Introduction
Esophageal cancer (EC) has a high incidence with
marked geographical delineations (1). Esophageal squamous cell carcinoma
(ESCC) is the most common pathological form of EC in China
(2). In 2015, the incidence and
mortality rates of EC were reported to be extremely high,
accounting for 25% of the total cancer-related mortality (3). Surgery, medication and radiation
therapy are the main treatment options for ESCC; however, survival
rates remain poor, rendering further effort for developing new
therapeutic strategies necessary (4,5).
Inflammation and immunity are among the many factors
that influence cancer initiation and development (6). Inflammasomes are multi-protein
platforms that activate procaspase-1 and mature interleukin (IL)-18
and IL-1β (7), the latter of which
is associated with carcinogenesis, when its expression is increased
(8). NLR pyrin family domain
containing 3 (NLRP3) is an important member of the inflammasome
family, which recruits apoptosis-associated speck-like protein
containing a CARD (ASC) and pro-caspase-1, and further
self-hydrolyzes into active caspase-1, leading to the maturation
and secretion of IL-1β and IL-18 (9).
The NLRP3 inflammasome, which could initiate
inflammatory responses, is considered to contribute to the
development of cancer-related inflammation and play a crucial role
in tumorigenesis (10,11). Recent evidence has also shown that
the malignant phenotype of human melanoma, as well as lung and
colon carcinoma, are associated with the excessive expression and
constitutive activation of the NLRP3 inflammasome (12–14).
The activated NLRP3 inflammasome may suppress the antitumor effect
of gemcitabine and 5-fluorouracil, thus promoting tumor growth
(15). By contrast, the expression
level of the NLRP3 inflammasome was found to be negatively
associated with human hepatic cell carcinoma (16). Therefore, the role of the NLRP3
inflammasome in tumor initiation and promotion remains
controversial and appears to be dependent on tumor type. However,
there are only a few reports on the association of the NLRP3
inflammasome with EC.
Therefore, the potential effect of the NLRP3
inflammasome on human ESCC tissues and cell lines was explored, in
order to identify a new clinical diagnostic and prognostic target
of ESCC.
Materials and methods
Patients and tumor samples
We obtained 84 paired ESCC and adjacent
non-cancerous tissues from patients who underwent surgery at the
First Clinical Hospital of Zhengzhou University (Zhengzhou, Henan,
China) from July 2015 to June 2017. Surgical patients had not
received any prior treatment. The Ethics Committee of Zhengzhou
University approved this project, and all patients signed informed
consent. Out of the 84 patient samples, 42 were used as the first
cohort for immunohistochemistry (IHC), which was used to evaluate
the relationship between the NLRP3 inflammasome and patient
clinical characteristics, and tissue microarrays. The other 42 were
used as the second cohort and kept in liquid nitrogen for gene and
protein expression detection.
IHC analysis
IHC was performed as previously described (17). Briefly, paraffin-embedded ESCC
tissues were dewaxed, hydrated, inactivated and blocked. The tissue
slides were then incubated with the primary antibodies: Rabbit
polyclonal NLRP3 antibody (cat. no. ab214185; dilution 1:200;
Abcam), rabbit polyclonal IL-1 beta antibody (cat. no. ab9722;
dilution, 1:500; Abcam), mouse monoclonal Ki-67 antibody (cat. no.
9449S; dilution, 1:200; Cell Signaling Technology, Inc.; CST),
polyclonal antibody specific for human ASC (cat. no. AL177;
dilution, 1:500; AdipoGen Life Sciences, Inc.) or rabbit monoclonal
caspase-1 antibody (cat. no. ab108362; dilution, 1:50; Abcam)
overnight at 4°C. They were then incubated with secondary
antibodies for 1 h at 37°C. The slides were photographed and
mounted, and the H-score was calculated to evaluate the expression
of NLRP3, Ki-67 and IL-1β in a semi-quantitative manner. The
H-score was calculated, according to the staining intensity and
percentage of occupancy, as the weak stain percentage multiplied by
the factor 1 plus the medium stain percentage multiplied by the
factor 2 plus the strong stain percentage multiplied by the factor
3; score range, 0–300 (18).
Hematoxylin and eosin (H&E)
staining
The ESCC tissues were fixed with 10% formaldehyde
and paraffin-embedded. The dried slices were dewaxed with xylene,
hydrated with anhydrous ethanol, 95, 85, 75 and 50% alcohol and
distilled water step by step, and then stained with H&E.
Pathological changes in the tissues were observed under a field of
×100 and ×400 magnification under optical microscopy.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA from ESCC tissues was isolated using
Trizol (cat. no. 15596026; Thermo Fisher Scientific, Inc.), and
quantified on a NanoDrop 2000 spectrometer (Thermo Fisher
Scientific, Inc.). The cDNA was reverse-transcribed from 1 µg of
total RNA using the Prime Script RT reagent kit (cat. no. 4472908;
Takara Bio), and then used as a template for RT-qPCR in the
SYBR-Green Master Mix kit (cat. no. A25779; Thermo Fisher
Scientific, Inc.). The thermal cycling conditions used were as
follows: Holding at 50°C for 2 min, pre-denaturation at 95°C for 2
min, 40 cycles of 95°C for 15 sec and 60°C for 1 min. The
2−ΔΔCT method was used to assess RNA levels normalized
to the β-actin level (19). The
primer sequences were designed and are listed in Table I.
 | Table I.Primer sequences for RT-qPCR. |
Table I.
Primer sequences for RT-qPCR.
| Gene | Forward
(5′-3′) | Reverse
(5′-3′) |
|---|
| β-actin |
GGGAAATCGTGCGTGACATT |
GGAAGGAAGGCTGGAAGAGT |
| NLRP3 |
GCTGCGATCAACAGGAGAGA |
GCTCACACTCTCACCCAGA |
| ASC |
AAGCCAGGCCTGCACTTTAT |
AGAGCTTCCGCATCTTGCTT |
|
Caspase-1 |
CATCCCACAATGGGCTCTGT |
TCACTCTTTCAGTGGTGGGC |
| IL-1β |
TGAGCTCGCCAGTGAAATGA |
TGAGCTCGCCAGTGAAATGA |
Western blot analysis
Tissues or cell proteins were extracted using RIPA
lysis buffer (Beijing Solarbio Science & Technology Co., Ltd.)
with a protease inhibitor cocktail (Beyotime Institute of
Biotechnology). First, equal amounts of proteins (50 µg) were
subjected to 10% sodium dodecyl sulfate-polyacrylamide gel
(SDS-PAGE) electrophoresis and then transferred onto polyvinylidene
fluoride membranes. Next, 5% fat-free milk was used to block the
membranes for 2 h, which were then incubated with primary
antibodies: Rabbit polyclonal NLRP3 antibody (cat. no. ab214185;
dilution, 1:2,000; Abcam), rabbit monoclonal caspase-1 antibody
(cat. no. ab108362; 1:1,000; Abcam), rabbit polyclonal IL-1β
antibody (cat. no. ab9722; dilution, 1:1,000; Abcam), polyclonal
antibody specific for human ASC (cat. no. AL177; dilution, 1:1,000;
AdipoGen Life Sciences, Inc.), and mouse monoclonal β-actin
antibody (cat. no. 60008-1; dilution, 1:5,000; ProteinTech Group,
Inc.) overnight at 4°C. Then the blots were incubated with
secondary antibodies, including HRP-conjugated AffiniPure goat
anti-rabbit IgG (H+L) (cat. no. SA00001-2; dilution, 1:5,000;
ProteinTech Group, Inc.) and HRP-conjugated AffiniPure goat
anti-mouse IgG (H+L) (cat. no. SA00001-1; dilution, 1:5,000;
ProteinTech Group, Inc.) at 37°C. After 2 h, immunoreactive bands
were visualized using an ECL system ((FluorChem E;
ProteinSimple).
Enzyme-linked immunosorbent assay
(ELISA)
To assess the content of tissue-derived IL-1β in
ESCC, frozen tissue samples were thawed and weighed, and 100 mg per
tissue was transferred into tubes on ice containing 900 ml PBS. The
samples were homogenized and then centrifuged at 5,000 × g at 4°C
for 10 min. A BCA kit (Beyotime Institute of Biotechnology) was
used to determine the total protein concentration in the
supernatant. A commercial ELISA kit (BioLegend) was used to measure
the levels of IL-1β, expressed as pg/ml.
Cell lines
The human KYSE-70, EC109, KYSE-510, TE1, TE13, EC1
and EC9706 ESCC cell lines and the normal esophageal squamous
epithelium HET-1A cell line were provided by Basic Medical Sciences
of Zhengzhou University (Zhengzhou, China). TE13 cell
authentication has been identified by STR profiling. Cells were
incubated in RPMI-1640 medium (Gibco; Thermo Fisher Scientific,
Inc.), containing 10% FBS (Corning Inc.) and 1% penicillin and
streptomycin with 5% CO2 at 37°C.
Cell transfection
To knockdown NLRP3, KYSE-70 and TE13 cells were
transfected with NLRP3-small interfering RNA (siRNA) and a
non-silencing siRNA (Guangzhou RiboBio Co., Ltd.), as negative
control. For RNA interference, 50 pmol siRNA was transiently
transfected into the cells using 3.75 µl Lipofectamine™
3000 Reagent (Thermo Fisher Scientific, Inc.) and then cells were
incubated for 48 h.
To overexpress NLRP3, KYSE-510 and EC9706 cells were
transfected with the plasmid pCDNA3.1 (+)-NLRP3 and pcDNA-3.1(+)
vector plasmid as negative control. Cells were then incubated for
18 h, and then transfection was conducted with
Lipofectamine™ 3000 Reagent and P3000™
reagent (Thermo Fisher Scientific, Inc.) following the
manufacturer's instructions.
Cell proliferation assay
In 96-well plates, 5×103 cells per well
were cultured for 6, 12, 24 and 48 h. At each time point, 10 µl
CCK-8 chemical compound was added to the cell supernatant. After 2
h, cell absorbance was determined at 450 nm using a
spectrophotometer (Thermo Fisher Scientific, Inc.).
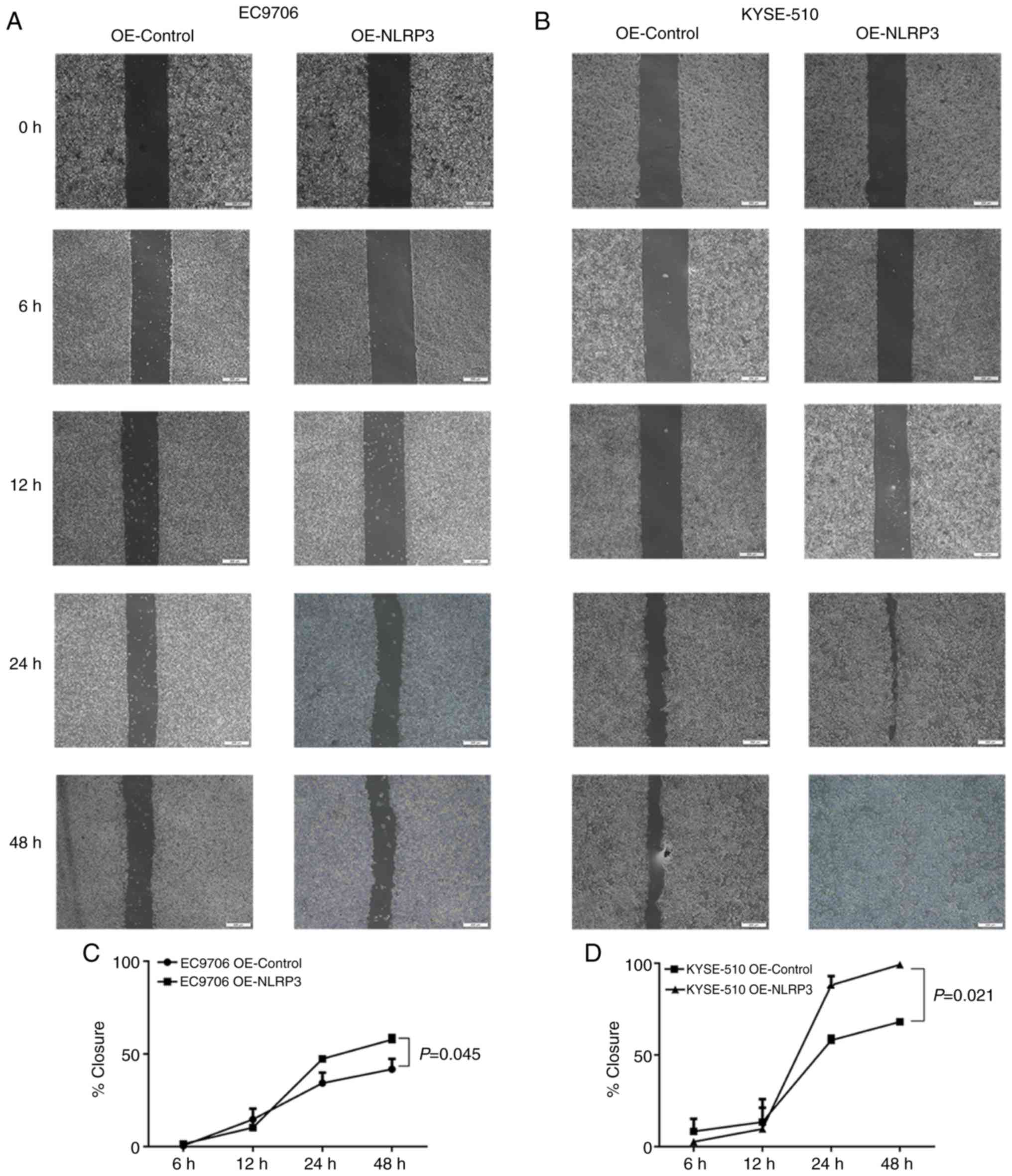
Wound healing assay
For the wound healing assay, 1×105 cells
per well were inoculated in 6-well plates. Cells were allowed to
grow until 90% confluency, then a gap was made using a 200-µl
pipette tip. Then, the cells were cultured with fresh RPMI-1640
medium contained 1% FBS. The distance of wound closure was measured
at 0, 6, 12, 24 and 48 h, respectively. The percent wound closure
was determined using the following equation:
(A0-At)/A0 × 100, where
A0 is the wound area at the hour of the wound
generation, and At is the wound area at the hour of
observation.
Cell migration and invasion
assays
Transwell assay was used to detect the ability of
cell migration and invasion, as previously described (20). Briefly, 1×104 cells were
inoculated in an inner Transwell chamber (8 µm; Corning Inc.),
which was pre-coated with or without 300 µg/ml Matrigel (BD
Biosciences) for 2 h. The outer chamber was filled with 20% FBS in
RPMI-1640 medium as an inducer. After 48 h, the migrated cells were
fixed, stained with 0.1% crystal violet and counted (magnification,
×200) in five different areas under an inverted fluorescence
microscope (Nikon Eclipse TS100-F; Nikon).
Statistical analysis
SPSS software version 3 (SPSS, Inc.) and GraphPad
Prism 5 (GraphPad software, Inc.) were used for statistical
analysis and plotting, respectively. One-way analysis of variance
(ANOVA) followed by Tukey's post hoc test was used to analyze the
difference for more than two groups. The two-tailed Student's
t-test was used for comparison of two groups. Repeated measurement
ANOVA was used to analyze cell viability and wound closure at
different time intervals for each group, separately. Comparison of
NLRP3 or IL-1β and patient clinical characteristics was performed
using χ2 or Fisher's exact test. The Spearman's
correlation analysis was used for the correlation between Ki-67 and
NLRP3. In this study, P<0.05 was considered statistically
significant. Data are presented as the mean ± SD. The data are
derived from cell experiments with 3 independently repeats.
Results
Increased levels of the NLRP3
inflammasome in human ESCC tissues
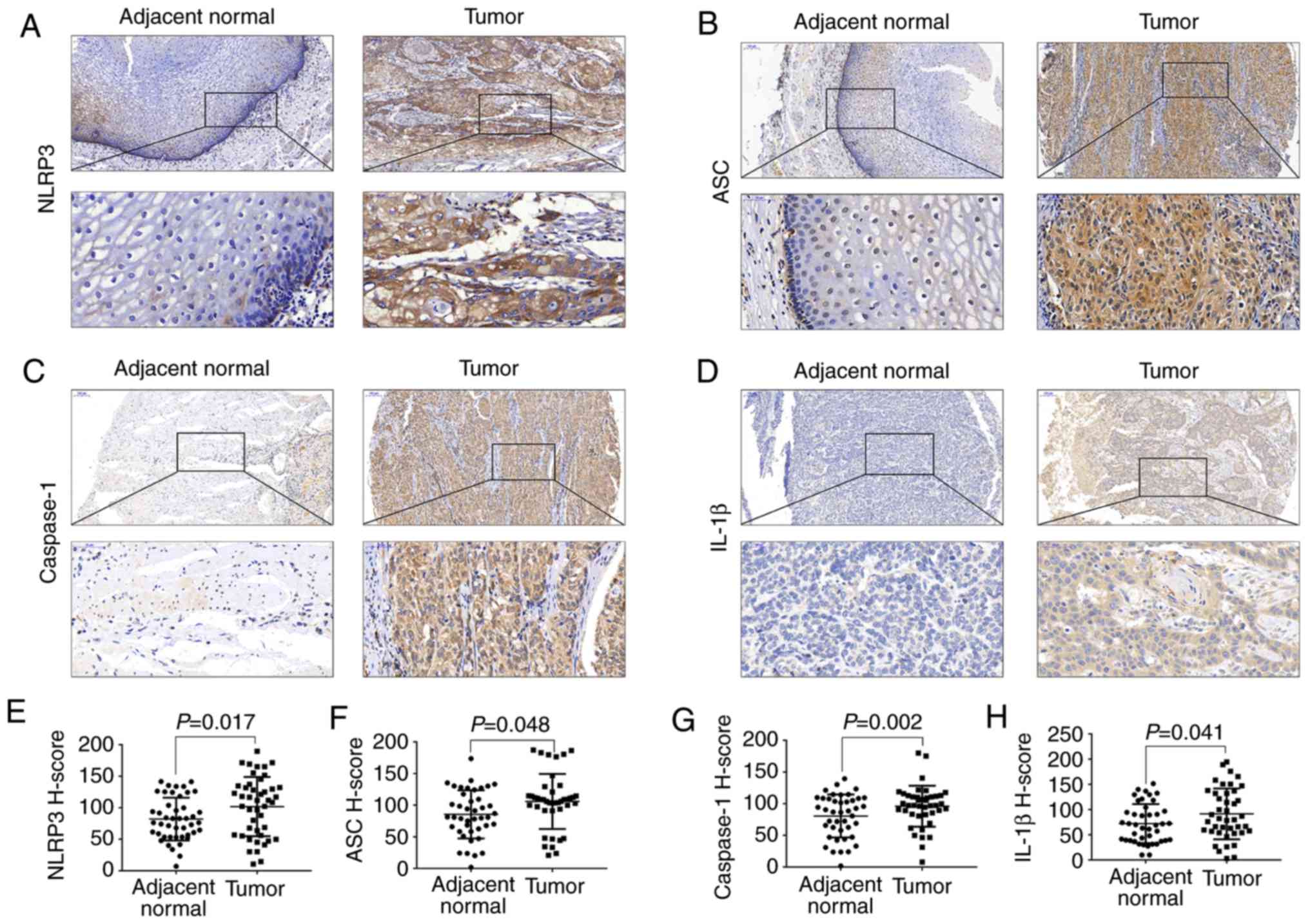
IHC of the first cohort containing 42 ESCC and
matched adjacent non-cancerous tissue samples showed that NLRP3 and
the three other main inflammasome components, ASC, caspase-1 and
IL-1β, were weakly expressed in adjacent control tissues but highly
expressed in the tumor tissues (Fig.
1A-D). We demonstrated that the H-scores of immunostaining
positive NLRP3, ASC, caspase-1 and IL-1β in tumor tissues were
significantly higher than these scores in the adjacent control
tissues (Fig. 1E-H).
Next, the mRNA and protein levels of NLRP3, ASC,
IL-1β and caspase-1 in the second cohort of 42 fresh ESCC samples
were assessed by RT-qPCR and western blot analysis. The mRNA levels
of NLRP3 (18/42, 42.86%; P=0.009), ASC (23/42,
54.76%; P=0.008), caspase-1 (21/42, 50.00%; P=0.003) and
IL-1β (27/42, 64.28%, P=0.001) were increased more than
two-fold in the tumors (Fig. 2A).
Western blot analysis confirmed higher expression levels of NLRP3,
ASC, caspase-1 and IL-1β in the tumors (Fig. 2B). Additionally, IL-1β secreted in
the tumors was higher than that in adjacent non-cancerous tissues
(n=32, P=0.001; Fig. 2C and D).
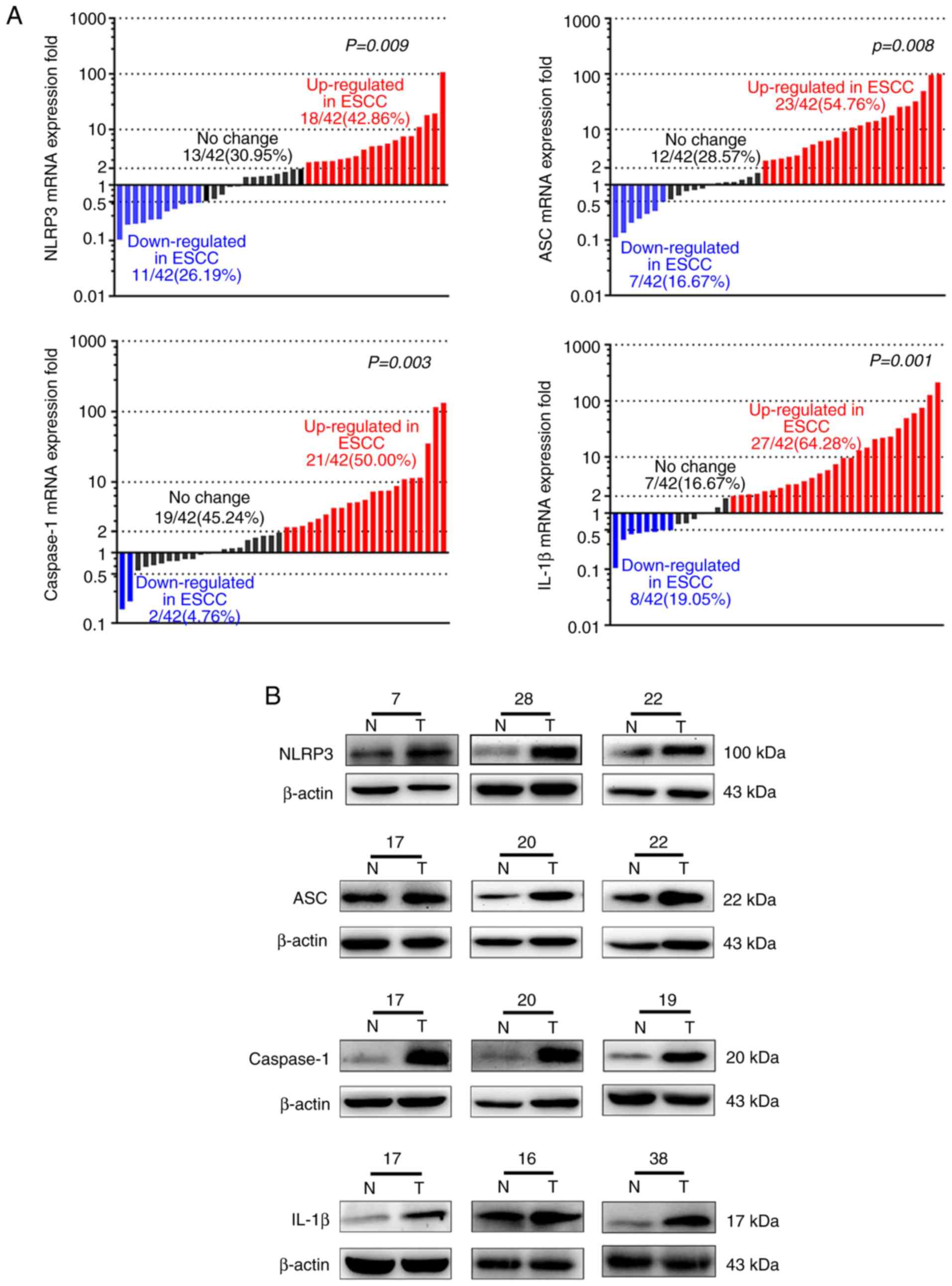 | Figure 2.Expression levels of NLRP3 and main
inflammasome components are increased in ESCC tissues from the
second cohort. (A) mRNA expression levels of NLRP3 (P=0.009), ASC
(P=0.008), caspase-1 (P=0.003) and IL-1β (P=0.001) in ESCC and
adjacent normal tissues were determined by RT-qPCR (n=42). These
bars represent the fold change of mRNA expression of ESCC compared
with adjacent noncancerous tissues. Red bars, ≥2-fold increase;
blue bars, ≥2-fold decrease; black bars, fold change of mRNA are
<2-fold. (B) The protein expression levels of NLRP3, ASC,
caspase-1 and IL-1β in ESCC tissues (T) and adjacent normal tissues
(N) were determined by western blot analysis (n=42). The number
above each western blot is the patient number. (C and D) IL-1β
expression in the tissues was detected by ELISA (n=32, P=0.001).
NLRP3, NLR pyrin family domain containing 3; ESCC, esophageal
squamous cell carcinoma; ASC, apoptosis-associated speck-like
protein containing a CARD; IL, interleukin; RT-qPCR, reverse
transcription-quantitative polymerase chain reaction; ELISA,
enzyme-linked immunosorbent assay. |
Association of the NLRP3 or IL-1β
expression and clinical characteristics
A higher NLRP3 protein expression was detected in 22
tumor samples (52.38%) of the first cohort, as compared with the
control tissues. An increased NLRP3 protein expression was found to
be associated with the T category and TNM stage (P=0.032 and
P=0.021, respectively), but not with the age, sex, lymph node
status and metastasis status of the patients (Table II). The NLRP3 protein expression
was higher in pathologic stage III–IV than I–II. Similarly, a
higher IL-1β protein expression was found to be significantly
associated with T category (P=0.014) and lymph node status
(P=0.005; Table II), as determined
by IHC. A high protein expression level of NLRP3 was found to be
associated with T category and TNM stage (P=0.030 and P=0.020,
respectively) in the second cohort (Table III), as determined by western blot
analysis.
 | Table II.Association of NLRP3 or IL-1β
expression and clinical characteristics of the ESCC patients
(n=42). |
Table II.
Association of NLRP3 or IL-1β
expression and clinical characteristics of the ESCC patients
(n=42).
| Clinical
characteristics | No. of
patients | No. of cases with
low levels of NLRP3 | No. of cases with
high levels of NLRP3 |
P-valuea | No. of cases with
low levels of IL-1β | No. of cases with
high levels of IL-1β |
P-valuea |
|---|
| Age (years) |
|
<60 | 18 | 7 | 11 | 0.327 | 7 | 11 | 0.856 |
|
≥60 | 24 | 13 | 11 |
| 10 | 14 |
|
| Sex |
|
|
|
|
|
|
|
|
Male | 27 | 12 | 15 | 0.580 | 9 | 18 | 0.206 |
|
Female | 15 | 8 | 7 |
| 8 | 7 |
|
| Metastasis |
|
|
|
|
|
|
|
| M0 | 39 | 19 | 20 | 0.607 | 16 | 23 | 0.794 |
|
M1-MX | 3 | 1 | 2 |
| 1 | 2 |
|
| Lymph node |
|
|
|
|
|
|
|
| N0 | 21 | 11 | 10 | 0.537 | 13 | 8 | 0.005b |
|
N1-NX | 21 | 9 | 12 |
| 4 | 17 |
|
| T category |
|
|
|
|
|
|
|
|
T1-T2 | 20 | 13 | 7 | 0.032b | 12 | 8 | 0.014b |
|
T3-T4 | 22 | 7 | 15 |
| 5 | 17 |
|
| TNM stage |
|
|
|
|
|
|
|
|
I–II | 26 | 16 | 10 | 0.021b | 10 | 16 | 0.735 |
|
III–IV | 16 | 4 | 12 |
| 7 | 9 |
|
 | Table III.Comparison of NLRP3 expression and
the clinical characteristics of the ESCC patients (n=42). |
Table III.
Comparison of NLRP3 expression and
the clinical characteristics of the ESCC patients (n=42).
| Clinical
characteristics | No. of
patients | No. of cases with
low levels of NLRP3 | No. of cases with
high levels of NLRP3 |
P-valuea |
|---|
| Age (years) |
|
<60 | 14 | 5 | 9 | 0.381 |
|
≥60 | 28 | 14 | 14 |
|
| Sex |
|
|
|
|
|
Male | 27 | 13 | 14 | 0.611 |
|
Female | 15 | 6 | 9 |
|
| Metastasis |
|
|
|
|
| M0 | 42 | 19 | 23 |
|
|
M1-MX | 0 | 0 | 0 |
|
| Lymph node |
|
|
|
|
| N0 | 20 | 11 | 9 | 0.226 |
|
N1-NX | 22 | 8 | 14 |
|
| T category |
|
|
|
|
|
T1-T2 | 21 | 13 | 8 | 0.030b |
|
T3-T4 | 21 | 6 | 15 |
|
| TNM stage |
|
|
|
|
|
I–II | 25 | 15 | 10 | 0.020b |
|
III–IV | 17 | 4 | 13 |
|
Association between NLRP3 and Ki-67
expression
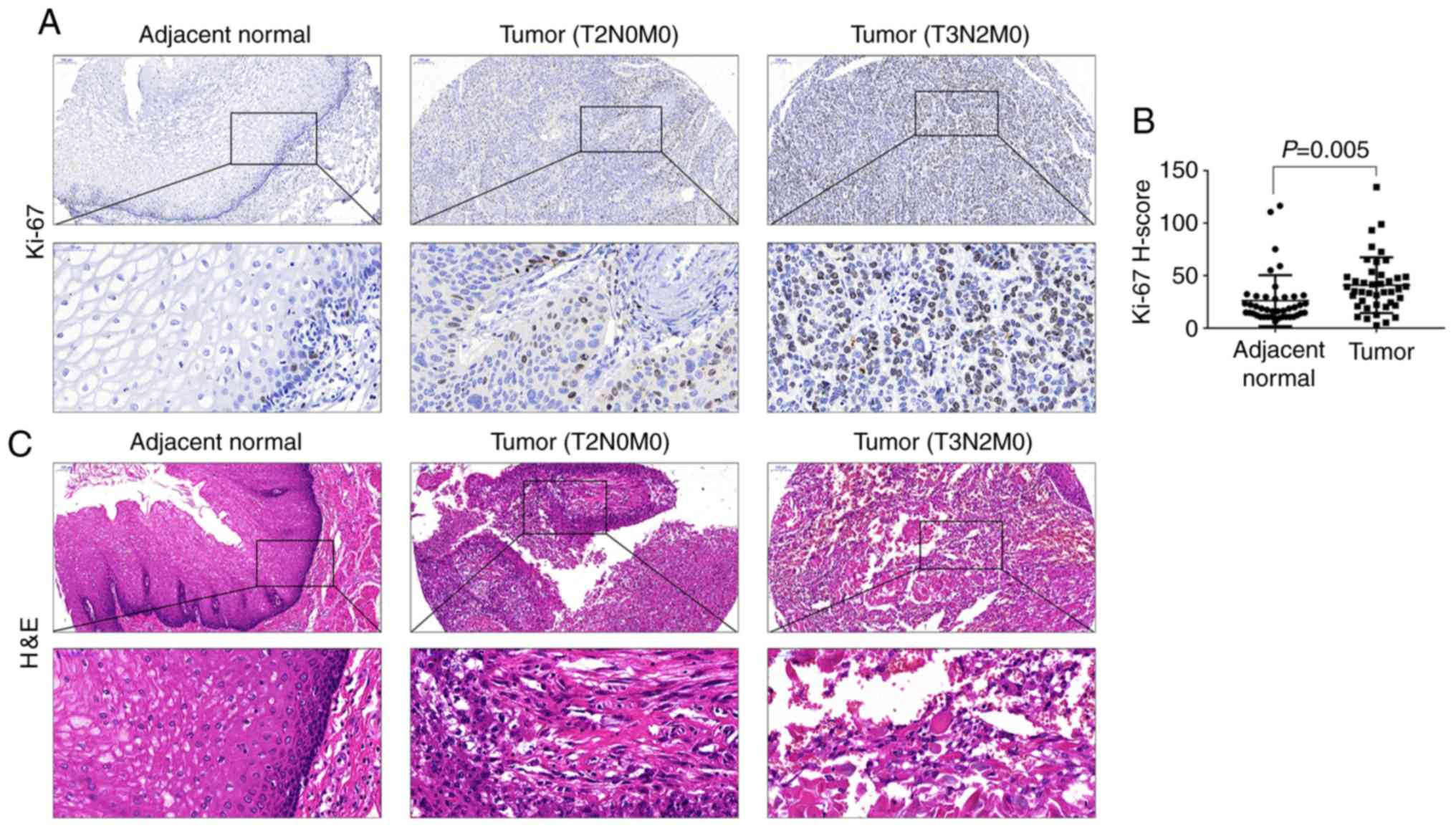
The expression of the proliferation marker Ki-67 was
explored and its association with NLRP3 was evaluated. The paraffin
sections of the first cohort of 42 ESCC samples were IHC-stained
for Ki-67 expression. The IHC results revealed that the Ki-67
expression was higher in the tumor tissues than that in the
adjacent non-cancerous tissues (Fig.
3A). H-score of Ki-67 immunostaining-positive cells was
significantly higher in cancer tissues than that in the normal
tissues (Fig. 3B). Organic tissues
were confirmed by pathological diagnosis following H&E
staining, and the results revealed a complete structure and orderly
arrangement of cells in normal tissues, but in cancer tissues
obvious atypia, diffuse distribution, vigorous growth, irregular
arrangement, large nuclei and dark cytoplasm (Fig. 3C). Patients were divided into a low
and high Ki-67 group, according to the relative expression of Ki-67
in cancer tissues, as compared to normal tissues. A positive
correlation between Ki-67 and NLRP3 was revealed by Spearman's
correlation analysis (P=0.014, r=0.355; Table IV). These data indicated that
Ki-67-positive patients tend to have a higher NLRP3 expression.
 | Table IV.Correlation between NLRP3 and Ki-67
expression (n=42). |
Table IV.
Correlation between NLRP3 and Ki-67
expression (n=42).
|
| Ki-67
expression |
|
|
|---|
|
|
|
|
|
|---|
|
| Low | High |
P-valuea | r |
|---|
| NLRP3
expression |
|
Low | 12 | 8 | 0.014b | 0.355 |
|
High | 5 | 17 |
|
|
NLRP3 silencing reduces cell
proliferation, migration and invasion
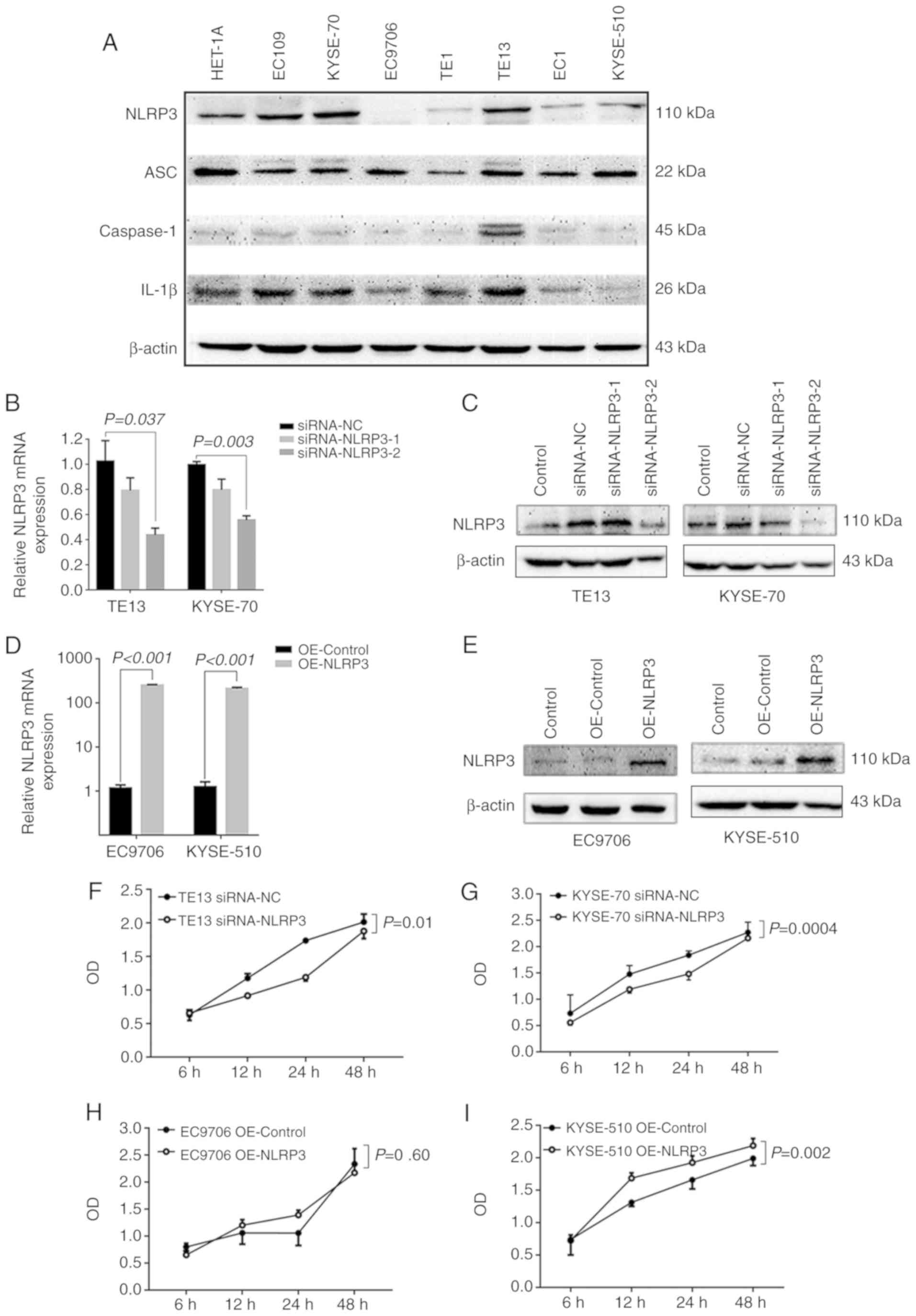
Seven ESCC cell lines were selected for western blot
analysis to study the expression of NLRP3. The results showed that
TE13 and KYSE-70 cells had the highest expression levels of NLRP3,
whereas EC9706 and KYSE-510 cells had the lowest (Fig. 4A).
Next, TE13 and KYSE-70 cells were transfected with
NLRP3-siRNAs (siRNA-NLRP3-1 and siRNA-NLRP3-2), respectively, and
siRNA knockdown efficacy was confirmed by RT-qPCR and western blot
analysis; siRNA-NLRP3-2 was selected for further experiments, due
to its more effective inhibition of NLRP3 in the TE13 (P=0.037) and
KYSE-70 cell lines (P=0.003) (Fig. 4B
and C). Silencing of NLRP3 strongly attenuated the cell
viability of TE13 (P=0.01) and KYSE-70 (P=0.0004) cells,
respectively (Fig. 4F and G),
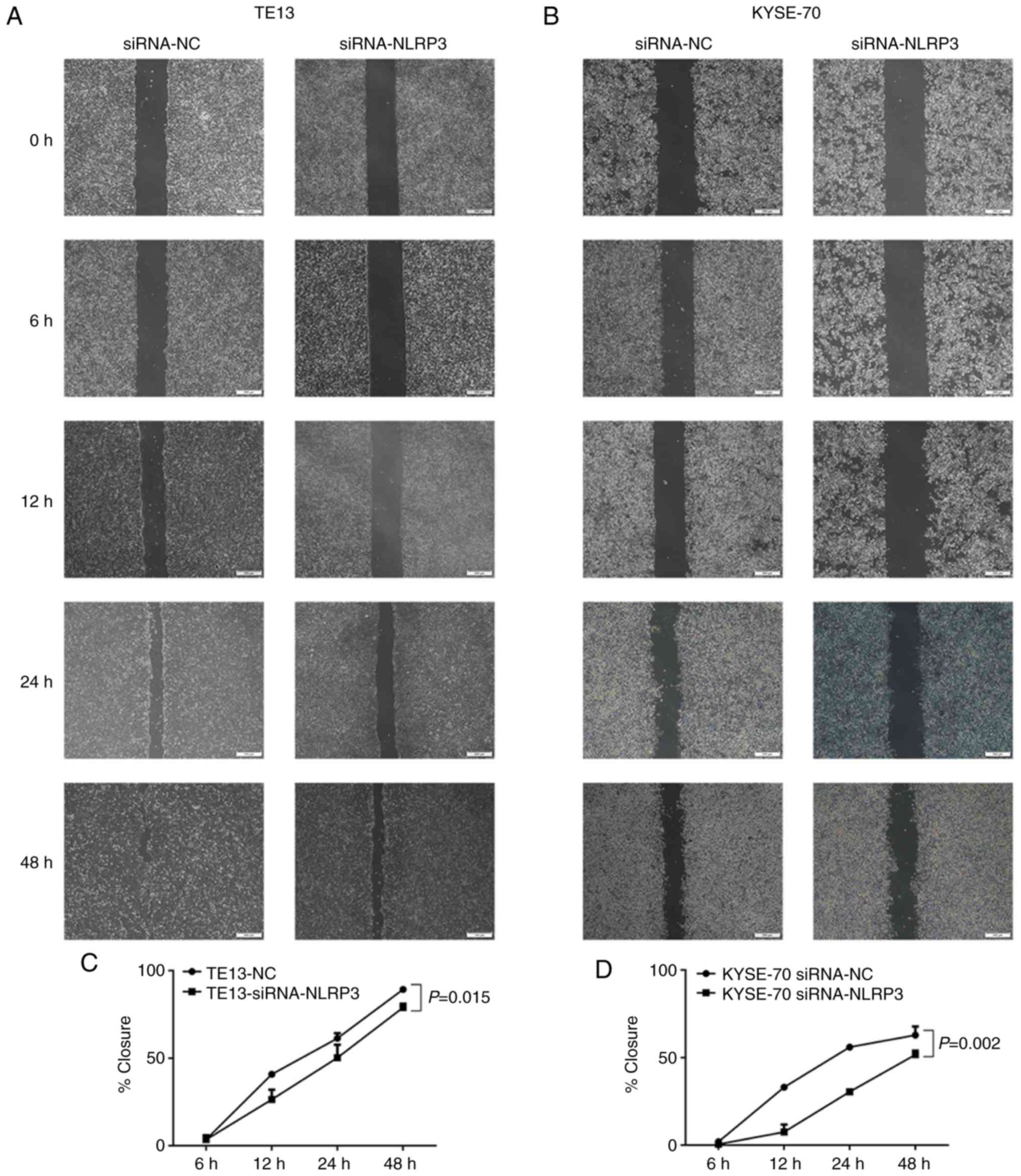
decreased cell mobility in the wound-healing assays in the TE13
(P=0.015, Fig. 5A and C) and
KYSE-70 (P=0.002, Fig. 5B and D)
cell lines. Moreover, silencing of NLRP3 significantly decreased
tumor cell invasion in the TE13 cells (P<0.0001) and KYSE-70
(P=0.0002) cells, as well as migration in the TE13 (P<0.0001)
and KYSE-70 (P=0.0037) cells (Fig.
5E-H).
Overexpression of NLRP3 promotes ESCC
biological function
pcDNA-3.1(+)-NLRP3 plasmid (OE-NLRP3) or the vector
pcDNA-3.1(+) (OE-Control) was transfected into EC9706 and KYSE-510
cells with low NLRP3 expression levels (P<0.001; Fig. 4D and E). As compared with the
pcDNA-3.1(+) plasmid-transfected cells, the CCK-8 assay results
showed that pcDNA-3.1(+)-NLRP3-transfected (OE-NLRP3) KYSE-510
cells had an increased viability (P=0.002), however, EC9706 cells
showed no significant increase in viability (P=0.60) (Fig. 4H and I), demonstrating that NLRP3
promotes cell proliferation. Consistently, the wound-healing
capability of the cells was also strongly increased in the EC9706
(P=0.045) and KYSE-510 (P=0.021) cells following NLRP3
overexpression (Fig. 6A-D).
Furthermore, NLRP3 upregulation increased the number of invasive
cells in the EC9706 (P<0.0001) and KYSE-510 cell lines
(P=0.0005), and enhanced tumor cell migration in the EC9706
(P<0.0001) and KYSE-510 (P<0.0001) cell lines (Fig. 6E-H). Overall, the present results
demonstrated that NLRP3 is positively correlated with tumor
progression by promoting cell proliferation, migration and
invasion.
Discussion
Inflammasomes have been reported to play an
important role in the progression of human cancer (21). The NLR pyrin family domain
containing 3 (NLRP3) inflammasome has been shown to have both
beneficial and adverse effects in multiple tumor types (22,23).
Allen et al found that the NLRP3 inflammasome could suppress
colitis-associated carcinoma development (24), whereas Huang et al
demonstrated that NLRP3 inflammasome promoted the development of
head and neck squamous cell carcinoma (25). Similarly, Li et al found that
the NLRP3 inflammasome accelerated the proliferation of epithelial
cells and gastric cancer carcinogenesis (26). A study concerning oral cavity
squamous cell carcinoma also showed that NLRP3 and interleukin
(IL)-1β not only influenced poor overall and disease-specific
survival but also were correlated with disease-free survival
(27). However, the effect of the
NLRP3 inflammasome on esophageal squamous cell carcinoma (ESCC)
progression is unclear.
The present results showed that the mRNA levels of
the components of the NLRP3 inflammasome [NLRP3,
apoptosis-associated speck-like protein containing a CARD
(ASC), caspase-1 and IL-1β] were all elevated in
human ESCC tissues, as compared with those of adjacent
non-cancerous tissues, although the degree of elevation varied
between patients. Following the evaluation of the pathological
characteristics of each patient, it was found that the high NLRP3
protein expression was associated with TNM stage and T category,
but not with patient lymph node status, metastasis status, sex or
age. Of note, a higher expression of NLRP3 was observed in patients
with a higher pathological stage (III–IV vs. I–II). These data
indicate that patients with a high NLRP3 expression may have a more
malignant clinical phenotype.
To obtain a further understanding of the correlation
of the NLRP3 with ESCC progression, the biological function of
NLRP3 was assessed in vitro. It was found that the knockdown
or overexpression of NLRP3 could markedly abolish or promote cell
migration and invasion and decrease or increase cell mobility,
respectively. These results suggest that NLRP3
inflammasome-mediated inflammation contributes to the development
and progression of ESCC. Therefore, targeting NLRP3 may be an
effective strategy for treating NLRP3-mediated diseases, such as
ESCC.
This study, however, had several limitations. The
upregulated NLRP3 inflammasome was only detected in some but not
all ESCC patients. Further investigation could help clarify the
reason for this apparent inconsistency. Alternatively, NLRP3
inflammasome undergoes post-translational revision. Therefore, the
phosphorylation or ubiquitination of NLRP3 may also affect
inflammasome activation (28).
In conclusion, it was found in the present study
that the NLRP3 inflammasome is upregulated in ESCC tissues and its
activation is significantly associated with Ki-67 levels, T
category and TNM stage. The NLRP3 inflammasome can stimulate ESCC
proliferation, migration and invasion in vitro. Additional
research is required to better understand the modulation of NLRP3
expression and target it accordingly.
Acknowledgements
The authors would like to thank Assistant Professor
Xinshou Ouyang (Yale University, New Haven, Connecticut, CT, USA)
for donating the pcDNA-3.1(+)-NLRP3 and pcDNA-3.1(+) vector
plasmids.
Funding
The present study was sponsored by the National
Natural Science Foundation in China (grant no. U1704282).
Availability of data and materials
This article included all study data.
Authors' contributions
All authors contributed equally in the present
study. YHW, SNH and LRZ designed the study. SY, JJY, SGL and CZH
carried out the experiments and analyzed the data. JXM analyzed the
data. NH carried out the western blot analysis. TLF and XNL
contributed to tissue collection. All authors read and approved the
manuscript and agree to be accountable for all aspects of the
research in ensuring that the accuracy or integrity of any part of
the work are appropriately investigated and resolved.
Ethics approval and consent to
participate
The Ethics Committee of First Clinical Hospital of
Zhengzhou University (Zhengzhou, Henan, China) approved this study,
and all patients provided informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Napier KJ, Scheerer M and Misra S:
Esophageal cancer: A review of epidemiology, pathogenesis, staging
workup and treatment modalities. World J Gastrointest Oncol.
6:112–120. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mimura K, Yamada L, Ujiie D, Hayase S,
Tada T, Hanayama H, Thar Min AK, Shibata M, Momma T, Saze Z, et al:
Immunotherapy for esophageal squamous cell carcinoma: A review.
Fukushima J Med Sci. 64:46–53. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
van Hagen P, Hulshof MC, van Lanschot JJ,
Steyerberg EW, van Berge Henegouwen MI, Wijnhoven BP, Richel DJ,
Nieuwenhuijzen GA, Hospers GA, Bonenkamp JJ, et al: Preoperative
chemoradiotherapy for esophageal or junctional cancer. N Engl J
Med. 366:2074–2084. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Grivennikov SI, Greten FR and Karin M:
Immunity, inflammation, and cancer. Cell. 140:883–899. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Broz P and Monack DM: Molecular mechanisms
of inflammasome activation during microbial infections. Immunol
Rev. 243:174–190. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gross O, Thomas CJ, Guarda G and Tschopp
J: The inflammasome: An integrated view. Immunol Rev. 243:136–151.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Fink SL and Cookson BT: Apoptosis,
pyroptosis, and necrosis: Mechanistic description of dead and dying
eukaryotic cells. Infect Immun. 73:1907–1916. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Karki R, Man SM and Kanneganti TD:
Inflammasomes and cancer. Cancer Immunol Res. 5:94–99. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zitvogel L, Kepp O, Galluzzi L and Kroemer
G: Inflammasomes in carcinogenesis and anticancer immune responses.
Nat Immunol. 13:343–351. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Okamoto M, Liu W, Luo Y, Tanaka A, Cai X,
Norris DA, Dinarello CA and Fujita M: Constitutively active
inflammasome in human melanoma cells mediating autoinflammation via
caspase-1 processing and secretion of interleukin-1beta. J Biol
Chem. 285:6477–6488. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang Y, Kong H, Zeng X, Liu W, Wang Z, Yan
X, Wang H and Xie W: Activation of NLRP3 inflammasome enhances the
proliferation and migration of A549 lung cancer cells. Oncol Rep.
35:2053–2064. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang H, Wang Y, Du Q, Lu P, Fan H, Lu J
and Hu R: Inflammasome-independent NLRP3 is required for
epithelial-mesenchymal transition in colon cancer cells. Exp Cell
Res. 342:184–192. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bruchard M, Mignot G, Derangère V, Chalmin
F, Chevriaux A, Végran F, Boireau W, Simon B, Ryffel B, Connat JL,
et al: Chemotherapy-triggered cathepsin B release in
myeloid-derived suppressor cells activates the Nlrp3 inflammasome
and promotes tumor growth. Nat Med. 19:57–64. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wei Q, Mu K, Li T, Zhang Y, Yang Z, Jia X,
Zhao W, Huai W, Guo P and Han L: Deregulation of the NLRP3
inflammasome in hepatic parenchymal cells during liver cancer
progression. Lab Invest. 94:52–62. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chen LC, Wang LJ, Tsang NM, Ojcius DM,
Chen CC, Ouyang CN, Hsueh C, Liang Y, Chang KP, Chen CC and Chang
YS: Tumour inflammasome-derived IL-1β recruits neutrophils and
improves local recurrence-free survival in EBV-induced
nasopharyngeal carcinoma. EMBO Mol Med. 4:1276–1293. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Pirker R, Pereira JR, von Pawel J,
Krzakowski M, Ramlau R, Park K, de Marinis F, Eberhardt WE,
Paz-Ares L, Störkel S, et al: EGFR expression as a predictor of
survival for first-line chemotherapy plus cetuximab in patients
with advanced non-small-cell lung cancer: Analysis of data from the
phase 3 FLEX study. Lancet Oncol. 13:33–42. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Guanen Q, Junjie S, Baolin W, Chaoyang W,
Yajuan Y, Jing L, Junpeng L, Gaili N, Zhongping W and Jun W:
miR-214 promotes cell metastasis and inhibits apoptosis of
esophageal squamous cell carcinoma via PI3K/AKT/mTOR signaling
pathway. Biomed Pharmacother. 105:350–361. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
de Martel C and Franceschi S: Infections
and cancer: Established associations and new hypotheses. Crit Rev
Oncol Hematol. 70:183–194. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
He Q, Fu Y, Tian D and Yan W: The
contrasting roles of inflammasomes in cancer. Am J Cancer Res.
8:566–583. 2018.PubMed/NCBI
|
|
23
|
Moossavi M, Parsamanesh N, Bahrami A,
Atkin SL and Sahebkar A: Role of the NLRP3 inflammasome in cancer.
Mol Cancer. 17:1582018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Allen IC, TeKippe EM, Woodford RM, Uronis
JM, Holl EK, Rogers AB, Herfarth HH, Jobin C and Ting JP: The NLRP3
inflammasome functions as a negative regulator of tumorigenesis
during colitis-associated cancer. J Exp Med. 207:1045–1056. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Huang CF, Chen L, Li YC, Wu L, Yu GT,
Zhang WF and Sun ZJ: NLRP3 inflammasome activation promotes
inflammation-induced carcinogenesis in head and neck squamous cell
carcinoma. J Exp Clin Cancer Res. 36:1162017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li S, Liang X, Ma L, Shen L, Li T, Zheng
L, Sun A, Shang W, Chen C, Zhao W and Jia J: miR-22 sustains NLRP3
expression and attenuates H. pylori-induced gastric carcinogenesis.
Oncogene. 37:884–896. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Mantovani A, Romero P, Palucka AK and
Marincola FM: Tumour immunity: Effector response to tumour and role
of the microenvironment. Lancet. 371:771–783. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Juliana C, Fernandes-Alnemri T, Kang S,
Farias A, Qin F and Alnemri ES: Non-transcriptional priming and
deubiquitination regulate NLRP3 inflammasome activation. J Biol
Chem. 287:36617–36622. 2012. View Article : Google Scholar : PubMed/NCBI
|