statistically significantly correlated with high
TCF12 expression. To the best of our knowledge, the present study
was the first to demonstrate that TCF12 is a direct target of
miR-26a by the dual luciferase reporter assay. The expression of
TCF12 in OC cells was inhibited by miR-26a at both the mRNA and
protein levels. Furthermore, the proliferation, migration and
invasion of SK-OV-3 and A2780 cells were inhibited by miR-26a
through suppression of TCF12, whereas apoptosis was induced.
Therefore, the findings of the present study suggest
that upregulation of miR-26a may inhibit metastasis and promote
apoptosis in OC cells by targeting TCF12, which may be a potential
strategy for OC treatment.
Acknowledgements
Not applicable.
Funding
The present study was supported by a grant from the
Science Foundation of Nantong City, Jiangsu Province, China (no.
MS12017008-2).
Availability of data and materials
The datasets generated and analyzed during the
present study are available from the corresponding author upon
reasonable request.
Authors' contributions
SG, TB and YZ contributed to conception and design
of this study; YL and TB collected the clinical samples and patient
information; SG, TB, MS and YL performed the experiments; SG, TB,
MS, YL and YZ acquired, analyzed and interpreted the data; SG was a
major contributor to drafting and writing the manuscript; TB, MS,
YL and YZ revised the manuscript. All authors have read and
approved the final version of the manuscript for publication.
Ethics approval and consent to
participate
Written informed consent was obtained from all the
patients and the study protocol was approved by the Ethics
Committee of the Affiliated Hospital of Nantong University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Vetter MH and Hays JL: Use of targeted
therapeutics in epithelial ovarian cancer: A review of current
literature and future directions. Clin Ther. 40:361–371. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Morris CR, Sands MT and Smith LH: Ovarian
cancer: Predictors of early-stage diagnosis. Cancer Causes Control.
21:1203–1211. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mallory AC and Bouché N: MicroRNA-directed
regulation: To cleave or not to cleave. Trends Plant Sci.
13:359–367. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lee RC, Feinbaum RL and Ambros V: The
C. elegans heterochronic gene lin-4 encodes small RNAs with
antisense complementarity to lin-14. Cell. 75:843–854. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chen K and Rajewsky N: The evolution of
gene regulation by transcription factors and microRNAs. Nat Rev
Genet. 8:93–103. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bell E and Taylor MA: Functional roles for
exosomal microRNAs in the tumour microenvironment. Comput Struct
Biotechnol J. 15:8–13. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mahdian-Shakib A, Dorostkar R, Tat M,
Hashemzadeh MS and Saidi N: Differential role of microRNAs in
prognosis, diagnosis, and therapy of ovarian cancer. Biomed
Pharmacother. 84:592–600. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Erdmann K, Kaulke K, Rieger C, Salomo K,
Wirth MP and Fuessel S: miR-26a and miR-138 block the G1/S
transition by targeting the cell cycle regulating network in
prostate cancer cells. J Cancer Res Clin Oncol. 142:2249–2261.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Rizzo M, Berti G, Russo F, Fazio S,
Evangelista M, D'Aurizio R, Pellegrini M and Rainaldi G:
Discovering the miR-26a-5p targetome in prostate cancer cells. J
Cancer. 8:2729–2739. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ding K, Wu Z, Wang N, Wang X, Wang Y, Qian
P, Meng G and Tan S: miR-26a performs converse roles in
proliferation and metastasis of different gastric cancer cells via
regulating of PTEN expression. Pathol Res Pract. 213:467–475. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Pastuszak-Lewandoska D, Kordiak J,
Czarnecka KH, Migdalska-Sęk M, Nawrot E, Domańska-Senderowska D,
Kiszałkiewicz JM, Antczak A, Górski P and Brzeziańska-Lasota E:
Expression analysis of three miRNAs, miR-26a, miR-29b and miR-519d,
in relation to MMP-2 expression level in non-small cell lung cancer
patients: A pilot study. Med Oncol. 33:962016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lin G, Liu B, Meng Z, Liu Y, Li X, Wu X,
Zhou Q and Xu K: miR-26a enhances invasive capacity by suppressing
GSK3β in human lung cancer cells. Exp Cell Res. 352:364–374. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Song Q, Liu B, Li X, Zhang Q, Cao L, Xu M,
Meng Z, Wu X and Xu K: miR-26a-5p potentiates metastasis of human
lung cancer cells by regulating ITGβ8- JAK2/STAT3 axis. Biochem
Biophys Res Commun. 501:494–500. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gong Y, Wu W, Zou X, Liu F, Wei T and Zhu
J: miR-26a inhibits thyroid cancer cell proliferation by targeting
ARPP19. Am J Cancer Res. 8:1030–1039. 2018.PubMed/NCBI
|
|
15
|
Liu P, Tang H, Chen B, He Z, Deng M, Wu M,
Liu X, Yang L, Ye F and Xie X: miR-26a suppresses tumour
proliferation and metastasis by targeting metadherin in triple
negative breast cancer. Cancer Lett. 357:384–392. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cabello P, Pineda B, Tormo E, Lluch A and
Eroles P: The antitumor effect of metformin is mediated by miR-26a
in breast cancer. Int J Mol Sci. 17:12982016. View Article : Google Scholar :
|
|
17
|
Zhao XX, Yuan QZ, Mu DP, Sun DW, Bo QA,
Pan GZ, Li GQ, Cui T, Ding PP, You FP, et al: MicroRNA-26a inhibits
proliferation by targeting high mobility group AT-hook 1 in breast
cancer. Int J Clin Exp Pathol. 8:368–373. 2015.PubMed/NCBI
|
|
18
|
Lin Y, Chen H, Hu Z, Mao Y, Xu X, Zhu Y,
Xu X, Wu J, Li S, Mao Q, et al: miR-26a inhibits proliferation and
motility in bladder cancer by targeting HMGA1. FEBS Lett.
587:2467–2473. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sun M, Zhao X, Liang L, Pan X, Lv H and
Zhao Y: Sialyltransferase ST3GAL6 mediates the effect of
microRNA-26a on cell growth, migration, and invasion in
hepatocellular carcinoma through the protein kinase B/mammalian
target of rapamycin pathway. Cancer Sci. 108:267–276. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ma Y, Deng F, Li P, Chen G, Tao Y and Wang
H: The tumor suppressive miR-26a regulation of FBXO11 inhibits
proliferation, migration and invasion of hepatocellular carcinoma
cells. Biomed Pharmacother. 101:648–655. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gao XM, Zhu Y, Li JH, Wang XY, Zhang XF,
Yi CH and Yang X: MicroRNA-26a induces a mitochondrial apoptosis
mediated by p53 through targeting to inhibit Mcl1 in human
hepatocellular carcinoma. Onco Targets Ther. 11:2227–2239. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Jinushi T, Shibayama Y, Kinoshita I,
Oizumi S, Jinushi M, Aota T, Takahashi T, Horita S, Dosaka-Akita H
and Iseki K: Low expression levels of microRNA-124-5p correlated
with poor prognosis in colorectal cancer via targeting of SMC4.
Cancer Med. 3:1544–1552. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liu B, Pan S, Xiao Y, Liu Q, Xu J and Jia
L: LINC01296/miR-26a/GALNT3 axis contributes to colorectal cancer
progression by regulating O-glycosylated MUC1 via PI3K/AKT pathway.
J Exp Clin Cancer Res. 37:3162018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
ParvizHamidi M, Haddad G, Ostadrahimi S,
Ostadrahimi N, Sadeghi S, Fayaz S and Fard-Esfahani P: Circulating
miR-26a and miR-21 as biomarkers for glioblastoma multiform.
Biotechnol Appl Biochem. 66:261–265. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gao J, Zeng K, Liu Y, Gao L and Liu L:
LncRNA SNHG5 promotes growth and invasion in melanoma by regulating
the miR-26a-5p/TRPC3 pathway. Onco Targets Ther. 12:169–179. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chen Z, Zhao L, Zhao F, Yang G and Wang J:
MicroRNA-26b regulates cancer proliferation migration and cell
cycle transition by suppressing TRAF5 in esophageal squamous cell
carcinoma. Am J Transl Res. 8:1957–1970. 2016.PubMed/NCBI
|
|
27
|
Thorsen K, Schepeler T, Øster B, Rasmussen
MH, Vang S, Wang K, Hansen KQ, Lamy P, Pedersen JS, Eller A, et al:
Tumor-specific usage of alternative transcription start sites in
colorectal cancer identified by genome-wide exon array analysis.
BMC Genomics. 12:5052011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
L'Abbate A, Tolomeo D, De Astis F, Lonoce
A, Lo Cunsolo C, Mühlematter D, Schoumans J, Vandenberghe P, Van
Hoof A, Palumbo O, et al: T(15;21) translocations leading to the
concurrent downregulation of RUNX1 and its transcription factor
partner genes SIN3A and TCF12 in myeloid disorders. Mol Cancer.
14:2112015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Labreche K, Simeonova I, Kamoun A, Gleize
V, Chubb D, Letouzé E, Riazalhosseini Y, Dobbins SE, Elarouci N,
Ducray F, et al: TCF12 is mutated in anaplastic oligodendroglioma.
Nat Commun. 6:72072015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Tang X, Hou Y, Yang G, Wang X, Tang S, Du
YE, Yang L, Yu T, Zhang H, Zhou M, et al: Stromal miR-200s
contribute to breast cancer cell invasion through CAF activation
and ECM remodeling. Cell Death Differ. 23:132–145. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chen YF, Yang CC, Kao SY, Liu CJ, Lin SC
and Chang KW: MicroRNA-211 enhances the oncogenicity of
carcinogen-induced oral carcinoma by repressing TCF12 and
increasing antioxidant activity. Cancer Res. 76:4872–4886. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee
DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, et al:
Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic
Acids Res. 33:e1792005. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hertz DL and McLeod HL: Integrated patient
and tumor genetic testing for individualized cancer therapy. Clin
Pharmacol Ther. 99:143–146. 2016. View
Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ichimura A, Ruike Y, Terasawa K and
Tsujimoto G: miRNAs and regulation of cell signaling. FEBS J.
278:1610–1618. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Svoronos AA, Engelman DM and Slack FJ:
OncomiR or tumor suppressor? The duplicity of microRNAs in cancer.
Cancer Res. 76:3666–3670. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Moles R: MicroRNAs-based therapy: A novel
and promising strategy for cancer treatment. Microrna. 6:102–109.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Deb B, Uddin A and Chakraborty S: miRNAs
and ovarian cancer: An overview. J Cell Physiol. 233:3846–3854.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Lu YY, Lin Y, Ding DX, Su S, Chi QQ, Zhang
YC, Sun J, Zhang X, Zhu HM, Huang QS, et al: miR-26a functions as a
tumor suppressor in ambient particulate matter-bound
metal-triggered lung cancer cell metastasis by targeting
LIN28B-IL6-STAT3 axis. Arch Toxicol. 92:1023–1035. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Chang L, Li K and Guo T: miR-26a-5p
suppresses tumor metastasis by regulating EMT and is associated
with prognosis in HCC. Clin Transl Oncol. 19:695–703. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Sun TY, Xie HJ, He H, Li Z and Kong LF:
miR-26a inhibits the proliferation of ovarian cancer cells via
regulating CDC6 expression. Am J Transl Res. 8:1037–1046.
2016.PubMed/NCBI
|
|
42
|
De Lima AB, Silva LM, Gonçales NG,
Carvalho MRS, da Silva Filho AL and da Conceição Braga L:
Three-dimensional cellular arrangement in epithelial ovarian cancer
cell lines TOV-21G and SKOV-3 is associated with apoptosis-related
miRNA expression modulation. Cancer Microenviron. 11:85–92. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Gao S, Bian T, Zhang Y, Su M and Liu Y:
TCF12 overexpression as a poor prognostic factor in ovarian cancer.
Pathol Res Pract. 215:1525272019. View Article : Google Scholar : PubMed/NCBI
|