Introduction
Lung cancer is a leading cause of cancer-related
deaths worldwide, accounting for 18% of cancer-related deaths in
2008 (1); The incidence ratio
between men and women is 2.1:1 (2,3).
Typically, lung cancer is classified into two main types, namely,
small-cell and non-small-cell cancers (4), and tobacco smoke has been revealed to
be the primary cause of lung cancer, causing ~85% of all cases of
lung cancer (5). Due to the lack of
observable symptoms at the early stages, the long-term prognosis of
lung cancer is poor, with a low 5-year relative survival of 6–14%
for men and 7–18% for women (6).
The Wnt/β-catenin pathway is normally inactive in
many tissues in adults (7), and
inappropriate activation is thought to be the initiating event in
intestinal epithelial cell transformation (8). Intracellularly, Wnt signaling is
transduced by disheveled (Dsh) proteins, leading to the
accumulation of β-catenin in the cytosol, which then translocates
to the nucleus to form complexes with transcription factors, such
as the T-cell factor family proteins (TCFs). These transcription
factors transactivate many target genes, such as the oncogenes
c-myc and cyclin D1, which regulate cell proliferation, development
and genes involved in tumorigenesis (8–10). A
previous study revealed that the Wnt/β-catenin signaling cascade
plays a key role in cancer (11),
and Wnt family genes have been shown to be upregulated in many
cancers, including lung cancer (12,13).
In addition, it has been revealed that the metastasis of lung tumor
cell lines was enhanced by increased Wnt/β-catenin signaling
(14).
Serine-threonine kinases (STKs) comprise a primary
family of kinases in the human kinase group, and their expression
has frequently been revealed to be altered in human cancers,
suggesting a key role for the STK family in cancer development
(15,16). STK31, which is a member of the STK
family, is a novel cancer/testis (CT)-related gene that is critical
in human cancers. It has been revealed that STK31 regulates the
cell cycle phases and is highly expressed in several types of
cancers, including lung and colorectal cancers (17). The dysregulated expression of cell
cycle kinases has been revealed to lead to uncontrolled cell
proliferation and genomic instability, both of which are hallmarks
of carcinogenesis (18). As a cell
cycle-regulated protein, STK31 has been reported to contribute to
the tumorigenicity of epithelial cancer, and overexpression of
STK31 promoted cell migration and invasion, whereas STK31 knockdown
induced apoptosis (17). In
addition, STK31 has been revealed to be a novel biomarker for the
risk of colorectal cancer metastasis (19,20).
However, the roles and underlying mechanisms of STK31 in lung
cancer cells remain unclear.
In the present study, analysis of the lung cancer
The Cancer Genome Atlas (TCGA) dataset revealed that STK31 was
highly expressed in lung cancer, and the Wnt/β-catenin pathway was
positively correlated with STK31 expression, which is consistent
with the high expression of STK31 and β-catenin that is typically
observed in lung cancer patients. Downregulation of STK31 in lung
cancer cells significantly suppressed cell proliferation by
blocking cell cycle progression, whereas upregulation of STK31
resulted in the opposite effect. In addition, a Wnt/β-catenin
inhibitor (XAV939) completely attenuated the effects of STK31 on
the lung cancer cells, and a c-myc inhibitor had an effect similar
to that of XAV939. A chromatin immunoprecipitation (ChIP) assay
confirmed that c-myc directly bound to the STK31 promoter. These
findings indicated that STK31 may be an oncogene in lung cancer and
involved in the Wnt/β-catenin pathway and c-myc expression.
Materials and methods
Tissue samples of lung cancer and
normal lung
After written informed consent was obtained, 30
patients with lung cancer who were treated at Linyi Central
Hospital (Shandong, China) aged 55–80 years old were enrolled in
the study. Tissue samples of lung cancer and normal lung were
collected and frozen in liquid nitrogen, followed by RNA and
protein extraction. The expression of STK31 and β-catenin in tissue
samples was determined by real-time PCR and western blot analysis.
All experiments performed in the present study were approved by the
Research Ethics Committee of Linyi Central Hospital of Shandong
(Linyi, China).
Cell culture
Seven human lung cancer cell lines (A549, NCIH1299,
CALU3, NCIH23, NCIH358, NCIH1650 and NCIH1975) and 16HBE, a normal
lung cell line, were purchased from the Cell Bank of the Chinese
Academy of Sciences (Shanghai, China). All cell lines were cultured
in an incubator at 37°C with 5% CO2 (Thermo Forma cat.
no. 3111; Thermo Fisher Scientific, Inc., Waltham, MA, USA) in
RPMI-1640 medium (cat. no. SH30809.01B; HyClone, Logan, UT, USA)
containing 10% fetal bovine serum (FBS; cat. no. 16000-044; Gibco;
Thermo Fisher Scientific, Inc.) and 1% antibiotic (penicillin and
streptomycin; cat. no. P1400-100; Beijing Solarbio Science &
Technology Co., Ltd., Shanghai, China). The medium was refreshed
every two days during culturing.
Construction of the lentivirus
Specific shRNAs targeting various sites of the STK31
gene (cat. no. AF285599.1; as shown in Table I) were synthesized, and then
inserted into the pLKO.1-puro vector using the restriction sites
AgelI and EcolI followed by transformation and
plasmid extraction. Meanwhile, through Genewiz, Inc. (South
Plainfield, NJ, USA), the coding DNA sequence (CDS) region of STK31
(containing EcoRI/BamHI sites, full-length 3,060 bp)
was synthesized, and then was inserted into the pLVX-Puro vector
using the EcoRI/BamHI restriction sites. After the
constructs were sequenced (Shanghai Majorbio Pharmaceutical
Technology Co., Ltd., Shanghai, China), pLKO.1-shSTK31 or
pLVX-Puro-STK31 was co-transfected into 293T cells with the viral
packaging plasmids psPAX2 and pMD2G (Addgene, Inc., Cambridge, MA,
USA) using Lipofectamine 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). The viral particles in the supernatant of the
medium were collected 48 h post-transfection.
 | Table I.STK31 interference sequences. |
Table I.
STK31 interference sequences.
| Name | Sequences |
|---|
| STK31 target site
1 |
GCTGCTGTGGATTTGACTA |
| (1045–1063) |
|
| STK31 target site
2 |
GGAGATAGCTCTGGTTGAT |
| (1713–1731) |
|
| STK31 target site
3 |
GCTGCGCAATAATGTCTTT |
| (2034–2052) |
|
Experimental grouping
In vitro, the expression of STK31 in lung
cancer cells was regulated by lentivirus-mediated RNA interference
or overexpression. NCIH1299 cells were infected with lentiviruses
carrying the STK31 overexpression vector (STK31), whereas NCIH358
and NCIH1975 cells were infected with lentiviruses carrying the
negative control (shNC)/STK31 interference (shSTK31-1, shSTK31-2
and shSTK31-3). Cells treated with RPMI-1640 medium alone served as
an additional control. After 48 h of infection, the efficiency of
the STK31/shSTK31 lentivirus infection was determined by real-time
PCR and western blot analysis.
Following infection with shNC, shSTK31-1 and
shSTK31-2, including medium-treated cells as a control, the cell
viability and proliferation of NCIH358 or NCIH1975 cells at 0, 24,
48 and 72 h were evaluated. The cell cycle and expression levels of
several related genes were also analyzed. In addition, NCIH1299
cells were treated with vector + dimethyl sulfoxide (DMSO), vector
+ 10 µmol/l XAV939 (a Wnt/β-catenin inhibitor; cat. no. S1180;
Selleck Chemicals, Houston, TX, USA), or STK31 + DMSO, STK31 + 10
µmol/l XAV939, and NCIH358 cells were treated with vector + DMSO,
vector + 100 µmol/l 10058-F4 (a c-myc inhibitor; cat. no. S7153;
Selleck Chemicals), STK31 + DMSO, or STK31 + 100 µmol/l 10058-F4.
After each treatment, the cell viability, proliferation, cell cycle
and expression levels of several related genes of the NCIH1299 or
NCIH358 cells were evaluated.
Real-time polymerase chain reaction
(RT-PCR) analysis
Using TRIzol reagent (1596–026; Invitrogen; Thermo
Fisher Scientific, Inc.), total RNA was extracted from tissues or
cells, and then quantified. Following the confirmation of the RNA
quality, reverse transcription was performed using a reverse
transcription kit (cat. no. K1622; Fermentas; Thermo Fisher
Scientific, Inc.) to obtain complementary DNA (cDNA). Using the
cDNA as a template, RT-PCR reactions were performed in triplicate
with a SYBR-Green PCR kit (cat. no. K0223; Thermo Fisher
Scientific, Inc.) on an ABI-7300 Real-Time PCR system (Applied
Biosystems; Thermo Fisher Scientific, Inc.). The thermocycling
program for the RT-PCR reactions was as follows: 95°C, 10 min
(95°C, 15 sec; 60°C, 45 sec) ×40 cycles; 95°C, 15 sec; 60°C, 1 min;
95°C, 15 sec; 60°C, 15 sec (21).
Subsequently, the mRNA expression of STK31 and β-catenin in tissues
and cells was calculated using the 2−ΔΔCq method
(22). Gene expression was
normalized to that of the internal reference, GAPDH. The primers
used were as follows: STK31 forward (F),
5′-TGAACTCTGGTGGTCTCCTTAC-3′ and reverse (R),
5′-CTTGGCTTCTGTGTCAACATCC-3′; β-catenin F,
5′-CGACACCAAGAAGCAGAGATG-3′ and R, 5′-GGGACAAAGGGCAAGATTTCG-3′;
transformation/transcription domain-associated protein (TRRAP)
forward, 5′-GCCCGACTTCCTCTACGACC-3′ and reverse,
5′-GCGACTCCTTCAGCATCTTCC-3′; and GAPDH forward,
5′-AATCCCATCACCATCTTC-3′ and reverse, 5′-AGGCTGTGTCATACTTC-3′.
Western blot analysis
Total protein was extracted from tissues or cells
using RIPA buffer (cat. no. R0010; Beijing Solarbio Science &
Technology Co., Ltd.) supplemented with protease and phosphatase
inhibitors. Following protein quantification using the BCA
quantification kit (cat. no. PICPI23223; Thermo Fisher Scientific,
Inc.), 25 µg protein was separated by 10 or 12% sodium dodecyl
sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and then
semi-transferred onto polyvinylidene fluoride (PVDF) membranes
(cat. no. HATF00010; EMD Millipore, Billerica, MA, USA).
Subsequently, the membranes were blocked at room temperature for 1
h in 5% skim milk (BYL40422; BD Biosciences, Franklin Lakes, NJ,
USA) followed by overnight incubation with primary antibodies
against STK31 (dilution 1:1,000; cat. no. ab69678; Abcam,
Cambridge, MA, USA), β-catenin (dilution 1:5,000; cat. no. ab32572;
Abcam), c-myc (dilution 1:1,000; cat. no. ab32072; Abcam), cyclin
D1 (dilution 1:10,000; cat. no. ab134175; Abcam) or GAPDH (dilution
1:1,000; cat. no. 5174; Cell Signaling Technology Inc., Danvers,
MA, USA) at 4°C with gentle shaking. Next, after 5–6 washes with
Tris-buffered saline with Tween-20 (TBST), the membranes were
incubated with HRP-labeled goat anti-rabbit (cat. no. A0208) and
goat anti-mouse (cat. no. A0216) secondary antibodies (dilution
1:1,000; Beyotime Institute of Biotechnology, Haimen, China) for 2
h at room temperature. Finally, the blots were developed with the
chemiluminescent reagent (cat. no. WBKLS0100; EMD Millipore) for 5
min after being washed with TBST, and the blots were exposed on an
ECL imaging system (Tanon 5200; Tanon Science and Technology Co.,
Ltd., Shanghai, China). Protein expression relative to GAPDH was
analyzed and calculated using ImageJ software version 1.47v (NIH;
National Institutes of Health, Bethesda, MD, USA).
Cell viability and proliferation
assay
A Cell Counting Kit-8 (CCK-8; CP002; SAB
Biotherapeutics, Inc., Sioux Falls, SD, USA) assay was used to
analyze the viability of the lung cancer cells. NCIH358, NCIH1975
and NCIH1299 cells were seeded at a density of 3,000 cells/well
into 96-well plates (TR4001; Trueline Inc., Romeoville, IL, USA)
and cultured overnight. The following day, the three types of cells
were treated as described above in the ‘experimental grouping’
section. After 0, 24, 48 and 72 h of treatment, 100 µl of CCK-8
solution (CCK-8: serum-free medium = 1:10) was added followed by 1
h of incubation at 37°C. Subsequently, using a microplate reader,
the absorbance value (OD) of the lung cancer cells at 450 nm
(CCK-8) was measured (DNM-9602; Beijing Perlong Medical Instrument
Ltd., Beijing, China). In addition, the proliferation of the lung
cancer cells was assessed by bromodeoxyuridine (BrdU)-enzyme-linked
immunosorbent assay (ELISA) using a BrdU Cell Proliferation ELISA
kit (cat. no. Ab126556; Abcam) according to the manufacturer's
protocol.
Cell cycle detection
NCIH358, NCIH1975 and NCIH1299 cells in the
logarithmic growth phase were trypsinized, resuspended at a density
of 500,000 cells/well and then inoculated in 6-well plates.
Following overnight incubation in a 5% CO2 incubator at
37°C, the three types of cells were divided and treated as
described above in the ‘Experimental grouping’ section.
After treatment, all the cells were collected and centrifuged for 5
min at 1,000 × g, followed by resuspension in 300 µl
phosphate-buffered saline (PBS) containing 10% FBS (16000-044;
Gibco; Thermo Fisher Scientific, Inc.). Next, the cells were fixed
in 700 µl absolute ethanol (precooled to 20°C) at 4°C for 24 h, and
then centrifuged at 1,000 × g for 5 min. After being washed with 1
ml PBS (precooled), the fixed cells were slowly and fully
resuspended in 100 µl RNase A solution (1 mg/ml; R8020-25; Beijing
Solarbio Science & Technology Co., Ltd.) and incubated at 37°C
for 30 min in the dark. Following a 10 min incubation in 400 µl
propidium iodide (PI) solution (50 µg/ml; C001-200; 7Sea Biotech,
Shanghai, China) in the dark, red fluorescence was detected at 488
nm (excitation wavelength) by flow cytometry (Accuri C6; BD
Biosciences), and then cell cycle progression was analyzed using
FlowJo software (version 7.6.1; Tree Star, Inc., Ashland, OR,
USA).
Online dataset analysis
The mRNA expression levels of STK31 in The Cancer
Genome Atlas (TCGA) Project for Lung Cancer (https://tcga-data.nci.nih.gov/tcga/; 488 lung cancer
tissues and 58 normal lung tissues) were analyzed.
To probe for STK31-associated pathways in lung
cancer, the STK31 expression data from the lung cancer TCGA dataset
were subjected to Gene Set Enrichment Analysis (GSEA) as previously
described (23).
ChIP assay
Cells (~10,000,000) were lysed in sodium dodecyl
sulfate (SDS containing lysis buffer) following a 10-min treatment
with 1% formaldehyde at room temperature. After sonification, the
cell lysates were centrifuged for 10 min at 8,000 × g, and then the
supernatant was collected and incubated with the anti-c-myc
antibody (1 µg/µl; cat. no. Ab56; Abcam) or the control IgG
overnight. Using protein A/G beads, the immune complex was
captured, followed by DNA purification. The binding of c-myc to
SKT31 was detected using SYBR-Green Real-Time PCR with the
following STK31 promoter primers: STK31 promoter forward (F),
5′-AGATTAGGAAAGACCACGAA-3′ and reverse (R),
5′-GACCCTGGACCCACATAC-3′.
Statistical analysis
All statistical analyses were performed with
GraphPad Prism 7.0 software (GraphPad Software, Inc., San Diego,
CA, USA). The data were presented as the mean ± standard deviation
(SD), and the experiments were performed in triplicate. A
two-tailed Student's t-test was used to examine the differences
between groups, and comparisons between multiple groups were
determined by one-way analysis of variance (ANOVA) followed by
Tukey's multiple comparison. Differences were considered
statistically significant at P<0.05.
Results
Analysis of the available STK31
expression data from the lung cancer TCGA dataset
The analysis of the available expression data from
488 cancer tissues and 58 normal samples in the lung cancer TCGA
dataset revealed that the expression of STK31 was significantly
increased in lung cancer tissues compared to normal tissues
(Fig. 1A), suggesting that STK31
may function as an oncogene in lung cancer. Furthermore, to probe
the STK31-associated pathways in lung cancer, we performed GSEA
using lung cancer samples from the TCGA dataset. As revealed in
Fig. 1B, the Wnt/β-catenin pathway
was identified as being positively correlated with STK31
expression.
The expression of STK31 and β-catenin
is significantly increased in lung cancer tissues
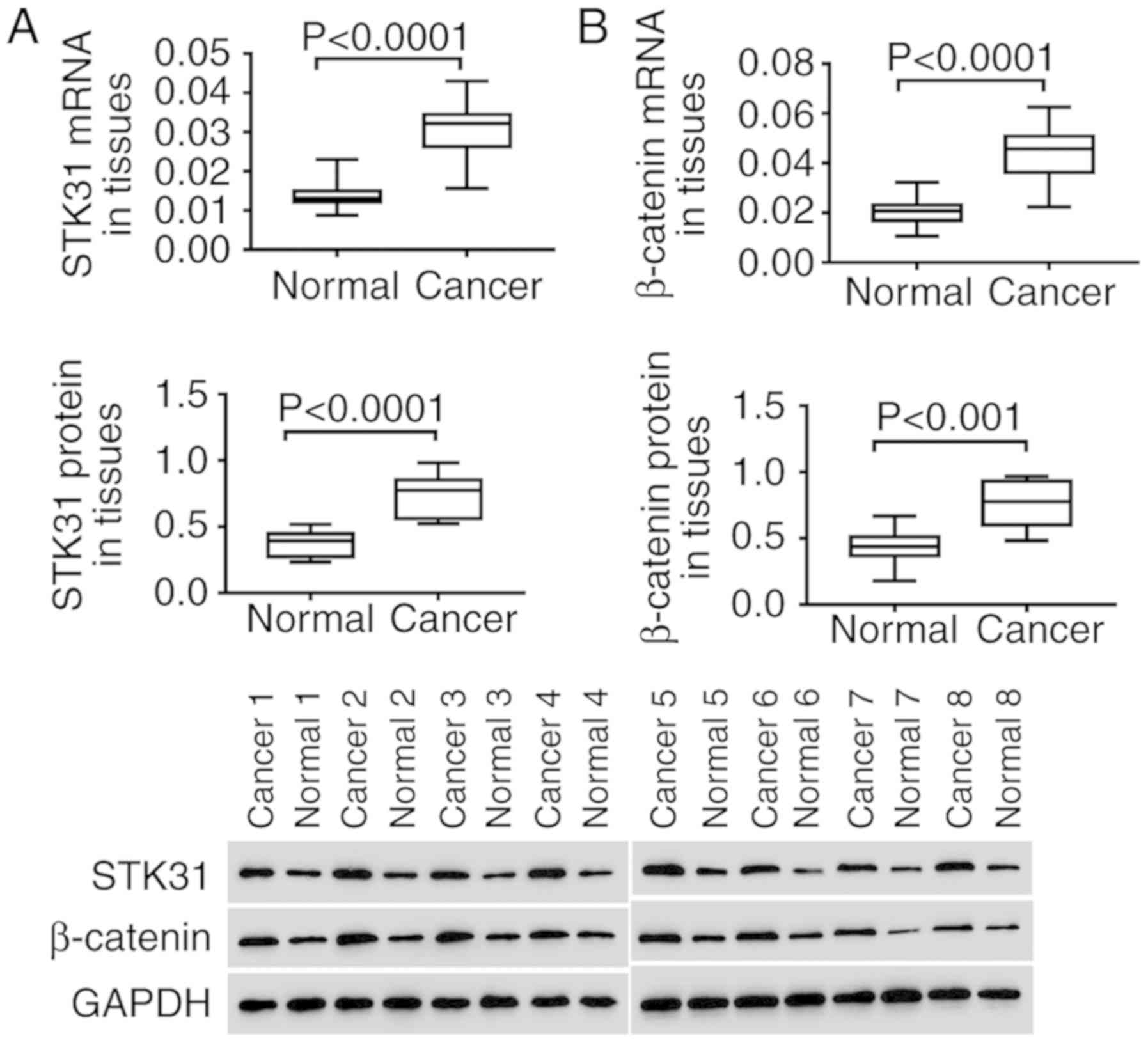
In vitro, the expression of STK31 and
β-catenin was analyzed in lung cancer and normal tissues. As
revealed in Fig. 2, both the mRNA
(upper) and protein (lower) expression of STK31 were significantly
and highly expressed in the tumors of 30 lung cancer patients
compared to normal lung tissues (Fig.
2A). In addition, high expression of β-catenin was also
observed in lung cancer tissues (Fig.
2B). These findings were consistent with the results of the
analysis of lung cancer from the TCGA dataset, further suggesting
the critical role of STK31 in lung cancer, which may involve the
Wnt/β-catenin pathway.
STK31 is increased in lung cancer
cells, and the expression of STK31 is regulated by lentiviral
infection
We examined the expression of STK31 in seven lung
cancer cell lines (A549, NCIH1299, CALU3, NCIH23, NCIH358, NCIH1650
and NCIH1975) and one lung epithelial cell line, 16HBE. The results
presented in Fig. 3A revealed a
high expression of STK31 in lung cancer cell lines, further
suggesting the important role of STK31 in the progression of lung
cancer. Compared with the other lung cancer cell lines, STK31
expression was much higher in NCIH358 and NCIH1975 cells but lower
in NCIH1299 cells. Therefore, in NCIH358, NCIH1975 and NCIH1299
cells, lentivirus-mediated STK31 interference or overexpression was
utilized to regulate STK31 expression. As revealed in Fig. 3B-D, STK31 expression in NCIH1299
cells was significantly upregulated by STK31 lentivirus infection
(Fig. 3B) at both the mRNA and
protein levels, whereas infection with shSTK31 lentivirus
(shSTK31-1, shSTK31-2 and shSTK31-3) significantly downregulated
STK31 expression in NCIH358 cells (Fig.
3C) and NCIH1975 cells (Fig.
3D). The vectors shSTK31-1 and shSTK31-2 resulted in better
downregulation efficiency and were therefore selected for the
following experiments.
 | Figure 3.STK31 is increased in lung cancer
cells, and STK31 expression is regulated by lentivirus infection.
(A) The mRNA (upper) and protein (lower) expression levels of STK31
in A549, NCIH1299, CALU3, NCIH23, NCIH358, NCIH1650, NCIH1975 and
16HBE cells were detected by RT-PCR and western blotting,
respectively. Among these lung cancer cell lines, NCIH358, NCIH1299
and NCIH1975 were infected with lentiviruses, and medium-treated
cells served as the control. (B) The upregulation efficiency of
STK31 lentivirus was analyzed NCIH1299 cells. (C and D) The
downregulation efficiency of shSTK31 lentivirus (shSTK31-1,
shSTK31-2 and shSTK31-3) was analyzed in NCIH358 and NCIH1975
cells. Data are presented as the mean ± SD, *P<0.05,
**P<0.01, ***P<0.001 and ****P<0.0001 compared to 16HBE,
vector or shNC. STK31, serine-threonine kinase 31. |
The downregulation of STK31 in lung
cancer cells suppresses proliferation by blocking cell cycle
progression
To investigate the effect of STK31 on cell
viability, proliferation and cell cycle progression in lung cancer
cells, NCIH358 and NCIH1975 cells were infected with the shSTK31
lentivirus. As revealed in Fig. 4,
the downregulation of STK31 significantly inhibited the cell
viability and proliferation of NCIH358 and NCIH1975 cells (Fig. 4A and C) and arrested the cell cycle
in the G1 phase, which reduced the proportion of cells in the S/G2
phase (Fig. 4B). Moreover, there
was a decrease in β-catenin, c-myc and cyclin D1 in STK31-silenced
lung cancer cells (Fig. 4D). These
results indicated that the downregulation of STK31 had a strong
inhibitory effect on the proliferation of lung cancer cells by
arresting the cell cycle. These findings may contribute to lung
cancer treatment.
 | Figure 4.The downregulation of STK31
expression in lung cancer cells suppresses proliferation by
blocking the cell cycle progression. NCIH358 and NCIH1975 cells
were infected with shNC, shSTK31-1 and shSTK31-2, and
medium-treated cells served as the control. (A) Cell viability was
assessed by CCK-8 assay at 0, 24, 48 and 72 h. (B) The proportion
of cells in each phase of the cell cycle was evaluated by flow
cytometry after 48 h of infection. (C) Cell proliferation was
assessed at 0, 24, 48 and 72 h by BrdU-ELISA. (D) The protein
expression of β-catenin, c-myc and cyclin D1 was analyzed by
western blotting. All data are presented as the mean ± SD,
*P<0.05, **P<0.01 and ***P<0.001 compared to shNC. STK31,
serine-threonine kinase 31; CCK-8, Cell Counting Kit-8; BrdU-ELISA,
bromodeoxyuridine-enzyme-linked immunosorbent assay. |
STK31 regulates proliferation and the
cell cycle in lung cancer cells via activation of the Wnt/β-catenin
pathway
It has been reported that activation of the
Wnt/β-catenin pathway is a critical oncogenic event during the
initiation and progression of tumors, and the oncogenes c-myc and
cyclin D1 are two downstream targets of the Wnt/β-catenin pathway
(24). In the present study, the
mechanism underlying the STK31-mediated regulation of lung cancer
cells was investigated. As revealed in Fig. 5, the upregulation of STK31
significantly increased the viability and proliferation of lung
cancer cells and promoted lung cancer cell entry into the S phase
from the G1 phase (Fig. 5A-C)
accompanied by an increase in the protein expression of β-catenin,
c-myc and cyclin D1 (Fig. 5D). In
contrast, treatment with XAV939, a Wnt/β-catenin inhibitor,
potently attenuated the effects of STK31 in lung cancer cells.
These findings were agreement with prior reports that the
activation of the Wnt/β-catenin pathway is frequently observed in
lung cancer and promotes lung cancer cell proliferation (14,25).
Therefore, it can be concluded that c-myc is a downstream target of
STK31/β-catenin and that STK31 likely regulates the proliferation
and cell cycle progression of lung cancer cells by activating the
Wnt/β-catenin pathway.
 | Figure 5.STK31 regulates proliferation and the
cell cycle in lung cancer cells via the activation of the
Wnt/β-catenin pathway. NCIH1299 cells were treated with vector +
DMSO, vector + 10 µmol/l XAV939 (a Wnt/β-catenin inhibitor), STK31
+ DMSO, or STK31 + 10 µmol/l XAV939. (A and B) Cell viability and
proliferation were assessed by CCK-8 and BrdU-ELISA assays,
respectively, at 0, 24, 48 and 72 h. (C) The proportion of cells in
each phase of the cell cycle was evaluated by flow cytometry after
48 h of treatment. (D) Proteins related to β-catenin, c-myc and
cyclin D1 protein levels were detected by western blotting. Data
are presented as the mean ± SD, **P<0.01 and ***P<0.001
compared to vector, ###P<0.001 compared to STK31.
STK31, serine-threonine kinase 31; DMSO, dimethyl sulfoxide; CCK-8,
Cell Counting Kit-8; BrdU-ELISA, bromodeoxyuridine-enzyme-linked
immunosorbent assay. |
STK31-mediated regulation of
proliferation and the cell cycle in lung cancer cells may be
achieved through positive feedback regulation by c-myc
Using LASAGNA-Search 2.0 (http://biogrid-lasagna.engr.uconn.edu/lasagna_search/),
a c-myc binding site was predicted in the STK31 promoter (−177 to
−167) (Fig. 6F). In the present
study, we used 10058-F4, a c-myc inhibitor, combined with the STK31
lentivirus to explore the role of c-myc in the regulation of STK31
in lung cancer cells. As revealed in Fig. 6, STK31-induced cell viability,
proliferation and cell cycle progression were significantly
counteracted by a c-myc inhibitor (Fig.
6A-C). Moreover, the mRNA and protein expression of STK31 were
markedly decreased by the c-myc inhibitor (Fig. 6D), indicating feedback regulation of
STK31 expression by c-myc. It is known that another protein, TRRAP,
interacts with myc and is required for the mitotic checkpoint and
normal cell cycle progression (26,27).
We found that the knockdown of TRRAP significantly inhibited STK31
expression (Fig. 6E). As shown in
Fig. 6F, a ChIP assay confirmed
that c-myc directly binds to the STK31 promoter. It could be
inferred that c-myc may also act upstream of STK31 and that STK31
potentially regulates proliferation and the cell cycle progression
in lung cancer cells through positive feedback regulation by
c-myc.
 | Figure 6.The STK31-mediated regulation of
proliferation and the cell cycle in lung cancer cells may act
through the positive feedback regulation by c-myc. NCIH358 cells
were treated with vector + DMSO, vector + 100 µmol/l 10058-F4 (a
c-myc inhibitor), STK31 + DMSO, or STK31 +100 µmol/l 10058-F4. (A
and B) Cell viability and proliferation were assessed by CCK-8 and
BrdU-ELISA assays, respectively, at 0, 24, 48 and 72 h. (C) The
proportion of cells in each phase of the cell cycle was evaluated
by flow cytometry after 48 h of treatment. (D) The levels of STK31
mRNA (left) and protein (right) were determined. (E) The expression
levels of TRRAP and STK31 were detected by RT-PCR and western
blotting. (F) The direct binding of c-myc to the STK31 promoter was
confirmed by a ChIP assay. The data are expressed as the mean ± SD,
*P<0.05, **P<0.01 and ***P<0.001, compared to vector or
shNC, ##P<0.01, ###P<0.001 compared to
STK31. STK31, serine-threonine kinase 31; DMSO, dimethyl sulfoxide;
CCK-8, Cell Counting Kit-8; TRRAP, transformation/transcription
domain-associated protein; ChIP, chromatin immunoprecipitation;
BrdU-ELISA, bromodeoxyuridine-enzyme-linked immunosorbent
assay. |
Discussion
Emerging research indicates that the STK family of
proteins plays a crucial role in the development of lung cancer.
STK15 is involved in the initiation and prognosis of lung cancer,
and its polymorphisms are reported to be closely associated with
the risk of lung cancer (28).
STK1, a cell cycle-dependent marker, has prognostic value and is a
monitoring factor for NSCLC (29).
STK11 is a tumor suppressor gene in lung cancer (30,31),
and STK39 acts as a tumor oncogene in NSCLC, promoting cell growth,
migration and invasion (32). The
results of the present study demonstrated high expression of STK31
and β-catenin in lung cancer tissues and cell lines, suggesting
that STK31 may function as an oncogene in lung cancer and may
contribute to the carcinogenesis of lung cancer. The downregulation
of STK31 in lung cancer cells exerted an inhibitory effect on cell
viability, cell proliferation and cell cycle progression. This
finding indicated that the downregulation of STK31 could suppress
the proliferation of lung cancer cells by blocking cell cycle
progression, which may be beneficial for lung cancer treatment.
We also investigated the potential molecular
mechanisms through which STK31 regulates lung cancer cell
proliferation. The Wnt/β-catenin pathway was identified as being
positively correlated with STK31 expression by GSEA using lung
cancer samples from the TCGA dataset. This finding was in agreement
with previous studies which revealed that lung cancer cell
proliferation was related to the activation of the Wnt/β-catenin
pathway (33,34). In lung cancer tissues and cells,
STK31 and β-catenin were significantly increased, and
downregulation of STK31 strongly suppressed the proliferation of
lung cancer cells by arresting the cell cycle in the G1 phase
concurrent with a decrease in the protein levels of β-catenin,
c-myc and cyclin D1, whereas upregulation of STK31 had the opposite
effects. c-myc and cyclin D1 are downstream of Wnt/β-catenin and
are reported to play crucial roles in cancer (35,36).
Overexpression of cyclin D1 was revealed to promote cell cycle
progression by inducing cells to enter the S phase (36,37).
Our findings demonstrated that STK31 may regulate lung cancer cell
proliferation via activation of the Wnt/β-catenin pathway, and
c-myc and cyclin D1 may be downstream of STK31/β-catenin.
Furthermore, we determined that STK31-induced proliferation, cell
cycle progression, and increases in β-catenin, c-myc and cyclin D1
levels in lung cancer cells were potently attenuated by a
Wnt/β-catenin inhibitor. These findings further confirmed that the
Wnt/β-catenin pathway is involved in the STK3-mediated regulation
of lung cancer cell proliferation. STK31 is a serine-threonine
kinase that can activate Wnt signaling by phosphorylation and
prevent GSK-3β activation, leading to the accumulation of stable
β-catenin, which regulates the expression of downstream genes, such
as c-myc and cyclin D1, thereby regulating cell proliferation and
cell cycle progression. In addition, the inhibition of c-myc
strongly suppressed the STK-induced proliferation of lung cancer
cells by promoting progression from the G to the S phase and by
significantly decreasing the expression of STK31. Likewise,
knockdown of TRRAP, a protein that interacts with myc, also
significantly decreased STK31 expression. In agreement with the
prediction of the software analysis, the ChIP assay confirmed that
c-myc directly binds to the STK31 promoter. These findings
indicated that c-myc may be an upstream transcription factor of
STK31. We inferred that positive feedback regulation of c-myc may
also be involved in STK31-mediated effects on the proliferation and
cell cycle progression of lung cancer cells, which was consistent
with a previous study that revealed that Wnt signaling-mediated
oncogenesis was associated with the induction of c-myc in cancer
(38). Collectively, these results
demonstrated that STK31 may act as an oncogene in lung cancer and
that downregulation of STK31 in lung cancer cells could suppress
cell proliferation by arresting the cell cycle in the G1 phase,
possibly by inactivating the Wnt/β-catenin pathway and by positive
feedback regulation by c-myc.
In conclusion, the present study demonstrated that
STK31 may be an oncogene in lung cancer. Downregulation of STK31
could potentially inhibit the proliferation of lung cancer cells by
blocking the cell cycle progression, possibly through inactivation
of the Wnt/β-catenin pathway and through the positive feedback
regulation by c-myc. Therefore, the expression of STK31 was
revealed to be closely associated with lung cancer risk and
targeting STK31 may likely provide a potential therapeutic option
for treating lung cancer.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during the present
study are included in this published article.
Authors' contributions
PY and JX conceived and designed the study. JX, SX,
ZD, LN, QX and PL performed the experiments. PY and JX wrote the
manuscript. All authors read and approved the final manuscript and
agree to be accountable for all aspects of the work in ensuring
that questions related to the accuracy or integrity of any part of
the work are appropriately investigated and resolved.
Ethics approval and consent to
participate
All experiments conducted in the present study were
approved by the Ethics Committee of Linyi Central Hospital
(Shandong, China), and written informed consent was obtained.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ferlay J, Shin H, Bray F and Forman D:
Cancer incidence and mortality worldwide: IARC cancer base no. 10.
Int J Cancer J Int Du Cancer. 136:E359–E386. 2015. View Article : Google Scholar
|
|
3
|
Bray F, Jemal A, Grey N, Ferlay J and
Forman D: Global cancer transitions according to the human
development index (2008–2030): A population-based study. Lancet
Oncol. 13:790–801. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hoffman PC, Mauer AM and Vokes EE: Lung
cancer. Lancet. 355:479–485. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fucito LM, Czabafy S, Hendricks PS, Kotsen
C, Richardson D and Toll BA; Association for the Treatment of
Tobacco Use and Dependence/Society for Research on Nicotine and
Tobacco Synergy Committee, : Pairing smoking-cessation services
with lung cancer screening: A clinical guideline from the
association for the treatment of tobacco use and dependence and the
society for research on nicotine and tobacco. Cancer.
122:1150–1159. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Youlden DR, Cramb SM and Baade PD: The
international epidemiology of lung cancer: Geographical
distribution and secular trends. J Thorac Oncol. 3:819–831. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Peifer M and Polakis P: Wnt signaling in
oncogenesis and embryogenesis-A look outside the nucleus. Science.
287:1606–1609. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Dihlmann S and von Knebel Doeberitz M:
Wnt/β-catenin-pathway as a molecular target for future anti-cancer
therapeutics. Int J Cancer. 113:515–524. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
He TC, Sparks AB, Rago C, Hermeking H,
Zawel L, da Costa LTD, Morin PJ, Vogelstein B and Kinzler KW:
Identification of c-MYC as a target of the APC pathway. Science.
281:1509–1512. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tetsu O and Mccormick F: Beta-catenin
regulates expression of cyclin D1 in colon carcinoma cells. Nature.
398:422–426. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Polakis P: Wnt signaling and cancer. Genes
Dev. 14:1837–1851. 2000.PubMed/NCBI
|
|
12
|
Chen G, Shukeir N, Potti A, Sircar K,
Aprikian A, Goltzman D and Rabbani SA: Up-regulation of Wnt-1 and
beta-catenin production in patients with advanced metastatic
prostate carcinoma: Potential pathogenetic and prognostic
implications. Cancer. 101:1345–1356. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang WM, Lo Muzio L, Rubini C and Yan G:
Effect of WNT-1 on β-catenin expression and its relation to Ki-67
and tumor differentiation in oral squamous cell carcinoma. Oncol
Rep. 13:1095–1099. 2005.PubMed/NCBI
|
|
14
|
Nguyen DX, Chiang AC, Zhang XHF, Kim JY,
Kris MG, Ladanyi M, Gerald WL and Massagu J: WNT/TCF signaling
through LEF1 and HOXB9 mediates lung adenocarcinoma metastasis.
Cell. 138:51–62. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Baselga J: Targeting tyrosine kinases in
cancer: The second wave. Science. 312:1175–1178. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Krause DS and Van Etten RA: Tyrosine
kinases as targets for cancer therapy. N Engl J Med. 353:172–187.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kuo PL, Huang YL, Hsieh CC, Lee JC, Lin BW
and Hung LY: STK31 is a cell cycle regulated protein that
contributes to the tumorigenicity of epithelial cancer cells. PLoS
One. 9:e933032014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhong L, Liu J, Hu Y, Wang W, Xu F, Xu W,
Han J and Biskup E: STK31 as novel biomarker of metastatic
potential and tumorigenicity of colorectal cancer. Oncotarget.
8:24354–24361. 2017.PubMed/NCBI
|
|
20
|
Watany MM, Elmashad NM, Badawi R and
Hawash N: Serum FBLN1 and STK31 as biomarkers of colorectal cancer
and their ability to noninvasively differentiate colorectal cancer
from benign polyps. Clin Chim Acta. 483:151–155. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hong JY, Kang B, Kim A, Hwang S, Ahn J,
Lee S, Kim J, Park JH and Cheon DS: Development of a highly
sensitive real-time one step RT-PCR combined complementary locked
primer technology and conjugated minor groove binder probe. Virol
J. 8:330. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCT method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES and Mesirov JP: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Vogelstein B and Kinzler KW: Cancer genes
and the pathways they control. Nature Med. 10:789–799. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lim JH, Park JW and Chun YS: Human arrest
defective 1 acetylates and activates β-Catenin, promoting lung
cancer cell proliferation. Cancer Res. 66:106772006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bouchard C, Dittrich O, Kiermaier A,
Dohmann K, Menkel A, Eilers M and Lüscher B: Regulation of cyclin
D2 gene expression by the Myc/Max/Mad network: Myc-dependent TRRAP
recruitment and histone acetylation at the cyclin D2 promoter.
Genes Dev. 15:2042–2047. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bhattacharya S and Ghosh MK: HAUSP
regulates c-MYC expression via de-ubiquitination of TRRAP. Cell
Oncol (Dordr). 38:265–277. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Gu J, Gong Y, Huang M, Lu C, Spitz MR and
Wu X: Polymorphisms of STK15 (Aurora-A) gene and lung cancer risk
in Caucasians. Carcinogenesis. 28:350–355. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li H, Lei DS, Wang XQ, Skog S and He Q:
Serum thymidine kinase 1 (STK1) is a prognostic and monitoring
factor in patients with non-small-cell lung cancer. Oncol Rep.
13:145–149. 2005.PubMed/NCBI
|
|
30
|
Carretero J, Medina PP, Pio R, Montuenga
LM and Sanchez-Cespedes M: Novel and natural knockout lung cancer
cell lines for the LKB1/STK11 tumor suppressor gene. Oncogene.
23:4037–4040. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Gill RK, Yang SH, Meerzaman D, Mechanic
LE, Bowman ED, Jeon HS, Roy Chowdhuri S, Shakoori A, Dracheva T,
Hong KM, et al: Frequent homozygous deletion of the LKB1/STK11 gene
in non-small cell lung cancer. Oncogene. 30:3784–3791. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li Z, Zhu W, Xiong L, Yu X, Chen X and Lin
Q: Role of high expression levels of STK39 in the growth, migration
and invasion of non-small cell type lung cancer cells. Oncotarget.
7:61366–61377. 2016.PubMed/NCBI
|
|
33
|
Gao Y, Song C, Hui L, Li CY, Wang J, Tian
Y, Han X, Chen Y, Tian DL, Qiu X and Wang E: Overexpression of
RNF146 in non-small cell lung cancer enhances proliferation and
invasion of tumors through the Wnt/β-catenin signaling pathway.
PLoS One. 9:e853772014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wan L, Zhang L, Fan K, Cheng ZX, Sun QC
and Wang JJ: Circular RNA-ITCH suppresses lung cancer proliferation
via inhibiting the Wnt/β-catenin pathway. Biomed Res Int.
2016:15794902016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Miller DM, Thomas SD, Islam A, Muench D
and Sedoris K: c-Myc and cancer metabolism. Clin Cancer Res.
18:5546–5553. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hall M and Peters G: Genetic alterations
of cyclins, cyclin-dependent kinases, and Cdk inhibitors in human
cancer. Adv Cancer Res. 68:67–108. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lukas J, Bartkova J, Rohde M, Strauss M
and Bartek J: Cyclin D1 is dispensable for G1 control in
retinoblastoma gene-deficient cells independently of cdk4 activity.
Mol Cell Biol. 15:2600–2611. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
You Z, Saims D, Chen S, Zhang Z, Guttridge
DC, Guan KL, Macdougald OA, Brown AM, Evan G, Kitajewski J and Wang
CY: Wnt signaling promotes oncogenic transformation by inhibiting
c-Myc-induced apoptosis. J Cell Biol. 157:429–440. 2002. View Article : Google Scholar : PubMed/NCBI
|