Introduction
Non-small cell lung cancer (NSCLC) is the most
common type of lung cancer and accounts for ~85% of all lung cancer
cases, and is one of the leading causes of cancer-associated
mortality worldwide (1–3). Adenocarcinoma is the fastest growing
subtype of NSCLC, and the 5-year survival rate is <20% (4), and 30–40% of patients with advanced
stage lung cancer develop bone metastases, resulting in
skeletal-related events which, in-turn cause hypercalcemia,
pathological fractures, spinal compression and bone pain, leading
to poor prognoses (5). Exploring
the underlying molecular mechanisms of lung cancer metastasis to
the bone has gained increasing interest, particularly for the
exploration of novel therapeutic targets.
Cullin 4A (CUL4A) is an 87-kDa protein
and is a member of the cullin family of proteins. CUL4A
forms part of the multifunctional ubiquitin ligase E3 complex
(6). The ubiquitin-proteasome
pathway serves an important role in the degradation of proteins,
including several well-defined tumor-suppressor genes, such as
p21, p27 and p53 (7).
Additionally, it has been reported that CUL4A is abnormally
expressed in various types of malignancies (7). Therefore, CUL4A may act as an
oncogene to promote tumor progression; however, the association
between CUL4A and metastasis of lung adenocarcinoma to the
bone has not been reported.
Epithelial-mesenchymal transition (EMT) is the
initial event in the tumor metastatic process, which promotes the
dissemination of tumor cells from the primary lesion to
colonization at distant sites (8).
Zinc finger E-box binding homeobox 1 (ZEB1) is a
transcriptional activator of EMT, and it represses expression of
epithelial genes by binding to the promoter regions of E-boxes,
inducing EMT and thus promoting cancer metastasis (9). Furthermore, ZEB1 has been
reported to promote metastasis of lung cancer to the bone in
vivo (10). Therefore,
clarifying the association between CUL4A and ZEB1 may
improve our understanding of metastasis of lung cancer to the
bone.
The present study revealed that CUL4A
overexpression promoted proliferation, migration and invasion of
lung adenocarcinoma cells in vitro and metastasis of lung
cancer to the bones in vivo. Knockdown of CUL4A had
the opposite effects on the biological behaviors of lung
adenocarcinoma cells in vitro. Mechanistically, CUL4A
induced EMT and promoted metastasis of lung adenocarcinoma to the
bone by regulating the transcriptional expression of ZEB1.
These results provide novel insight into the mechanistic role of
CUL4A in metastasis of lung adenocarcinoma to the bone,
suggesting that CUL4A may serve as a potential therapeutic
target for patients with advanced lung adenocarcinoma.
Materials and methods
Cell lines and cell culture
The human lung adenocarcinoma cell lines A549, H1299
and H460 were purchased from the American Type Culture Collection
(ATCC) and have been preserved in our laboratory in a liquid
nitrogen storage tank. Cells were grown in culture flasks with
RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.) containing
10% FBS (Biochrom, Ltd.) with 5% CO2 at 37°C in an
incubator.
Reverse transcription-quantitative
(RT-q)PCR
Total RNA was extracted from cells using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) and reverse-transcribed into cDNA using a QuantiNova™ Reverse
Transcription kit (Qiagen GmbH) according to the manufacturer's
protocol. The quantification of gene transcripts was determined
using qPCR using a QuantiNova™ SYBR Green PCR kit (Qiagen GmbH) on
a Mx3005P qPCR system (Agilent Technologies, Inc.). The
thermocycling conditions were: Pre-denaturation at 95°C for 2 min;
followed by 40 cycles of denaturation at 95°C for 10 sec and
annealing at 60°C for 30 sec. Gene expression was quantified using
the 2−ΔΔCq method (11).
The sequences of the PCR primers were as follows: CUL4A
forward, GGCTCCAAGAAGCTGGTCAT and reverse, CTGATGGAGGTGCTGCTCTG;
GAPDH forward, GAAGGTGAAGGTCGGAGTC and reverse,
GAAGATGGTGATGGGATTTC. GAPDH was used as the internal
control.
Protein extraction and western blot
analysis
Total protein was extracted from cells using RIPA
lysis buffer (Beyotime Institute of Biotechnology Biotechnology),
and the protein concentration was quantified using a bicinchoninic
acid protein assay kit (Pierce; Thermo Fisher Scientific, Inc.).
Subsequently, 20 µg of protein was loaded on a 10% SDS gel and
resolved using SDS-PAGE. Resolved proteins were transferred to PVDF
membranes, and the membranes were blocked in 5% fat-free milk for 1
h at room temperature and incubated overnight at 4°C with specific
primary antibodies against CUL4A (dilution 1:500; cat. no.
14851-1-AP; ProteinTech Group, Inc.), β-actin (dilution 1:10,000;
cat. no. 051M4892; Sigma-Aldrich; Merck KGaA), E-cadherin (dilution
1:1,000; cat. no. MABT26; Merck KGaA), Vimentin (dilution 1:1,000;
cat. no. MABT26; Merck KGaA), ZEB1 (dilution 1:500; cat. no.
21544-1-AP; ProteinTech Group, Inc.). After incubation with the
primary antibodies, the membranes were incubated with horseradish
peroxidase-conjugated goat anti-mouse/rabbit secondary antibody
(dilution 1:2,500; Beijing Zhongshan Jinqiao Biotechnology Co.,
Ltd.) for 1 h at room temperature, and signals were visualized
using chemiluminescence reagent (Pierce; Thermo Fisher Scientific,
Inc.) and analyzed using AlphaImager 2200 software version 3.2.1.2
(Alpha Innotech Corporation). β-actin was used as the loading
control.
Stable transfection
Short hairpin (sh)RNAs (CUL4A-shRNA28399-1,
5′-GCAGAACTGATCGCAAAGCAT-3′; CUL4A-shRNA28400-1,
5′-CCAGAATATCTTAACCATGTA-3′; CUL4A-shRNA28402-1,
5′-GCAGGTGTATAAAGATTCATT-3′) targeting CUL4A
(CUL4A-GV248-RNAi NM_001008895, target sequence:
GCAGAACTGATCGCAAAGCAT), control shRNA (NC-GV248, target sequence:
TTCTCCGAACGTGTCACGT), recombinant CUL4A lentivirus (homo;
NM_001008895), CUL4A-NC lentivirus (Ubi-MCS-3FLAG-SV40-puromycin)
and Luciferin-LV (Ubi-MCS-Luc-IRES-Puromycin) were synthesized by
Shanghai GeneChem Co., Ltd. (Shanghai, China), with virus titers of
1×109, 1×109, 1×109,
1×109 and 5×108 TU/ml. The CUL4A
overexpression lentiviral vector (CUL4A-LV) was respectively
infected into A549 and H1299 cells at a multiplicity of infection
(MOI) of 80 and 40 with complete medium containing ENi.S and
Polybrene (Shanghai GeneChem Co., Ltd.). Similarly, H460 cells were
infected with the CUL4A-shRNA lentivirus at a MOI of 100 to knock
down CUL4A expression, and NC-shRNA was used as the negative
control. Transfections were performed according to the
manufacturer's instructions. The infected cells were selected for 2
weeks using a medium with a concentration of 2.0 µg/ml puromycin to
obtain stably transfected cells. Then the puromycin level in the
culture medium was maintained at 1 µg/ml. Luciferin-LV virus was
used to infect both A549-CUL4A and A549-NC cells for viewing the
distribution of tumor cells in vivo. The efficiency of
knockdown or overexpression was assessed using RT-qPCR and western
blotting.
MTT assay
Cells in the logarithmic growth phase were collected
and seeded into 96-well plates at a density of 2×103
cells/well. A total of eight 96-well plates were cultured as
described above. On the following days, a 96-well plate was taken
out at a fixed daily time every 24 h, MTT solution (5 mg/ml) was
added (20 µl/well), and the plate was incubated at 37°C for a
further 4 h. The medium was carefully discarded and 150 µl of DMSO
was added. The 96-well plate was agitated for 10 min to dissolve
the formamidine completely, and the absorbance value was measured
at a wavelength of 490 nm on a microplate reader (Multiskan MK3;
Thermo Fisher Scientific, Inc.). This assay was performed in
triplicate.
Colony formation assay
The cells in the logarithmic growth phase were
harvested and plated in a 6-well plate at a density of 200
cells/well, and the plate was incubated as described above for two
weeks. Subsequently, the cells were stained with 0.25% crystal
violet for 20 min at room temperature. Subsequently, cell colonies
(>50 cells) were counted manually. This assay was performed in
triplicate.
Wound-healing assay
The cells in the logarithmic growth phase were
plated in a 6-well plate at a density of 5×105
cells/well and cultured in RPMI-1640 medium supplemented with 10%
FBS, for 24 h until the cells reached ~90% confluence. The cell
monolayer was scratched using a 200-µl pipette tip, and the cell
debris was washed away with PBS. Then, the cells were cultured in
serum-free RPMI-1640 medium for 24 h. The wounds were imaged at ×4
magnification using an inverted light microscope at 0 and 24 h
after the scratch was made. The distance of the migration relative
to the initial distance was calculated, and the migration distance
was analyzed using ImageJ (version 1.8.0; ImageJ, Inc.). This assay
was performed in triplicate.
Cell invasion assay
Transwell inserts (8.0-µm pore size) were coated
with 70 µl Matrigel (1:8 dilution; both from Corning Inc.). Cells
in the logarithmic growth phase were harvested and resuspended to a
density of 1×105 cells/ml in serum-free RPMI-1640
medium. The single-cell suspension was plated into the upper
chamber (200 µl/well). A total of 500 µl RPMI-1640 medium
supplemented with 10% FBS was added to the bottom chamber. The
chambers were incubated as described above for 24 h. Subsequently,
the non-invading cells in the upper chamber were gently wiped off
using cotton swabs, whereas cells that had invaded through the
Matrigel were fixed in 95% ethyl alcohol for 5 min at room
temperature and stained with 0.5% crystal violet for 20 min at room
temperature. Subsequently, 10 randomly selected fields were imaged
using a light microscope at ×200 magnification, and the number of
invaded cells were counted. This assay was performed in
triplicate.
In vivo metastasis
All experiments involving animals were performed in
accordance with the protocol approved by the Laboratory Animal Care
of the Air Force Military Medical University (Xi'an, China). In the
present study, 10 4-week-old female NOD-SCID mice weighing 15–17 g
were obtained from Hunan SJA Laboratory Animal Co., Ltd.
(http://zzx0251.bioon.com.cn/). Mice were
randomly divided into two groups, each group consisting of 5 mice,
and fed in a special pathogen-free grade animal facility at the Air
Force Military Medical University. The mice were housed with a 12-h
light/12-h dark cycle environment at 22°C; ventilation rate, 15/h;
the food was sterilized with Cobalt-60 irradiation and water was
autoclaved; and the mice had ad libitum access to food.
A549-CUL4A and A549-NC cells in logarithmic growth phase were
harvested with PBS to a single cell suspension with a density of
1.5×107 cells/ml. Single cell suspensions
(3×106 cells/200 µl) were injected into the mice via the
tail vein. The health and progression of the tumor mass in the mice
was examined weekly from the fifth week after injection onwards.
When the experimental mice began to develop symptoms such as
lameness, joint stiffness, decreased exercise capacity, paraplegia,
or the experiment reached 42 days, the experiment was immediately
terminated. D-Luciferin solution (150 µl) (20 mg/ml) was
intraperitoneally injected into the mice. After 10 min, the mice
were sacrificed humanely in a transparent euthanasia device
(ventilated 3% isoflurane for induction of anaesthesia and
subsequent ventilated 1.5% isoflurane for maintenance of
anaesthesia) and placed in a prone position on the in vivo
Imaging system (Carestream Health, Inc.) to capture X-ray images
and biofluorescence imaging of the mice for examination of
metastasis to the bone.
Statistical analysis
A Student's t-test or one-way ANOVA was used to
analyze statistical differences of the effect of CUL4A on
cell proliferation, colony formation, migration and invasion and
data are presented as the mean ± standard deviation of three
replicates. A Wilcoxon rank sum test was used to analyze the bone
metastasis data in vivo. Statistical tests were performed
using SPSS (version 13.0.0; SPSS, Inc.). P<0.05 was considered
to indicate a statistically significant difference.
Results
Expression of CUL4A in human lung
adenocarcinoma cell lines
CUL4A expression was determined in A549,
H1299 and H460 cells. The results showed that the expression level
of CUL4A in the H460 cells was higher when compared with the
A549 and H1299 cells (Fig. 1A and
B). Furthermore, transfection efficiency of CUL4A was
analyzed using RT-qPCR and western blotting. CUL4A mRNA
(Fig. 1C) and protein (Fig. 1D) expression levels in A549-CUL4A
and H1299-CUL4A cells were both stably increased compared with the
respective control cells, and the expression levels of CUL4A
in H460-shCUL4A cells were stably decreased compared with the
parental H460-NC cells (Fig. 1E and
F).
CUL4A increases the proliferative
capacity of lung adenocarcinoma cells
The effect of CUL4A on the proliferative
capacity of lung adenocarcinoma cells was determined using an MTT
assay (Fig. 2A, C and E) and colony
formation assays (Fig. 2B, D and
F). Compared with the respective vector-only controls, both
A549-CUL4A and H1299-CUL4A cells exhibited significantly increased
cell proliferation and colony formation. Conversely, silencing of
CUL4A expression in the H460 cells significantly reduced
cell proliferation and colony formation compared with the control
H460 cells.
CUL4A increases the migratory and
invasive capacity of lung adenocarcinoma cells in vitro
The effect of CUL4A on cell migration was
first assessed using a wound-healing assay (Fig. 3A). Both A549-CUL4A (0.48±0.025) and
H1299-CUL4A (0.40±0.020) cells had significantly faster wound
closure rates compared with the respective controls (0.25±0.050 and
0.33±0.025, respectively), and conversely the wound closure rate of
the H460-shCUL4A cells (0.10±0.020) was slower compared with the
respective control cells (0.18±0.029). Additionally, A549-CUL4A
(156±21.08) and H1299-CUL4A (137±13.53) cells showed a greater
degree of invasion in the Matrigel invasion assays compared with
the respective control cells (84±12.77 and 68±16.65, respectively;
Fig. 3B and C). In contrast,
silencing of CUL4A expression in H460 cells significantly
reduced the invasive capacity of H460 cells (82±11.00 and
155±17.69, respectively; Fig. 3D).
These results indicate that CUL4A promotes the migratory and
invasive capacity of lung adenocarcinoma cells.
CUL4A overexpression facilitates bone
metastasis in vivo
To explore the biological role of CUL4A
overexpression in the metastasis of lung adenocarcinoma to the
bone, an experimental bone metastatic mouse model was constructed.
A549-CUL4A cells were injected into NOD/SCID mice through the tail
vein and the mice were assayed for the development of bone
metastatic lesions. Compared with the control group, the injection
of A549-CUL4A cells resulted in a significant increase in bone
metastatic lesions (Fig. 4;
Table I). Taken together, these
results suggested that CUL4A overexpression had the
potential to promote the metastatic ability of lung adenocarcinoma
bone metastasis in vivo.
 | Table I.Incidence of bone metastases and the
number of metastatic lesions formed in the NOD-SCID mice. |
Table I.
Incidence of bone metastases and the
number of metastatic lesions formed in the NOD-SCID mice.
| Cell line | Incidence | No. of bone
metastases |
|---|
| A549-NC | 1/5 | 0.2±0.45 |
| A549-CUL4A | 4/5 |
1.6±1.14a |
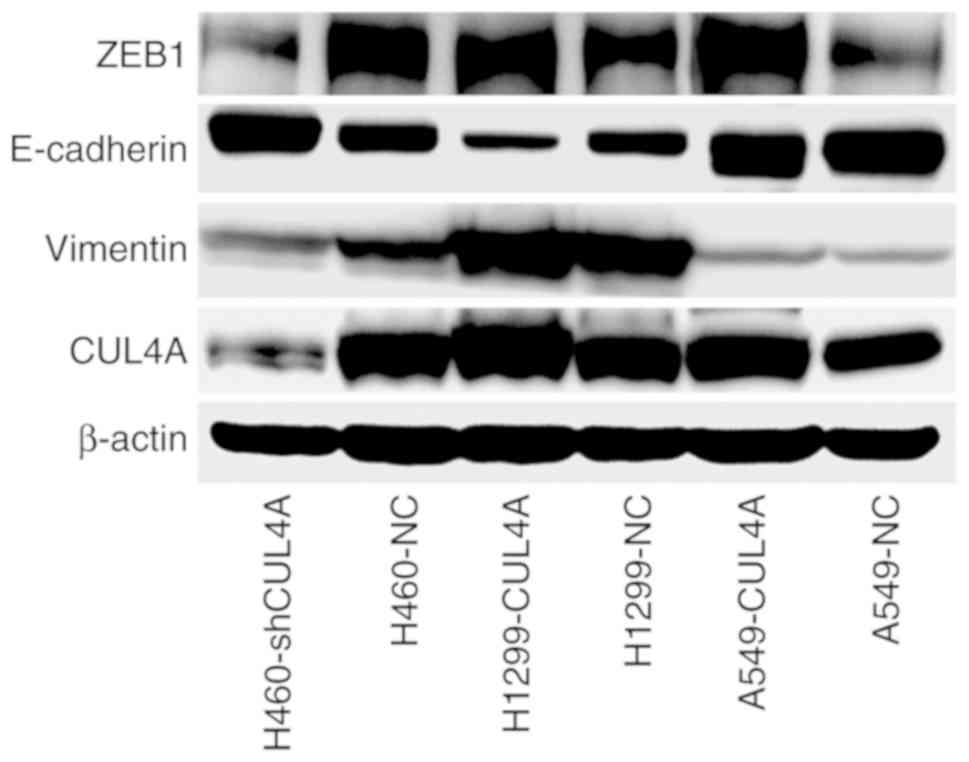
CUL4A promotes metastasis of lung
adenocarcinoma to the bone via ZEB1-mediated induction of EMT
In order to gain insight into the mechanism by which
CUL4A promotes bone metastasis of A549 cells, western blot
analysis was used to examine expression of the EMT-activator
ZEB1 and well-characterized EMT markers in lung
adenocarcinoma cells (Fig. 5). The
results showed that expression of the EMT-activator ZEB1,
and the mesenchymal marker vimentin, were markedly increased
compared with the control, and expression of the epithelial marker
E-cadherin was markedly decreased in lung adenocarcinoma cells
overexpressing CUL4A, compared with the respective control
cells. However, the expression levels of these proteins were
reversed in the CUL4A-silenced lung adenocarcinoma cells.
Therefore, these findings suggest that CUL4A may promote
metastasis of lung adenocarcinoma to the bone via
ZEB1-mediated induction of EMT.
Discussion
In the present study it was shown that CUL4A
expression was associated with metastasis of lung adenocarcinoma.
Upregulation of CUL4A expression in lung adenocarcinoma
cells increased proliferation, migration and invasion in
vitro and increased metastasis to the bone in vivo.
Conversely, silencing of CUL4A resulted in the opposite
effects in the H460 cells. Mechanistically, the transcriptional
expression levels of ZEB1 were associated with CUL4A
expression.
CUL4A is a member of the evolutionarily
conserved cullin family of proteins, which consists of
seven-related cullins (Cul1, Cul2, Cul3, Cul4A, Cul4B, Cul5,
and Cul7) (12).
CUL4A forms part of the ubiquitin ligase E3 complex, and
serves a crucial role in DNA replication, cell cycle regulation and
genomic instability (13–17). Previous studies have demonstrated
that CUL4A acts as an oncogene in various types of tumors
and promotes cancer development, including lung cancer, breast
cancer, prostate cancer and other types of cancer (7,18–20)
and its upregulation is associated with less favorable outcomes
(21), which further supports the
results of the present study. Therefore, CUL4A may be a
potential anticancer target due to the fact that several well-known
tumor-suppressor genes, including p21, p27 and p53,
are ubiquitinated and degraded by the CUL4A-mediated E3
ubiquitin proteasome system (17,22,23).
In a CUL4A-defcient mouse model of skin cancer,
significantly increased resistance to UV-induced skin cancer was
observed (24). In addition, a
recent study reported that CUL4A modulates invasion and
metastasis of lung cancer through regulation of ANXA10
(25). Similarly, in the present
study, it was demonstrated that CUL4A overexpression served
a significant role in promoting development of lung adenocarcinoma.
The present study highlights a novel function of CUL4A in
metastasis of lung adenocarcinoma to the bone through
transcriptional upregulation of the EMT-activator ZEB1.
EMT is considered as the initial event during the
development of metastasis of cancer, and is crucial for
dissemination of tumor cells from primary sites and to colonize at
distant tissues (26,27). ZEB1, a transcriptional
repressor, is an essential inducer of EMT, and physiologically is
required for the regulation of skeletal morphogenesis. Kerstin
et al (28) reported that
ZEB1 stimulates BMP-inhibitor mediated osteoclast
differentiation and promotes metastasis of breast cancer to the
bone. Studies have suggested that ZEB1 may serve an import
role in progression of lung adenocarcinoma (29,30)
and significantly increase metastasis of lung cancer to t he bone
(31), suggesting that ZEB1
is a critical regulator of bone migration of lung cancer cells.
Additionally, it has also been reported that CUL4A
transcriptionally activates ZEB1 through modulation of
histone H3K4me3, inducing EMT and promoting metastasis of breast
cancer (32). CUL4A is
associated with lung cancer cell proliferation and expression is
associated with resistance to chemotherapy (20). However, the detailed mechanisms
underlying CUL4A-mediated lung adenocarcinoma bone
metastasis remain unknown. Therefore, in the present study, the
means by which CUL4A induces EMT and promotes metastasis was
examined, and it was demonstrated that CUL4A is associated
with ZEB1 expression in lung adenocarcinoma cells,
highlighting a potentially novel therapeutic target for prevention
of bone metastasis in patients with lung cancer. The results of the
present study showed that lung adenocarcinoma cells overexpressing
CUL4A exhibited aggressive behaviors, including increased
proliferation, migration and invasive capacities in vitro.
Silencing of CUL4A reversed these biological functions.
In vivo, it was demonstrated that CUL4A
overexpression was significantly positively associated with
increased bone metastatic lesions compared with the control group.
These results are supported by Yang et al (31), Wang et al (32) and Kerstin et al (28), where it was demonstrated that
ZEB1 expression is positively associated with CUL4A
expression, and upregulation of ZEB1 expression promotes
bone metastasis of lung and breast cancer. Therefore, CUL4A
may serve as a novel therapeutic target for prevention of
metastasis of lung cancer to the bone.
Taken together, aberrant upregulation of
CUL4A expression upregulates the expression levels of
ZEB1, which in-turn increases expression of EMT-associated
proteins and increases invasion and metastasis. This may underlie
the mechanism by which CUL4A increases metastasis of lung
adenocarcinoma to the bone.
Acknowledgements
The authors are grateful for all the colleagues of
the Oncology Research Center for their comments on earlier versions
of this manuscript.
Funding
The present study was supported by the National
Natural Science Foundation of China (nos. 81572251, 81572814 and
81902318).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
PPC and HLZ designed the study. PPC and WJC
performed all the in vitro experiments and collected the
data. HLP, WWS, PX, LD and YXX conducted the animal experiments.
HLP, WWS, PX, LD, YXX and LLL analyzed the data and performed the
relative statistical analysis. LLL provided guidance during the
study. PPC contributed to the writing of the manuscript. All
authors have read and approved the final version of this manuscript
and agree to be accountable for all aspects of the research in
ensuring that the accuracy or integrity of any part of the work are
appropriately investigated and resolved.
Ethics approval and consent to
participate
All animal studies strictly abided by the
Regulations on Animal Experimentation formulated by the Laboratory
Animal Center of the Air Force Military Medical University (Xi'an,
China) and this study was approved by the Animal Experimental
Ethical Inspection Committee of this Center (no. 20181101).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Lemjabbar-Alaoui H, Hassan OU, Yang YW and
Buchanan P: Lung cancer: Biology and treatment options. Biochim
Biophys Acta. 1856:189–210. 2015.PubMed/NCBI
|
|
2
|
Reck M and Rabe KF: Precision diagnosis
and treatment for advanced non-small-cell lung cancer. N Engl J
Med. 377:849–861. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2019. CA Cancer J Clin. 69:7–34. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Allemani C, Matsuda T, Di Carlo V,
Harewood R, Matz M, Nikšić M, Bonaventure A, Valkov M, Johnson CJ,
Estève J, et al: Global surveillance of trends in cancer survival
2000-14 (CONCORD-3): Analysis of individual records for 37 513 025
patients diagnosed with one of 18 cancers from 322 population-based
registries in 71 countries. Lancet. 391:1023–1075. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Coleman RE: Clinical features of
metastatic bone disease and risk of skeletal morbidity. Clin Cancer
Res. 12:6243s–6249s. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hannah J and Zhou P: Distinct and
overlapping functions of the cullin E3 ligase scaffolding proteins
CUL4A and CUL4B. Gene. 573:33–45. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sharma P and Nag A: CUL4A ubiquitin
ligase: A promising drug target for cancer and other human
diseases. Open Biol. 4:1302172014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
De Craene B and Berx G: Regulatory
networks defining EMT during cancer initiation and progression. Nat
Rev Cancer. 13:97–110. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Thiery JP, Acloque H, Huang RY and Nieto
MA: Epithelial- mesenchymal transitions in development and disease.
Cell. 139:871–890. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu Y, Zhang N, Wang Y, Xu M, Liu N, Pang
X, Cao J, Ma N, Pang H, Liu L and Zhang H: Zinc finger E-box
binding homeobox 1 promotes invasion and bone metastasis of small
cell lung cancer in vitro and in vivo. Cancer Sci. 103:1420–1428.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhong W, Feng H, Santiago FE and Kipreos
ET: CUL-4 ubiquitin ligase maintains genome stability by
restraining DNA-replication licensing. Nature. 423:885–889. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lee J and Zhou P: Pathogenic role of the
CRL4 ubiquitin ligase in human disease. Front Oncol. 2:212012.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sugasawa K: The CUL4 enigma: Culling DNA
repair factors. Mol Cell. 34:403–404. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Han J, Zhang H, Zhang H, Wang Z, Zhou H
and Zhang Z: A Cul4 E3 ubiquitin ligase regulates histone hand-off
during nucleosome assembly. Cell. 155:817–829. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hu J and Xiong Y: An evolutionarily
conserved function of proliferating cell nuclear antigen for Cdt1
degradation by the Cul4-Ddb1 ubiquitin ligase in response to DNA
damage. J Biol Chem. 281:3753–3756. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li B, Jia N, Kapur R and Chun KT: Cul4A
targets p27 for degradation and regulates proliferation, cell cycle
exit, and differentiation during erythropoiesis. Blood.
107:4291–4299. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xu Y, Wang Y, Ma G, Wang Q and Wei G:
CUL4A is overexpressed in human pituitary adenomas and regulates
pituitary tumor cell proliferation. J Neurooncol. 116:625–632.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Melchor L, Saucedo-Cuevas LP, Muñoz-Repeto
I, Rodríguez- Pinilla SM, Honrado E, Campoverde A, Palacios J,
Nathanson KL, García MJ and Benítez J: Comprehensive
characterization of the DNA amplification at 13q34 in human breast
cancer reveals TFDP1 and CUL4A as likely candidate target genes.
Breast Cancer Res. 11:R862009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang Y, Zhang P, Liu Z, Wang Q, Wen M,
Wang Y, Yuan H, Mao JH and Wei G: CUL4A overexpression enhances
lung tumor growth and sensitizes lung cancer cells to erlotinib via
transcriptional regulation of EGFR. Mol Cancer. 13:2522014.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Birner P, Schoppmann A, Schindl M, Dinhof
C, Jesch B, Berghoff AS and Schoppmann SF: Human homologue for
Caenorhabditis elegans CUL-4 protein overexpression is associated
with malignant potential of epithelial ovarian tumours and poor
outcome in carcinoma. J Clin Pathol. 65:507–511. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Nishitani H, Shiomi Y, Iida H, Michishita
M, Takami T and Tsurimoto T: CDK inhibitor p21 is degraded by a
proliferating cell nuclear antigen-coupled Cul4-DDB1Cdt2 pathway
during S phase and after UV irradiation. J Biol Chem.
283:29045–29052. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Nag A, Bagchi S and Raychaudhuri P: Cul4A
physically associates with MDM2 and participates in the proteolysis
of p53. Cancer Res. 64:8152–8155. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liu L, Lee S, Zhang J, Peters SB, Hannah
J, Zhang Y, Yin Y, Koff A, Ma L and Zhou P: CUL4A abrogation
augments DNA damage response and protection against skin
carcinogenesis. Mol Cell. 34:451–460. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hung MS, Chen YC, Lin P, Li YC, Hsu CC,
Lung JH, You L, Xu Z, Mao JH, Jablons DM and Yang CT: Cul4A
modulates invasion and metastasis of lung cancer through regulation
of ANXA10. Cancers (Basel). 11(pii): E6182019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Prudkin L, Liu DD, Ozburn NC, Sun M,
Behrens C, Tang X, Brown KC, Bekele BN, Moran C and Wistuba II:
Epithelial-to-mesenchymal transition in the development and
progression of adenocarcinoma and squamous cell carcinoma of the
lung. Mod Pathol. 22:668–678. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cowin P, Rowlands TM and Hatsell SJ:
Cadherins and catenins in breast cancer. Curr Opin Cell Biol.
17:499–508. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Mock K, Preca BT, Brummer T, Brabletz S,
Stemmler MP and Brabletz T: The EMT-activator ZEB1 induces bone
metastasis associated genes including BMP-inhibitors. Oncotarget.
6:14399–14412. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Takeyama Y, Sato M, Horio M, Hase T,
Yoshida K, Yokoyama T, Nakashima H, Hashimoto N, Sekido Y, Gazdar
AF, et al: Knockdown of ZEB1, a master epithelial-to-mesenchymal
transition (EMT) gene, suppresses anchorage-independent cell growth
of lung cancer cells. Cancer Lett. 296:216–224. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gemmill RM, Roche J, Potiron VA, Nasarre
P, Mitas M, Coldren CD, Helfrich BA, Garrett-Mayer E, Bunn PA and
Drabkin HA: ZEB1-responsive genes in non-small cell lung cancer.
Cancer Lett. 300:66–78. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yang X, Li L, Huang Q, Xu W, Cai X, Zhang
J, Yan W, Song D, Liu T, Zhou W, et al: Wnt signaling through
Snail1 and Zeb1 regulates bone metastasis in lung cancer. Am J
Cancer Res. 5:748–755. 2015.PubMed/NCBI
|
|
32
|
Wang Y, Wen M, Kwon Y, Xu Y, Liu Y, Zhang
P, He X, Wang Q, Huang Y, Jen KY, et al: CUL4A induces
epithelial-mesenchymal transition and promotes cancer metastasis by
regulating ZEB1 expression. Cancer Res. 74:520–531. 2014.
View Article : Google Scholar : PubMed/NCBI
|