Introduction
Liver cancer is one of the most common cancers and
is a serious threat to human health. After surgery, the incidence
of tumor recurrence and metastasis is high, and the prognosis is
poor. Due to the development of chemotherapy and novel treatments,
such as immune checkpoint inhibitors (1) and chimeric antigen receptor T cells
(2), rates of recurrence and
metastasis have decreased and the 5-year survival rate has
improved. However, the mechanisms underlying the metastasis and
recurrence of liver cancer have not been fully elucidated.
Therefore, the discovery of new regulatory molecules to develop
more effective liver cancer treatment strategies remains
crucial.
Long non-coding RNAs (lncRNAs), non-coding RNAs with
>200 nucleotides, are involved in the transcriptional and
post-transcriptional regulation of various biological processes
involved in the development of tumors, and are associated with
tumor prognosis (3,4). Increasing studies have reported that
lncRNAs are implicated in liver cancer.
In previous studies, various lncRNAs, such as the
PVT1 oncogene (5), colorectal
neoplasia differentially expressed (CRNDE) (6) and CDKN2B antisense RNA 1 (CDKN2B-AS1)
(7) have been found to regulate HCC
cell migration, invasion, proliferation, apoptosis and other
important biological processes. DiGeorge syndrome critical region
gene 5 (DGCR5) was found to play a role in HCC by inhibiting the
progression of HCC through inactivation of the Wnt signaling
pathway (8). DGCR5 was found to
repress the development of HCC by targeting the miR-346/KLF14 axis
(9). Hepatocellular carcinoma
upregulated long non-coding RNA (HULC) was found to regulate the
drug resistance of HCC by triggering autophagy by stabilizing
Sirtuin 1 (10). Although thousands
of lncRNAs have been identified to be associated with liver cancer,
various aspects warrant further investigation and examination,
including the regulatory mechanisms of lncRNAs in liver cancer, the
underlying mechanisms of the association between lncRNAs and liver
cancer, and novel molecular markers that may be discovered.
lncRNA-BC200, also known as BCYRN1, is a
brain-specific small cytoplasmic RNA with a length of 200
nucleotides that is transcribed by RNA polymerase III. Human
lncRNA-BC200 is located on chromosome 2p16. lncRNA-BC200 consists
of a monomeric Alu, an A-rich central region with a unique C-rich
region, which can be divided into three domains. The 5′ end is
homologous to the high copy number Alu repeater in the primate
genome. The 3′ end is unique and has no apparent similarity to
known human DNA sequences, but is similar to several short elements
of rodent BC1 RNA. lncRNA-BC200 has RHAU helicase activity and
binds to RHAU, exerting regulatory functions by unwinding the
four-chain cytosine-rich tetra-chain at the 3′ end (11). lncRNA-BC200 can also bind to a
variety of proteins, such as poly(A)-binding protein, heterodimeric
signal recognition particles, fragile X mental retardation protein
and synaptic cytoplasmic interactions protein. Previous studies
have shown that lncRNA-BC200 serves an important role in certain
human diseases, such as asthma, Alzheimer's disease, and various
common tumors (12,13). Singh et al (14) showed that lncRNA BC200 is expressed
to a higher degree in estrogen receptor-positive breast cancer
compared with estrogen receptor-negative breast cancer, and low
expression of BC200 can inhibit the proliferation of
estrogen-dependent breast cancer tumor cells in vitro and
in vivo, by promoting the protein expression of the
apoptotic factor Bcl-xS. Zhao et al (15) showed that lncRNA BC200 is
significantly increased in esophageal squamous cell carcinoma and
is an independent risk factor for disease-free survival and overall
survival in patients with esophageal squamous cell carcinoma.
Notably, a high expression level of lncRNA BC200 may be associated
with poor prognosis. Recent studies have shown that lncRNA BC200 is
highly expressed in non-small cell lung cancer. In addition,
c-MYC can bind to the promoter region of the lncRNA BC200
gene. In vitro cell transfer assays have shown that lncRNA
BC200 regulates cell migration and invasion (16). Recent studies have shown that BC200
is highly expressed in hepatocellular carcinoma (HCC) and is an
effective independent prognostic marker. In addition, T3/TR
(thyroid hormone/its receptor) has a negative regulatory effect on
the expression of BC200 (17).
Lin et al (17) silenced BC200 in J7 and SK-Hep1 cells
by shRNA technology, and overexpressed BC200 in Hep3B and Huh7
cells, and further studied the function of BC200 in vitro
and in vivo. The results showed that BC200 promoted cell
growth and transformation in vitro and in vivo. In
the present study, we knocked out BC200 by using CRISPR/Cas9
technology and simultaneously knocked out and overexpressed BC200
in the same cell line, and investigated the expression, function
and potential mechanism of BC200 in liver cancer in vitro
and in vivo. According to a previous study, BC200 affects
cancer cell survival and proliferation (13). BC200 lncRNA is considered to be a
potential predictor of poor prognosis in esophageal squamous cell
carcinoma (15). However, there are
few reports on the effect of BC200 on liver cancer. In the present
study, the expression, function and potential mechanisms of BC200
in liver cancer were investigated in vitro and in
vivo.
Materials and methods
Collection of HCC tissue
specimens
In total, 45 pairs of matched HCC tissues and
adjacent tissues were obtained from patients at the Affiliated
Tumor Hospital of Guangxi Medical University between December 2016
and March 2017 (Table I). All
patients had no history of anticancer therapy. All tissues were
maintained in liquid nitrogen. Informed consent was obtained by
each patient. The present study was approved by The Ethics
Committee of the Affiliated Tumor Hospital of Guangxi Medical
University.
 | Table I.Characteristics of HCC patients. |
Table I.
Characteristics of HCC patients.
| Patients | Sex | Age (years) |
|---|
| 1 | Male | 43 |
| 2 | Male | 60 |
| 3 | Male | 57 |
| 4 | Male | 47 |
| 5 | Male | 70 |
| 6 | Male | 43 |
| 7 | Male | 55 |
| 8 | Male | 54 |
| 9 | Male | 60 |
| 10 | Male | 61 |
| 11 | Male | 63 |
| 12 | Male | 48 |
| 13 | Male | 63 |
| 14 | Male | 41 |
| 15 | Male | 45 |
| 16 | Male | 42 |
| 17 | Male | 43 |
| 18 | Male | 50 |
| 19 | Female | 41 |
| 20 | Male | 45 |
| 21 | Female | 63 |
| 22 | Male | 49 |
| 23 | Male | 69 |
| 24 | Male | 68 |
| 25 | Male | 68 |
| 26 | Male | 35 |
| 27 | Male | 37 |
| 28 | Male | 66 |
| 29 | Male | 63 |
| 30 | Male | 43 |
| 31 | Male | 49 |
| 32 | Male | 67 |
| 33 | Male | 64 |
| 34 | Male | 44 |
| 35 | Male | 58 |
| 36 | Male | 60 |
| 37 | Male | 55 |
| 38 | Male | 47 |
| 39 | Male | 33 |
| 40 | Male | 67 |
| 41 | Female | 24 |
| 42 | Male | 31 |
| 43 | Female | 69 |
| 44 | Male | 20 |
| 45 | Male | 42 |
RNA extraction and reverse
transcription-quantitative (RT-q)PCR
RNA was extracted from tissue or HepG2 cells using
TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.).
NanoDrop 2000 (Thermo Fisher Scientific, Inc.) instrument was used
to determine the concentration and quality of RNA. The RNA was
reverse transcribed using the Moloney murine leukemia virus reverse
transcriptase (Vazyme). Subsequently, qPCR was performed using the
ChamQ™SYBR®qPCR Master Mix (High ROX Premixed; Vazyme)
according to the manufacturer's instructions. The relative gene
expression was calculated using the 2−ΔΔCq method
(18). The primers used in the
present study were the following: GAPDH forward,
AACGGATTTGGTCGTATTG and reverse, GGAAGATGGTGATGGGATT; c-MYC
forward, TATCCCTAACTCTACATCAACC and reverse, TCAAATCTCGCTTCCACTT;
BC200 forward, GCCTGTAATCCCAGCTCTCA and reverse,
GTTGCTTTGAGGGAAGTTACGCT.
Cell culture
HepG2 cells were purchased from The Chinese Academy
of Sciences Cell Bank. The culture conditions of the cells were:
10% FBS DMEM (BI, Biological Industries), 5% CO2 and
37°C incubator.
Plasmid transfection
Knockout (KO) of BC200 was performed using
CRISPR/Cas9 technology as previously described (19). An empty plasmid (Plasmid-con) and
the plasmid for BC200 KO (Plasmid-BC200 KO) were obtained from the
University of Mississippi (Department of Pharmacology/Toxicology
Cancer Institute University of Mississippi Medical Center, Jackson,
MS, USA). The plasmid carries green fluorescence and puromycin
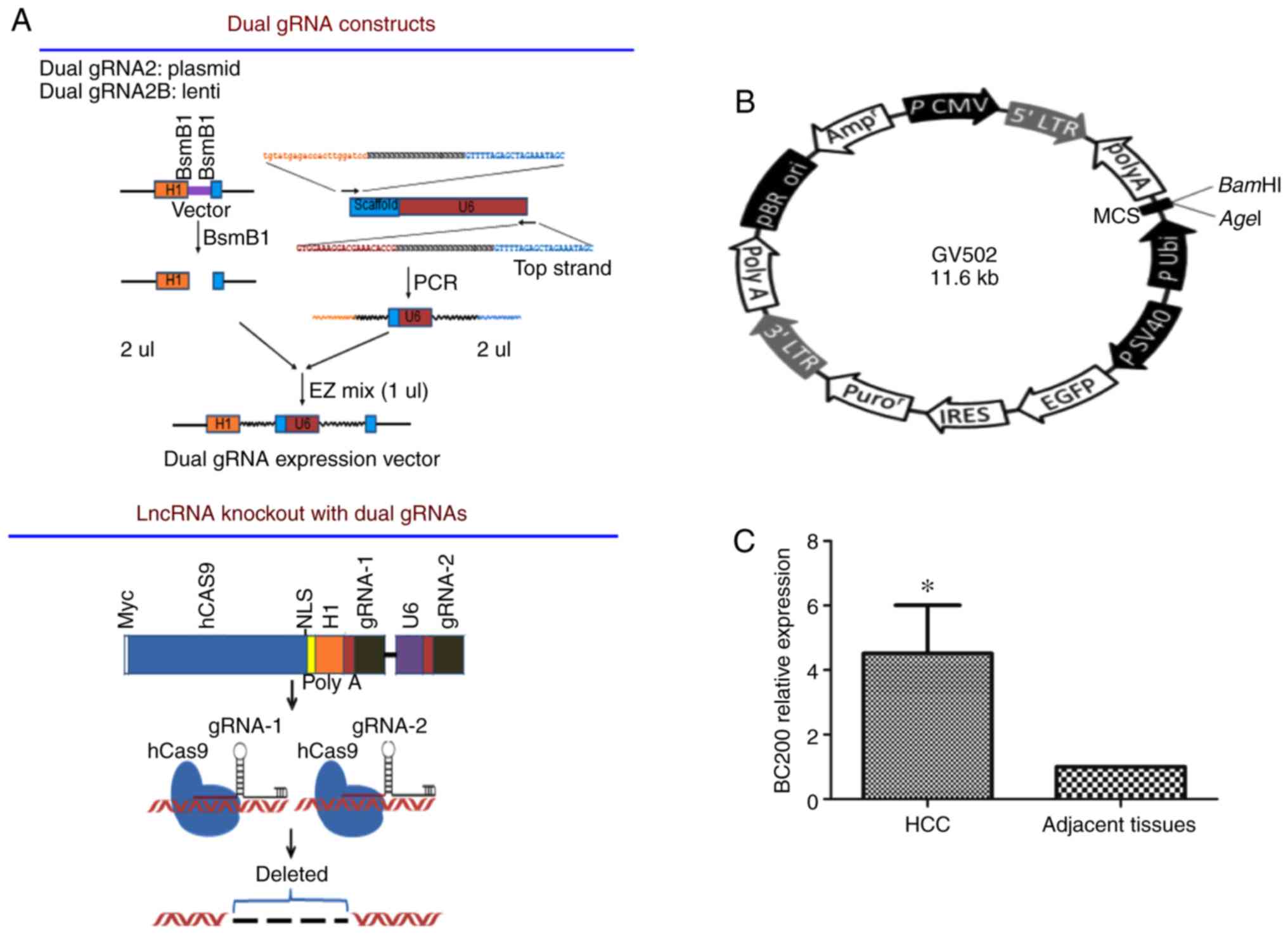
resistance, and the plasmid construction process is shown in
Fig. 1A. Plasmids were transfected
into HepG2 cells with Lipofectamine 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.), and cells were divided into: i) Negative control
(NC) group; ii) Plasmid-con group; and iii) Plasmid-BC200 KO group.
Cells were transfected with the indicated plasmid, and 1–3 days
after, stable cell lines were screened using 1 µg/ml puromycin.
Lentivirus (LV) infection
The lentiviral vectors were purchased from Shanghai
Jikai Gene Co., Ltd. The target gene vector was (polyA-MCS-UBI)
RV-SV40-EGFP- IRES-puromycin, and the negative control virus vector
was Ubi-MCS-SV40-EGFP-IRES-puromycin (Fig. 1B). Vectors were infected into HepG2
cells according to the manufacturer's instructions. Cells were
grouped into: i) NC group; ii) LV-con group; and iii) LV-BC200
group. After 1–3 days, stable cell lines were screened using 1
µg/ml puromycin.
Cell proliferation assay
Cells were seeded into 96-well plates at a density
of 2,000 cells/well. The assay was performed every 24 h. Cell
Counting Kit-8 [CCK-8; Multisciences (Lianke) Biotech Co., Ltd.]
solution was added 1 h before testing, The plates were incubated
for 5 days. Detection of absorbance at 450 nm was performed using a
microplate reader.
Cell migration assay
Transwell chambers (Corning, Inc.) were used for
Transwell migration assay. The cells were seeded at a density of
3×104/ml. In total, 200 µl of single-cell suspension was
added to the upper Transwell chamber, and the lower chamber was
filled with DMEM supplied with 10% FBS, while the upper chamber
contained serum-free DMEM. After 20 h, the cells were stained using
10% Giemsa staining for 30 min (Beijing Solarbio Science &
Technology Co., Ltd.), and observed under a microscope (Olympus
Corp.). A total of 10 high power fields of view were
randomly-selected, cells were counted and the mean cell number was
calculated.
Western blot analysis
RIPA buffer was used to lyse cells and extract
cellular proteins. Electrophoresis was used for protein separation.
Protein samples (150 µg) (including protein samples from HepG2
cells from the NC group, control group, BC200 KO group and LV-BC200
group) were transferred to a nitrocellulose membrane following
separation by 10% SDS-PAGE. The membranes were incubated with
various primary antibodies. Anti-Bax (cat. no. WL01637; rabbit mAb;
Wanleibio Co., Ltd.), anti-c-Myc (D3N8F) (cat. nο 13987; rabbit
mAb; Cell Signaling Technology, Inc.) and anti-Bcl-xL (54H6) (cat.
no. 2764; rabbit mAb; Cell Signaling Technology, Inc.) GAPDH (cat.
no. 5174; Cell Signaling Technology, Inc.) was used as the loading
control. Secondary antibody was HRP-labeled goat anti-rabbit IgG
(H+L) (cat. no. A0208; Beyotime Institute of Biotechnology). The
dilutions were carried out using Primary Antibody dilution buffer
(cat. no. A1810; Beijing Solarbio Science & Technology Co.,
Ltd.) and Secondary Antibody dilution buffer (cat. no. P0023D;
Beyotime Institute Biotechnology). The primary or secondary
antibodies were diluted at a ratio of 1:1,000. Protein bands were
analyzed using Gel Doc XR+ Analyzer Software (Bio-Rad Laboratories,
Inc.).
Experimental animals
The Experimental Animal Ethics Committee of Guangxi
Medical University approved the present study. In total, 18 male
BALB/c nude mice (weight, 16–20 g; age, 4–6 weeks) were purchased
from the Laboratory Animal Center of Guangxi Medical University.
Animals were housed at the animal facility of the Laboratory Animal
Center of Guangxi Medical University. The experimental conditions
of the mice included: A specific pathogen-free environment, 12-h
light/dark cycle, and free access to food and water. In order to
establish a subcutaneous tumor model, cultured HepG2 cells were
diluted using PBS at a final concentration of 2×106/ml.
In total, 200 µl of the cell suspension was subcutaneously injected
into the right leg of the mice. Nude mice were divided into: i) NC
group (n=6); ii) BC200 KO group (n=6); and iii) LV-BC200 group
(n=6). The weight of the nude mice and subcutaneous tumor volume
were measured every other week. Mice were sacrificed by cervical
dislocation on day 60. Subsequently, the subcutaneous solid tumor
was collected for measuring the tumor weight. Tumor volume was
calculated as follows: Tumor volume=length ×
(width)2/2.
Statement of authentication
HepG2 cell line has been authenticated by STR
profiling.
Statistical analysis
All data are presented as the mean ± SD. Differences
between groups were evaluated by Student's t-test, one-way ANOVA or
repeated measures ANOVA using SPSS 17.0 software. P<0.05 was
considered to indicate a statistically significant difference.
Results
BC200 is upregulated in HCC
To investigate the expression pattern of BC200 in
human HCC, the mRNA expression level of BC200 was examined in 45
matched pairs of adjacent tissue and HCC tissue specimens using
RT-qPCR. The levels of BC200 were measured and compared between HCC
and healthy samples. A higher expression level of BC200 was
detected in the HCC samples compared to the level noted in the
normal adjacent liver tissues (Fig.
1C).
Verification of transfection
efficiency
In order to downregulate the expression of BC200, a
BC200 KO plasmid was transfected into HepG2 cells to further
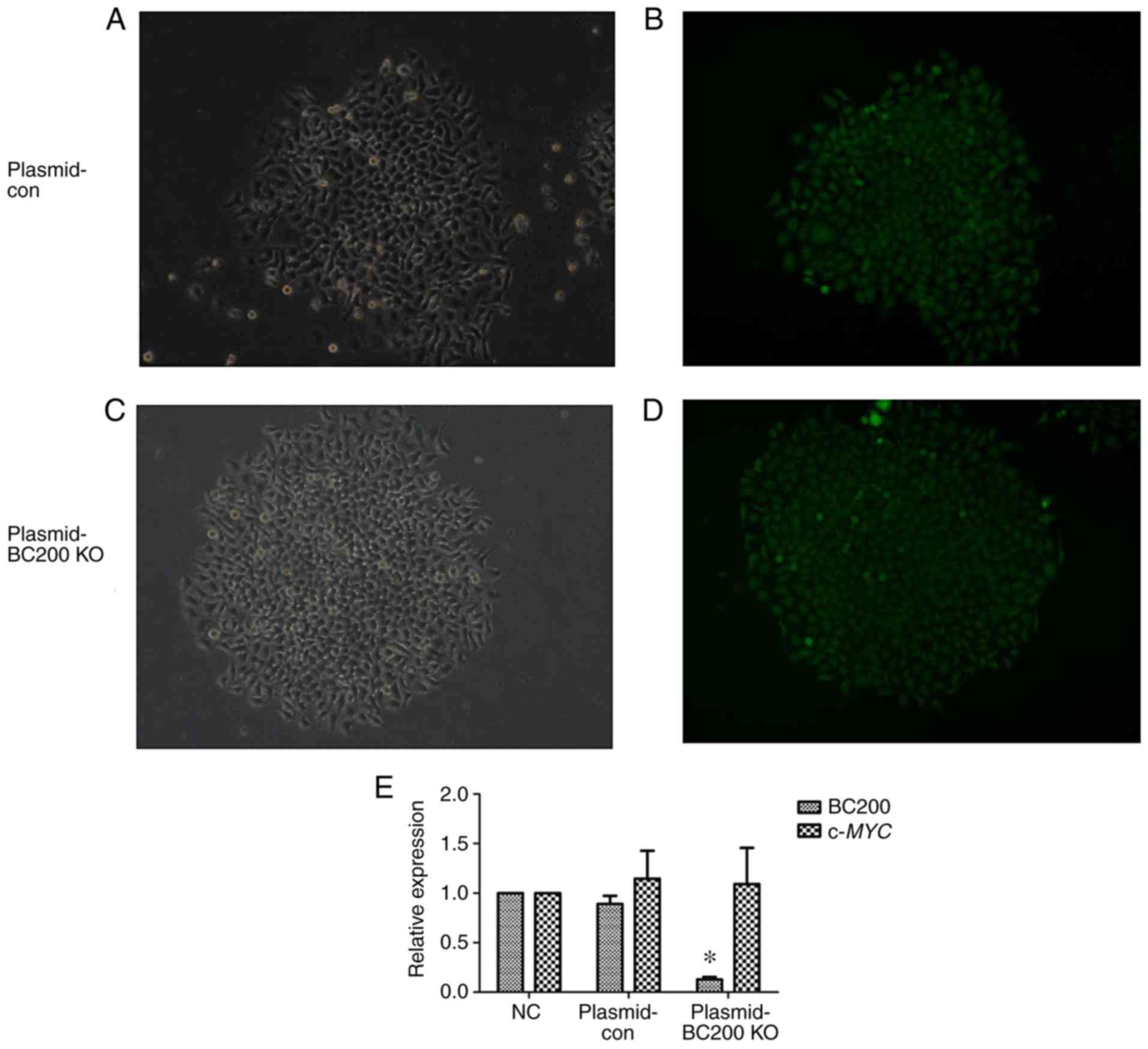
examine the functional role of BC200 in liver cancer. Fluorescence
of HepG2 cells after plasmid transfection was assessed under a
fluorescence microscope (Fig.
2A-D). The qPCR results showed a significant decrease in the
expression level of BC200 in the BC200-KO (Plasmid-BC200 KO) group
compared to the control (Plasmid-con) and NC groups (Fig. 2E). The RNA expression level of
c-MYC did not change significantly among the NC, Plasmid-con
and Plasmid-BC200-KO groups (Fig.
2E).
Verification of infection
efficiency
Cells were observed after 72 h of lentiviral
infection. It was found that >90% of cells expressed
fluorescence and presented a normal morphology (Fig. 3A-D). The expression level of BC200
was detected by qPCR in HepG2 cells from the NC group, LV-control
and LV-BC200 groups. In the LV-BC200 group, the expression level of
BC200 was significantly higher than levels noted in the NC and
LV-control groups (P<0.05; Fig.
3E). The mRNA expression level of c-MYC was not
significantly altered among the NC, LV-con and LV-BC200 groups
(Fig. 3E).
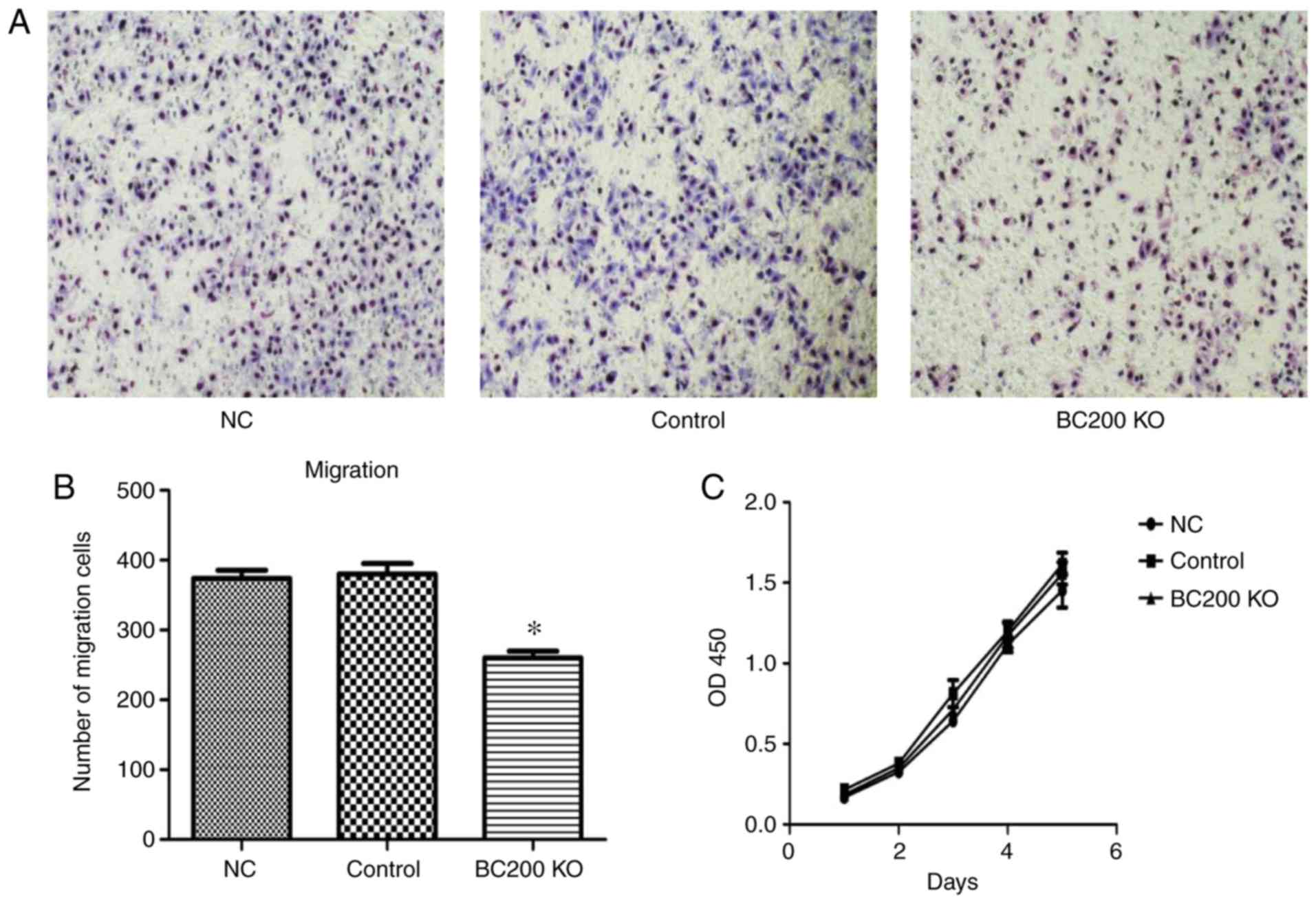
Effect of BC200 KO on the migration
and proliferation of HepG2 cells
After transfection of plasmid-BC200 KO into HepG2
cells, the effect of BC200 KO on proliferation and migration was
investigated. As shown in Fig. 4,
Transwell assay demonstrated that the migration rate of HepG2 cells
was significantly reduced after BC200 KO transfection (Fig. 4A and B). CCK-8 experiments showed
that there were no significant differences in the proliferation of
HepG2 cells among the BC200 KO, NC and control groups. The
proliferation curve was drawn according to the optical density
value (Fig. 4C).
Effect of overexpression of BC200 on
the migration and proliferation of HepG2 cells
After infection of LV-BC200 into HepG2 cells, the
influence of LV-BC200 on the proliferation and migration of HepG2
cells was investigated. As shown in Fig. 5, migration of HepG2 cells infected
with LV-BC200 was significantly promoted at 20 h. Transwell
experiments showed that the migration rate of HepG2 cells in the
LV-BC00 group was significantly higher than that in the NC and
control groups (Fig. 5A and B). The
proliferation of HepG2 cells transfected with LV-BC200 was not
significantly increased compared with the NC and control groups
during the 5 day period. The proliferation curve was drawn
according to the optical density value (Fig. 5C).
Effect of BC200 on subcutaneous tumor
formation in nude mice
To examine the effect of BC200 on tumor growth in
vivo, stable cell lines were transfected with a plasmid
overexpressing BC200 and BC200 KO and selected by puromycin
resistance. The present results showed that the volume and weight
of tumors in the BC200 KO group (0.53±0.45 cm3 and
0.70±0.61 g) were significantly decreased compared with the
LV-BC200 group (1.85±0.42 cm3 and 2.0±0.16 g; P<0.05;
Fig. 6A-D). The increase in the
body weight of the mice in the LV-BC200 group was lower than that
in the BC200 KO group (Fig.
6E).
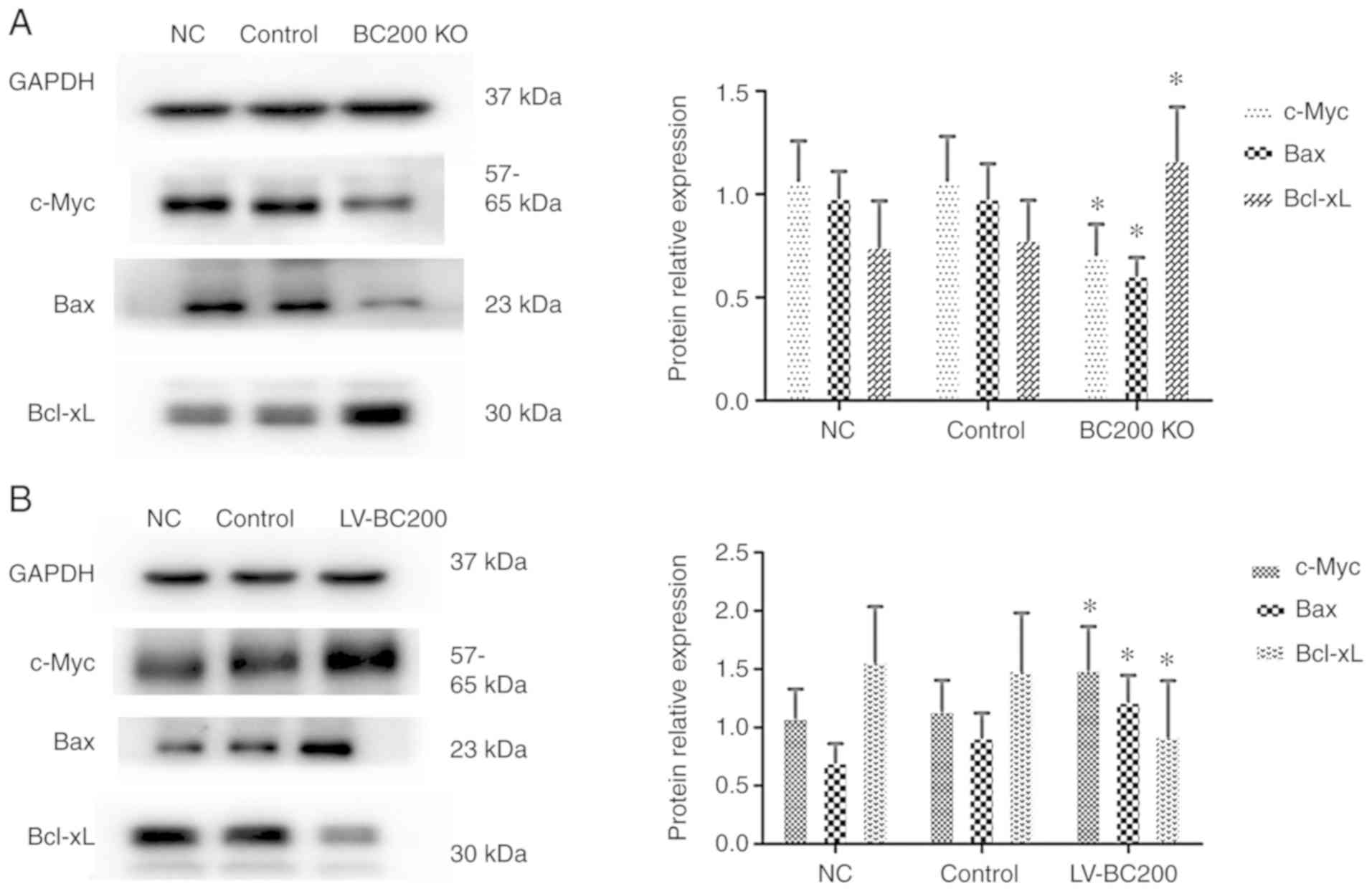
BC200 levels affect the protein
expression levels of c-Myc, Bax and Bcl-xL in HepG2 cells
In order to examine the exact mechanism of BC200,
western blotting was performed to detect the protein expression
levels of various factors in HepG2 cells. After knockout of BC200
the protein expression levels of c-Myc and Bax were decreased in
the BC200 KO group cells compared with the NC and control groups,
and Bcl-xL was increased (Fig. 7A).
Conversely, the protein expression levels of c-Myc and Bax were
increased in the LV-BC200 group compared with the NC and control
groups, while Bcl-xL was decreased (Fig. 7B). The present results suggested
that BC200 function is associated with c-Myc, Bax and Bcl-xL
proteins.
Discussion
During the past two decades, the role of long
non-coding RNAs in cancer has received increased attention
(20). In total, more than 60,000
lncRNAs have been identified, accounting for ~60% of the
transcriptome (21). In recent
years, research on lncRNAs in liver cancer has increased. lncRNAs
serve a variety of roles in the development of liver cancer, by
acting as an anti-apoptotic factor (22), promoting HCC metastasis (23), acting as prognostic biomarkers
(24), and promoting cell
proliferation, migration and invasion (7). However, the mechanisms underlying the
role of lncRNAs in liver cancer are not fully understood, and
further investigation is required.
lncRNA-BC200 was found to be expressed in the
cytoplasm, particularly in dendritic neuronal cells, and
transcribed by RNA polymerase III (25–27).
In addition, high expression of BC200 has been reported in various
tumor types. Chen et al (28) used northern hybridization technology
to detect the expression of BC200 RNA in various human tumor
tissues and found that BC200 is highly expressed in various tumor
tissues. A recent study identified that BC200 has a low expression
level relative to GAPDH in the liver (13). The present results demonstrated that
BC200 expression level in HCC tissues was higher than that in
adjacent tissues. However, the present data are in contrast with a
previous study by Chen et al (28) that suggested that BC200 RNA is not
detectable in liver carcinoma tissues. However, the present study
analyzed BC200 expression by qPCR in 45 matched pairs of HCC
tissues and adjacent tissues. The study by Chen et al
(28) investigated four patients
with liver carcinoma. Importantly, there were differences in the
detection method and in the number of samples, which may have
affected the experimental results. Lin et al (17) used RT-qPCR and In situ
hybridization (ISH) methods and found that BC200 expression was
significantly upregulated in HCC tissues compared with that noted
in benign and adjacent normal tissues. Moreover, Kaplan-Meier
survival analysis showed the association of high BC200 expression
with the poor overall survival (OS) rate of HCC patients. In the
TCGA database (http://cancergenome.nih.gov/), patients with high
expression of BC200 showed lower OS and disease-free survival than
patients with low expression of BC200.
To the best of our knowledge, the present study is
the first to detect the expression of BC200 in HepG2 cells. In
addition, various cell models were established, including BC200 KO
and BC200 overexpression cells, and RT-qPCR was used to verify that
the in vitro models were successfully constructed. The
present results showed that BC200 KO inhibited the migration of
HepG2 cells, and high expression of BC200 promoted the migration of
HepG2 cells. The present results are in line with a recent study
indicating that knockdown of BC200 RNA expression reduces cell
migration and invasion (29).
In the present study, BC200 was found to influence
cell proliferation by CCK-8 assay. The present results showed that
BC200 did not affect the proliferation of HepG2 cells, either
following knockout of BC200 or overexpression of BC200. These
results are in line with a recent study showing that the growth
rate was not affected in HCT116 cells transfected with BC200 small
interfering RNA (30). In addition,
the present study investigated the role of BC200 in vivo,
and overexpression of BC200 was found to promote the growth of
subcutaneous tumors in nude mice, whereas the knockout of BC200
inhibited the growth of subcutaneous tumors. These results are in
line with a previous study that showed that BC200 KO significantly
reduced tumor growth in female nude mice (14). Collectively, the present results
indicated that under in vitro conditions, where the cell
environment is relatively simple, changing the expression level of
BC200 did not affect cell proliferation. However, in a complex
in vivo environment, the expression level of BC200 affected
the growth of tumors in nude mice, indicating that the regulation
of cell proliferation by BC200 is context-dependent. The
environment of nude mice is more similar to the human body
environment compared with in vitro conditions, and in
vivo experiments can better reflect the role of BC200 in
humans.
c-Myc protein was previously identified to interact
with the BC200 promoter, and high expression of BC200 is due to the
binding of c-Myc to its promoter (16). Research by Hu and Lu (16). provided new ideas for our research.
Therefore, we conversely aimed to ascertain whether altering the
expression of BC200 affects the expression of c-Myc. In the present
study, the association between BC200 and c-MYC was
investigated at the RNA and protein levels following knockout and
overexpression of BC200 in HepG2 cells. The present results showed
that knockout of BC200 or overexpression of BC200 had no
significant effects on c-MYC mRNA levels, whereas c-Myc
protein expression was significantly decreased in the BC200 KO
group. Conversely, c-Myc protein was increased in the LV-BC200
group. This effect may have been caused by the lack of lncRNA-BC200
effects on the transcription level of the c-MYC gene. By
contrast, lncRNA-BC200 may have an effect only on the translation
rate of c-MYC mRNA. c-MYC is an oncogene that plays
an important role in cell proliferation, differentiation, apoptosis
and cell cycle. c-MYC is a transcription factor that
requires dimerization with another protein, such as Myc-associated
protein X, to become transcriptionally active. The mechanism by
which c-MYC oncogene/oncoprotein participates in cell
proliferation, apoptosis and cycle may vary depending on the
context and is regulated by a variety of factors (31).
The Bcl-2 family is divided into two classes,
pro-apoptotic proteins and anti-apoptotic proteins; Bcl-xL is an
anti-apoptotic protein, whereas Bcl-xS and Bax are pro-apoptotic
proteins (32,33). Singh et al (14) showed that knockout of BC200
increased Bcl-xS protein expression, whereas Bcl-xL protein was not
altered significantly. The regulation of Bcl-xS by BC200 is of
great significance for the pathogenesis of breast cancer. Gu et
al (34) showed that BC200 can
affect the expression of Bax and Bcl-2, suggesting that BC200 is
involved in the apoptosis of colorectal cancer (CRC) cells. It has
been previously reported that the ratio of Bax/Bcl-xL can be used
to measure the level of apoptosis (35). In the present study, following BC200
KO, Bax protein expression was decreased and Bcl-xL protein
expression was increased. Conversely, in the LV-BC200 group, Bax
protein expression was increased and Bcl-xL protein expression was
decreased. Collectively, the present results suggested that
lncRNA-BC200 may affect both apoptotic and anti-apoptotic proteins.
However, due to the balancing effects of anti-apoptotic Bcl-xL and
pro-apoptotic Bax, cell apoptotic rates may not be affected. Our
study is the first to study the relationship between BC200 and
Bcl-xL and Bax protein in liver cancer. The results of this study
are inconsistent with the results in other cancers, probably due to
other regulatory networks in liver cancer. However, there is no
relevant research to prove our guess, and more research is needed
to support it in the future. A review of the literature also found
a corresponding example, in neuroblastoma cells. Inhibition of the
expression of long noncoding RNA KCNQ1OT1, found that the
expression of Bax was reduced (36). In diabetic retinopathy (DR), Bax
expression was found to be increased by inhibiting the expression
of long noncoding RNA KCNQ1OT1 (37). This shows that there are different
regulatory effects in different cancers.
Acknowledgements
Thanks to Professor Yin-Yuan Mo from the University
of Mississippi Medical Center (Jackson, MS, USA), 56FD (Department
of Pharmacology/Toxicology Cancer Institute University of
Mississippi Medical Center, Jackson, MS, USA), for providing the KO
BC200 plasmid and technical support.
Funding
The present study was supported in part by grants
from the Natural Science Foundation of Guangxi Province (grant no.
2017GXNSFAA198015), The Technology Development and Promotion
Foundation of Guangxi Medical and Health Appropriate (grant no.
S2017104), Guangxi Key Research and Development Program (grant no.
guikeAB19110007).
Availability of data and materials
The datasets used and analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
NT, CO and BZ conceived and designed the experiment.
NT and HS performed the experiments. YFT and JRW collected and
analyzed the data. MF, CRL and ZQC collected the clinical samples
and analyzed the clinical data. NT wrote the manuscript. NT, CO and
HS reviewed/edited the manuscript. All authors read and approved
the manuscript and agree to be accountable for all aspects of the
research in ensuring that the accuracy or integrity of any part of
the work are appropriately investigated and resolved.
Ethics approval and consent to
participate
The patient study was approved by the Academic
Committee of the Ethics Committee of the Affiliated Tumor Hospital
of Guangxi Medical University. All patients and healthy volunteers
provided written informed consent prior to their inclusion within
the study. The animal study was approved by The Experimental Animal
Ethics Committee of Guangxi Medical University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Kudo M: Immune checkpoint inhibition in
hepatocellular carcinoma: Basics and ongoing clinical trials.
Oncology. 92:50–62. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yu S, Li A, Liu Q, Li T, Yuan X, Han X and
Wu K: Chimeric antigen receptor T cells: A novel therapy for solid
tumors. J Hematol Oncol. 10:782017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhu L, Huang F, Wan T, Xu H and Zhao Q:
Overexpression of long noncoding RNA LINC00882 is associated with
poor prognosis in hepatocellular carcinoma. Onco Targets Ther.
11:5209–5217. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Huang Y, Xiang B, Liu Y, Wang Y and Kan H:
LncRNA CDKN2B-AS1 promotes tumor growth and metastasis of human
hepatocellular carcinoma by targeting let-7c-5p/NAP1L1 axis. Cancer
Lett. 437:56–66. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Xu Y, Luo X, He W, Chen G, Li Y, Li W,
Wang X, Lai Y and Ye Y: Long non-coding RNA PVT1/miR-150/HIG2 axis
regulates the proliferation, invasion and the balance of iron
metabolism of hepatocellular carcinoma. Cell Physiol Biochem.
49:1403–1419. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ji D, Jiang C, Zhang L, Liang N, Jiang T,
Yang B and Liang H: LncRNA CRNDE promotes hepatocellular carcinoma
cell proliferation, invasion, and migration through regulating
miR-203/BCAT1 axis. J Cell Physiol. 234:6548–6560. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Huang D, Bi C, Zhao Q, Ding X, Bian C,
Wang H, Wang T and Liu H: Knockdown long non-coding RNA ANRIL
inhibits proliferation, migration and invasion of HepG2 cells by
down-regulation of miR-191. BMC Cancer. 18:9192018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang XL, Shi M, Xiang T and Bu YZ: Long
noncoding RNA DGCR5 represses hepatocellular carcinoma progression
by inactivating Wnt signaling pathway. J Cell Biochem. 120:275–282.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang YG, Liu J, Shi M and Chen FX: LncRNA
DGCR5 represses the development of hepatocellular carcinoma by
targeting the miR-346/KLF14 axis. J Cell Physiol. 234:572–580.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xiong H, Ni Z, He J, Jiang S, Li X, He J,
Gong W, Zheng L, Chen S, Li B, et al: LncRNA HULC triggers
autophagy via stabilizing Sirt1 and attenuates the chemosensitivity
of HCC cells. Oncogene. 36:3528–3540. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Booy EP, McRae EK, Howard R, Deo SR, Ariyo
EO, Dzananovic E, Meier M, Stetefeld J and McKenna SA: RNA helicase
associated with AU-rich element (RHAU/DHX36) interacts with the
3′-Tail of the long non-coding RNA BC200 (BCYRN1). J Biol Chem.
291:53552016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang XY, Zhang LX, Tian CJ, Tang XY, Zhao
LM, Guo YL, Cheng DJ, Chen XL, Ma LJ and Chen ZC: LncRNAs BCYRN1
promoted the proliferation and migration of rat airway smooth
muscle cells in asthma via upregulating the expression of transient
receptor potential 1. Am J Transl Res. 8:3409–3418. 2016.PubMed/NCBI
|
|
13
|
Booy EP, McRae EK, Koul A, Lin F and
McKenna SA: The long non-coding RNA BC200 (BCYRN1) is critical for
cancer cell survival and proliferation. Mol Cancer. 16:1092017.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Singh R, Gupta SC, Peng WX, Zhou N,
Pochampally R, Atfi A, Watabe K, Lu Z and Mo YY: Regulation of
alternative splicing of Bcl-x by BC200 contributes to breast cancer
pathogenesis. Cell Death Dis. 7:e22622016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhao RH, Zhu CH, Li XK, Cao W, Zong H, Cao
XG and Hu HY: BC200 LncRNA a potential predictive marker of poor
prognosis in esophageal squamous cell carcinoma patients. Onco
Targets Ther. 9:2221–2226. 2016.PubMed/NCBI
|
|
16
|
Hu T and Lu YR: BCYRN1, a c-MYC-activated
long non-coding RNA, regulates cell metastasis of non-small-cell
lung cancer. Cancer Cell Int. 15:1–8. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lin YH, Wu MH, Huang YH, Yeh CT, Chi HC,
Tsai CY, Chuang WY, Yu CJ, Chung IH, Chen CY and Lin KH: Thyroid
hormone negatively regulates tumorigenesis through suppression of
BC200. Endocr Relat Cancer. 25:967–979. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ho TT, Zhou N, Huang J, Koirala P, Xu M,
Fung R, Wu F and Mo YY: Targeting non-coding RNAs with the
CRISPR/Cas9 system in human cell lines. Nucleic Acids Res.
43:e172015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bartonicek N, Maag JL and Dinger ME: Long
noncoding RNAs in cancer: Mechanisms of action and technological
advancements. Mol Cancer. 15:432016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Iyer MK, Niknafs YS, Malik R, Singhal U,
Sahu A, Hosono Y, Barrette TR, Prensner JR, Evans JR, Zhao S, et
al: The landscape of long noncoding RNAs in the human
transcriptome. Nat Genet. 47:199–208. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cui X, Zhao C, Yao X, Qian B, Su C, Ren Y,
Yao Z, Gao X and Yang J: SND1 acts as an anti-apoptotic factor via
regulating the expression of lncRNA UCA1 in hepatocellular
carcinoma. RNA Biol. 2018:1–12. 2018.
|
|
23
|
Zhang YT, Li BP, Zhang B, Ma P, Wu QL,
Ming L and Xie LM: LncRNA SBF2-AS1 promotes hepatocellular
carcinoma metastasis by regulating EMT and predicts unfavorable
prognosis. Eur Rev Med Pharmacol Sci. 22:6333–6341. 2018.PubMed/NCBI
|
|
24
|
Lee YR, Kim G, Tak WY, Jang SY, Kweon YO,
Park JG, Lee HW, Han YS, Chun JM, Park SY and Hur K: Circulating
exosomal non-coding RNAs as prognostic biomarkers in human
hepatocellular carcinoma. Int J Cancer. 15:1444–1452. 2018.
|
|
25
|
Tiedge H, Chen W and Brosius J: Primary
structure, neural- specific expression, and dendritic location of
human BC200 RNA. J Neurosci. 13:2382–2390. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Martignetti JA and Brosius J: BC200 RNA: A
neural RNA polymerase III product encoded by a monomeric Alu
element. Proc Natl Acad Sci USA. 90:11563–11567. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kim Y, Lee J, Shin H, Jang S, Kim SC and
Lee Y: Biosynthesis of brain cytoplasmic 200 RNA. Sci Rep.
7:68842017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chen W, Bocker W, Brosius J and Tiedge H:
Expression of neural BC200 RNA in human tumours. J Pathol.
183:345–351. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Shin H, Lee J, Kim Y, Jang S and Lee Y,
Kim S and Lee Y: Knockdown of BC200 RNA expression reduces cell
migration and invasion by destabilizing mRNA for calcium-binding
protein S100A11. RNA Biol. 14:1418–1430. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li P, Yang B, Xia SY, Chen L, Ning N, Ma
B, Liu Q, Yang H, Zhang D and Du XH: BC200 RNA is over-expressed in
colorectal cancer and promotes migration and invasion of HCT116
cells. Int J Clin Exp Pathol. 9:1481–1486. 2016.
|
|
31
|
Robson S, Pelengaris S and Khan M: C-Myc
and downstream targets in the pathogenesis and treatment of cancer.
Recent Pat Anticancer Drug Discov. 1:305–326. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Opferman JT and Kothari A: Anti-apoptotic
BCL-2 family members in development. Cell Death Differ. 25:37–45.
2018. View Article : Google Scholar
|
|
33
|
de Jong Y, Monderer D, Brandinelli E,
Monchanin M, van den Akker BE, van Oosterwijk JG, Blay JY, Dutour A
and Bovée JVMG: Bcl-xl as the most promising Bcl-2 family member in
targeted treatment of chondrosarcoma. Oncogenesis. 7:742018.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Gu L, Lu L, Zhou D and Liu Z: Long
noncoding RNA BCYRN1 promotes the proliferation of colorectal
cancer cells via up-regulating NPR3 expression. Cell Physiol
Biochem. 48:2337–2349. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Bergandi L, Mungo E, Morone R, Bosco O,
Rolando B and Doublier S: Hyperglycemia promotes chemoresistance
through the reduction of the mitochondrial DNA damage, the
Bax/Bcl-2 and Bax/Bcl-XL ratio, and the cells in sub-G1 phase due
to antitumoral drugs induced-cytotoxicity in human colon
adenocarcinoma cells. Front Pharmacol. 9:8662018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li M, Liu XH, Zhao YC, Ma XY, Zhou YC,
Zhao YX and Liu XY: Long noncoding RNA KCNQ1OT1 promotes apoptosis
in neuroblastoma cells by regulating miR-296-5p/Bax axis. FEBS J.
21:150472019. View Article : Google Scholar
|
|
37
|
Shao J, Pan X, Yin X, Fan G, Tan C, Yao Y,
Xin Y and Sun C: KCNQ1OT1 affects the progression of diabetic
retinopathy by regulating miR-1470 and epidermal growth factor
receptor. J Cell Physiol. 234:17269–17279. 2019. View Article : Google Scholar : PubMed/NCBI
|