Introduction
With approximately 442,000 new cases diagnosed and
440,000 associated deaths worldwide in 2013, esophageal carcinoma
is ranked as the eighth most common human malignancy and the sixth
leading cause of cancer-related deaths worldwide (1). Esophageal squamous cell carcinoma
(ESCC), as the predominant histopathological type, accounts for
approximately 90% of esophageal cancer cases (2). Despite advances in diagnosis and
multimodal therapy, the prognosis remains less than satisfactory,
with 5 year overall survival rates ranging between 15 and 50%
(3–5). Lymph node metastasis (LNM) is the
single most important prognostic factor and remains the major cause
of mortality after complete resection in ESCC patients (6,7). In
fact, it has been revealed that the number of metastatic nodes is
closely related to survival, and the 5 year survival rate of
patients with 0, 1–2, ≥3 positive lymph nodes was 59.8%, 33.4% and
9.4%, respectively (3). Overall,
there is an urgent need to elucidate the underlying mechanisms of
LNM by identifying key molecules that may contribute to the
development of reasonable treatment plans for different patients
and improve the outcomes of ESCC patients.
Runt-related transcription factor 3 (RUNX3), located
on human chromosome 1p36.1, is a member of the runt-domain family
of transcription factors (8,9). RUNX3
has been identified as a potential tumor suppressor gene in a
variety of malignancies, especially in gastrointestinal cancers,
such as gastric cancer, hepatocellular cancer, colorectal cancer,
and ESCC (8,10–13).
RUNX3 inactivation is involved in tumor development and metastasis
through different processes, including the cell cycle, apoptosis,
angiogenesis, EMT, invasion and migration (14). Although researchers have
investigated the expression profile and roles of RUNX3 in ESCC
progression (15), the molecular
mechanisms of RUNX3 in EMT and ESCC metastasis have not yet been
investigated and need to be elucidated.
Tumor metastasis is a multicellular process that
involves cell-cell adherence, invasion, migration, angiogenesis,
extracellular matrix (ECM) degradation and EMT (16,17).
Among these processes, EMT is thought to have crucial functions.
EMT is a dynamic process in which epithelial cells typically lose
their epithelial characteristics, including cell polarity and
cell-cell contact, and acquire a spindle shape migration phenotype
(18,19). In addition, the expression of the
epithelial markers E-cadherin and claudin-1 is reduced, whereas
that of the mesenchymal markers N-cadherin and vimentin is
increased (20).
The transforming growth factor β (TGF-β) pathway
plays a complex dual role in tumor development. It acts as a tumor
suppressor pathway during the early stages of epithelial neoplasia
by inhibiting tumor cell proliferation and inducing apoptosis. At
later stages of carcinogenesis, TGF-β signaling contributes to
tumor invasion and metastasis by inducing EMT (18,21,22).
TGF-β/Smad signaling, the main pathway in EMT, is initiated by
transforming growth factor β1 (TGF-β1) binding to its type II
receptor (TβRII). TβRII forms a heterodimer with the TGF-β type I
receptor (TβRI); activated TβRI phosphorylates R-Smad (Smad2 and
Smad3) at the distal C-terminal SXS motif, after which pSmad2/3 and
Co-Smad (Smad4) form a transcription complex that transduces the
signal to the nucleus. In the nucleus, the Smad complex directly or
indirectly binds to various transcription factors, thus providing
additional regulation of target genes that mediate EMT (23–25).
On the basis of our previous studies, the aim of the
present study was to investigate the potential roles and molecular
mechanisms of RUNX3 in ESCC metastasis and EMT.
Materials and methods
Ethics statement
The present study was approved by the Research
Ethics Committee of Shandong Provincial Hospital Affiliated with
Shandong University. Written informed consent for use of the
tissues and data analysis was obtained from every patient or their
relatives.
Patients and specimens
In total, 102 ESCC tissues and 30 adjacent normal
tissues (>5 cm from the margin of the tumor) were harvested from
patients who underwent Ivor-Lewis esophagectomy with two-field
lymphadenectomy at the Department of Thoracic Surgery, Provincial
Hospital Affiliated to Shandong University from May 2017 to
December 2018. All patients underwent esophagectomy with complete
resection, and none received neoadjuvant radio-/chemotherapy.
Postoperative staging was based on the eighth edition of the
International Union Against Cancer (UICC) tumor-node-metastasis
(TNM) classification criteria published in 2017. Among the 102 ESCC
patients, 45 had LNM, and 57 did not have LNM. The detailed
clinical data for these patients are presented in Table I.
 | Table I.Correlations between RUNX3 expression
and clinicopathological features of 102 ESCC patients. |
Table I.
Correlations between RUNX3 expression
and clinicopathological features of 102 ESCC patients.
|
|
| RUNX3 expression
(%) |
|
|---|
|
|
|
|
|
|---|
| Parameters | Cases (n=102) | Positive
(n=23) | Negative
(n=79) | P-value |
|---|
| Sex |
|
|
| 0.475 |
|
Male | 45 | 12 (26.7) | 33 (73.3) |
|
|
Female | 57 | 11 (19.3) | 46 (80.7) |
|
| Age (years) |
|
|
| 0.810 |
|
≤50 | 41 | 10 (24.4) | 31 (75.6) |
|
|
>50 | 61 | 13 (21.3) | 48 (78.7) |
|
| Tumor size
(cm) |
|
|
| 0.347 |
|
<3 | 44 | 12 (27.3) | 32 (72.7) |
|
| ≥3 | 58 | 11 (19.0) | 47 (81.0) |
|
| T status |
|
|
| 0.027 |
|
T1-2 | 63 | 19 (30.2) | 44 (69.8) |
|
|
T3-4 | 39 | 4
(10.3) | 35 (89.7) |
|
| Differentiation
degree |
|
|
| 0.158 |
|
Low | 54 | 9
(16.7) | 45 (83.3) |
|
|
Mid-high | 48 | 14 (29.2) | 34 (70.8) |
|
| LNM |
|
|
| 0.017 |
| No | 57 | 18 (31.6) | 39 (68.4) |
|
|
Yes | 45 | 5
(11.1) | 40 (88.9) |
|
Cell culture and transfection
Five human ESCC cell lines (TE-1, Eca109, KYSE150,
KYSE450, and EC9706) were purchased from the Cell Bank of the
Shanghai Institute in China. All cells were cultured in Roswell
Park Memorial Institute (RPMI)-1640 medium supplemented with 1%
penicillin/streptomycin and 10% fetal bovine serum (FBS; Gibco;
Thermo Fisher Scientific, Inc.). For activation of the TGF-β
pathway, cells were treated with 10 ng/ml recombinant TGF-β1 (cat.
no. AF-100-21C-10; PeproTech, Inc.). Cell culture plates were
maintained at 37°C in a humidified 5% CO2 incubator.
Human RUNX3 overexpression plasmids (NM-004350) were chemically
synthesized and packaged into lentiviruses (Shanghai GeneChem Co.,
Ltd.). Puromycin was used at 5 µg/ml for 1 week to select stably
transfected cells. Cells were labeled BC (without lentivirus
transfection), vector (transfected with the control vector), and
RUNX3 (transfected with the RUNX3 overexpression plasmid).
Immunohistochemical (IHC)
analysis
IHC analysis was performed to detect the expression
level of RUNX3 as well as the correlation between the expression of
RUNX3 and EMT-related markers using the streptavidin-peroxidase
(SP) method. Sections were stained with anti-RUNX3 (dilution 1:100;
cat. no. ab40278), anti-N-cadherin (dilution 1:100; cat. no.
ab76011), and anti-Snail (dilution 1:100; cat. no. ab180714; all
from Abcam), and anti-E-cadherin (dilution 1:100; cat. no.
20874-1-AP; ProteinTech Group, Inc.) antibodies at 4°C overnight.
The primary antibodies were replaced with phosphate-buffered saline
(PBS) as a negative control to rule out nonspecific binding. The
secondary antibody and avidin-biotin peroxidase complex methods
were performed according to the standard protocols provided by the
manufacturer (ZSGB Biotech Beijing; OriGene technologies, Inc.).
All sections were evaluated by two pathologists who were blinded to
the clinical data.
Western blot analysis
Protein was extracted from tissue samples and tumor
cells using radioimmunoprecipitation (RIPA) lysis buffer (Beyotime
Institute of Biotechnology), and the concentration was determined
using a bicinchoninic acid (BCA) kit. Equal amounts of protein (30
µg) were separated by 8% or 10% SDS-PAGE and transferred to
polyvinylidene difluoride (PVDF) membranes. Briefly, 5% nonfat dry
milk was used to block nonspecific binding. The membranes were
incubated overnight at 4°C with primary antibodies. Following three
washes, the membranes were incubated with the corresponding
horseradish peroxidase (HRP)-conjugated secondary antibodies
(dilution 1:5,000; goat anti-mouse cat. no. ZB-2305 and goat
anti-rabbit cat. no. ZB-2301; both from ZSGB Biotech) for 1 h at
room temperature. Finally, the protein levels were quantified using
an enhanced chemiluminescence (ECL) detection system (Amersham
imager 600; General Electric). The antibodies used in the present
study were as follows: anti-RUNX3 (dilution 1:500; cat. no.
ab40278), anti-N-cadherin (dilution 1:1,000; cat. no. ab76011), and
anti-Snail (dilution 1:1,000; cat. no. ab180714; all from Abcam),
anti-matrix metallopeptidase 9 (MMP-9) (dilution 1:1,000; cat. no.
10375-2-AP), anti-GAPDH, (dilution 1:1,000; cat. no. 10494-1-AP),
and anti-E-cadherin (dilution 1:1,000; cat. no. 20874-1-AP;
ProteinTech Group, Inc.), anti-Smad2 (dilution 1:1,000; cat. no.
ab40855) and anti-Smad3 (dilution 1:1,000; cat. no. ab40854; both
from Abcam), anti-pSmad2ser465/467 (dilution 1:1,000;
cat. no. 3108T; Cell Signaling Technology, Inc.), and
anti-pSmad3s423+s425 antibodies (dilution 1:1,000; cat.
no. ab52903; Abcam).
RNA extraction and real-time PCR
Total RNA was isolated from cultured cells using
RNAiso Plus (Takara Biotechnology Co., Ltd.) according to the
manufacturer's instructions. All RNA samples were treated with
RNase-free DNase (Takara Biotechnology Co., Ltd.) and stored at
−80°C. One microgram of RNA was subjected to reverse transcription
using a reverse transcriptase reagent kit with gDNA Eraser (Takara
Biotechnology Co., Ltd.) to obtain cDNA. GAPDH was used for
normalization. Relative transcript quantities were calculated using
the 2−ΔΔCq method (26).
The thermocycling conditions were as follows: 95°C for 10 min and
then 40 cycles of 95°C for 15 sec and annealing/extension at 60°C
for 1 min. The experiments were performed in triplicate. The primer
sequences used were as follows: RUNX3, 5′-TCAACGACCTTCGCTTCGTG-3′
(forward primer) and 5′-ACCTTGATGGCTCGGTGGTA-3′ (reverse primer);
GAPDH, 5′-GCACCGTCAAGGCTGAGAAC-3′ (forward primer) and
5′-TGGTGAAGACGCCAGTGGA-3′ (reverse primer).
Transwell assay
For both the migration and invasion assays, cells
were pre-incubated in FBS-free medium for 24 h. Uncoated Transwells
were used for the migration assay, whereas 40 µl of Matrigel (1:4;
BD Biosciences) was used to pre-coat the upper surface of the
Transwells in the invasion assay. Next, 1.5×105
serum-starved cells were seeded in the upper Transwells chamber in
a 24-well plate (8 µm pore size polycarbonate membrane; EMD
Millipore) with 200 µl of FBS-free medium; 600 µl of medium with
15% FBS was added to the lower chamber. The cells were incubated
for 24 h for the migration assay and for 48 h for the invasion
assay. Finally, the cells migrating to the lower surface of the
membrane were fixed with 4% paraformaldehyde for 20 min and stained
with hematoxylin for 5 min at room temperature; cells from three
randomly selected fields were counted. The data are presented as
the mean ± SD.
Wound-healing assay
Cells were plated into 6-well plates and cultured in
RPMI-1640 with 10% FBS until they reached 90% confluence. The
confluent cell monolayers were scratched using a 10 µl pipette tip
and incubated in culture medium with 1% FBS. Images were captured
using a LEICA DMi8 inverted microscope every 12 h (magnification
×100).
Immunofluorescence analysis
Following fixation in 4% formaldehyde for 20 min,
permeabilization with 0.5% Triton X-100 for 20 min and blocking
with 10% normal goat serum for 1 h, cells were incubated with
rabbit anti-pSmad2 (dilution 1:200) and rabbit anti-pSmad3
(dilution 1:100) antibodies at 4°C overnight. The cells were then
incubated with Alexa Fluor 594-conjugated goat anti-rabbit IgG
(dilution 1:100) for 1 h at 37°C, and the nuclei were stained with
4′-6-diamidino-2-phenylindole (DAPI) for 5 min at room temperature.
Images were captured using an Olympus BX43 fluorescence microscope
(magnification ×200).
Statistical analysis
SPSS 22.0 software (IBM Corp.) was used for all
statistical analyses. Quantitative data was expressed as the mean ±
standard deviation (SD). The χ2 test was used to analyze
the association between RUNX3 expression and clinicopathological
variables. A paired Student's t-test was used to compare RUNX3 in
paired ESCC tumor tissues and adjacent normal tissues, and for all
other comparisons between two groups, unpaired Student's t-tests
were performed. The significance of differences among more than two
groups was calculated by one-way analysis of variance (ANOVA)
followed by Tukey's post hoc test. Pearson's correlation analysis
was used to analyze correlations between RUNX3 and EMT-related
marker expression. A statistically significant difference was
defined as a P-value <0.05.
Results
RUNX3 expression is decreased in
ESCC
The expression profile of RUNX3 in 30 pairs of ESCC
tumor tissues and adjacent normal tissues by was first assessed
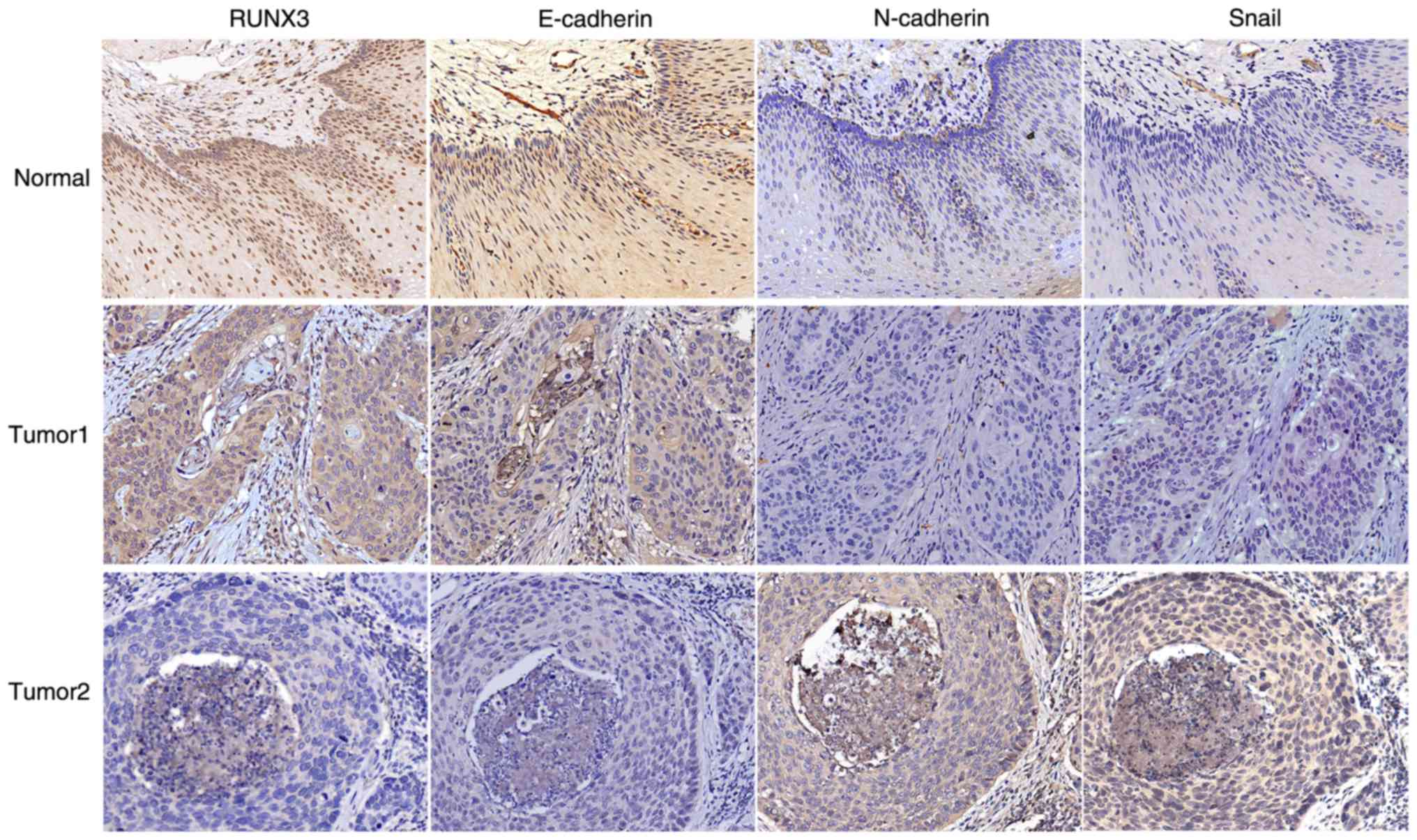
with IHC analysis. Significant staining was readily observed in the
nuclei of the noncancerous tissue; weak immunostaining was observed
in tumor cells (Fig. 1).
Furthermore, RUNX3 expression levels were verified through western
blot analysis and it was revealed that RUNX3 expression in tumor
tissues was significantly lower than that in adjacent normal
tissues (Fig. 2A and B;
RUNX3/GAPDH: 0.50±0.20 vs. 0.83±0.16; P<0.001).
RUNX3 expression is associated with T
status and LNM
Tissues were considered RUNX3-negative when nuclear
staining was present in <5% of the cells, with or without
cytoplasmic staining. According to these criteria, all 102 ESCC
patient samples were divided into two groups: 23 cases (22.55%)
were categorized as the positive expression group, and 79 cases
(77.45%) were categorized as the negative expression group. The
chi-square test was next used to investigate the relationship
between RUNX3 expression and clinicopathological parameters
(including age, sex, tumor size, T status, differentiation degree,
and LNM). As revealed in Table I,
negative RUNX3 expression was more common in T3-4 than in T1-2
cases (89.7% vs. 69.8%, respectively; P=0.027) and more prevalent
in node-positive than in node-negative cases (88.9% vs. 68.4%,
respectively; P=0.017). Thus, it was surmised that decreased RUNX3
expression may play a vital role in ESCC metastasis. No significant
differences were revealed between RUNX3 expression and other
clinicopathological parameters.
RUNX3 expression is negatively
correlated to Snail and N-cadherin expression and positively
correlated to E-cadherin expression in ESCC tissues
EMT has been reported to contribute to tumor
invasion, migration and metastasis in various cancers (27). To elucidate the mechanisms of RUNX3
in ESCC LNM, the correlation between RUNX3 and EMT-related marker
expression in LNM tissues and non-LNM tissues (Fig. 1) was evaluated. IHC results revealed
RUNX3 expression to be negatively correlated to the expression of
the mesenchymal marker N-cadherin (r=−0.429; P<0.01) and
transcription factor Snail (r=−0.364; P<0.01) and
positively correlated to the expression of the epithelial marker
E-cadherin (r=0.580; P<0.01) in ESCC tissues (Table II). These results indicated that
RUNX3 expression was associated with ESCC EMT.
 | Table II.Correlations between expressions of
RUNX3 and EMT related markers in 102 ESCC patients. |
Table II.
Correlations between expressions of
RUNX3 and EMT related markers in 102 ESCC patients.
|
| E-cadherin | N-cadherin | Snail |
|---|
|
|
|
|
|
|---|
| Parameters | Positive | Negative | Positive | Negative | Positive | Negative |
|---|
| RUNX3 |
| Positive | 17 | 6 | 7 | 16 | 11 | 12 |
| Negative | 10 | 69 | 62 | 17 | 67 | 12 |
| r | 0.580 |
| −0.429 |
| −0.364 |
|
| P-value | <0.01 |
| <0.01 |
| <0.01 |
|
Cell transfection
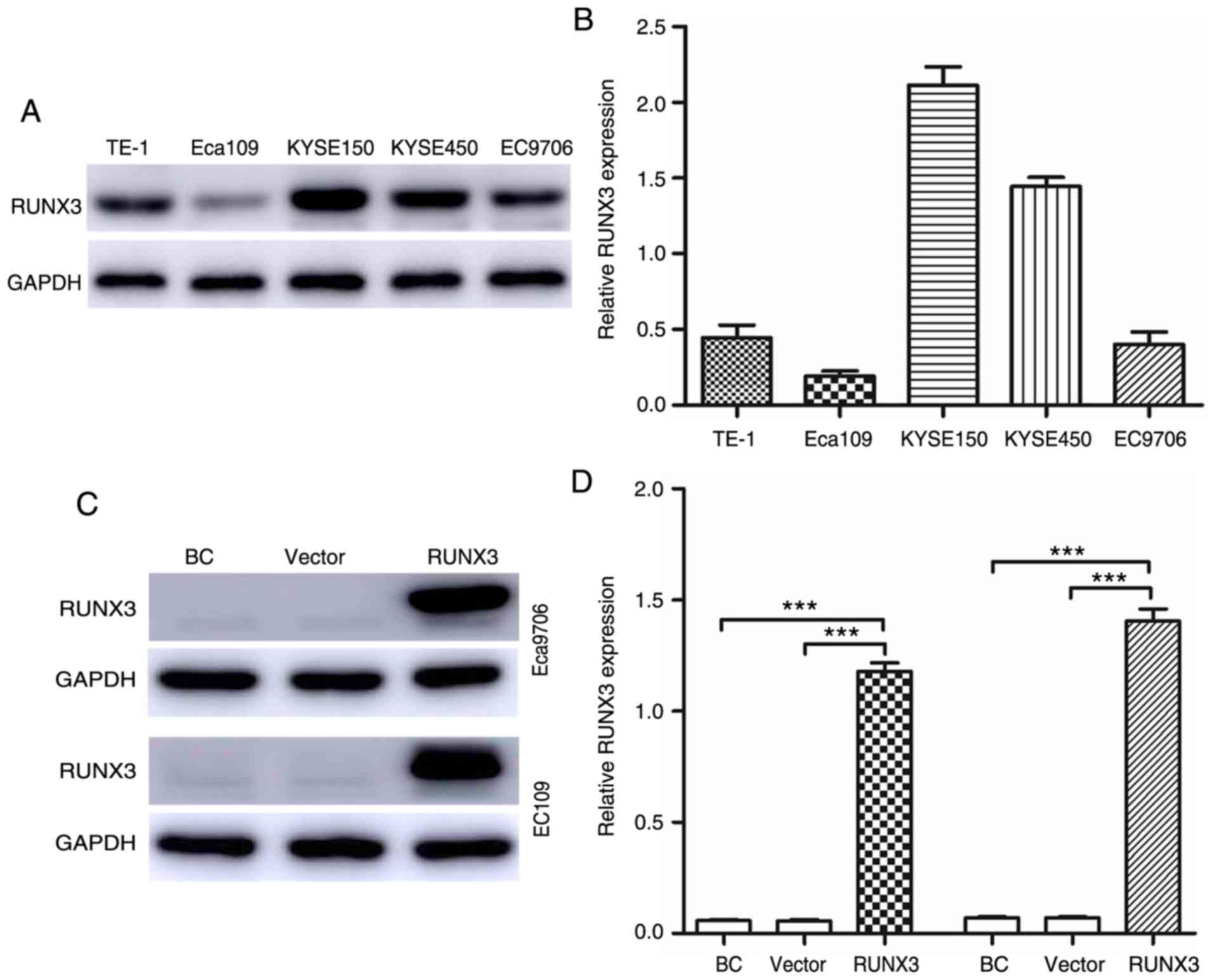
The expression levels of RUNX3 in five ESCC cell
lines were detected via qRT-PCR and western blot analyses.
According to the results, Eca109 and EC9706 cells exhibited the
lowest expression of RUNX3 at both the protein (Fig. 3A) and mRNA (Fig. 3B) levels. Thus, Eca109 and EC9706
cells were used to analyze the upregulation of RUNX3 expression.
Compared with the blank control and vector groups, the RUNX3 group
exhibited significantly upregulated RUNX3 expression, although
there was no difference in RUNX3 expression between the blank
control and vector groups (Fig. 3C and
D).
Upregulation of RUNX3 significantly
decreases the invasion and migration abilities of ESCC cells as
well as MMP-9 expression
Since RUNX3 expression was negatively correlated
with T status and LNM in ESCC patients, the effect of RUNX3 on cell
migration and invasion in ESCC cells was further investigated using
wound-healing and Transwell assays. In the Transwell assays, the
number of cells in the RUNX3 group that traversed the membrane was
significantly decreased compared with that in the vector group
(Fig. 4A and B; P<0.01).
Moreover, the number of cells that invaded Matrigel was also
significantly attenuated in the RUNX3 group (Fig. 4C and D; P<0.01). The data from
the wound-healing assay further supported this finding, as
upregulating expression of RUNX3 suppressed cell migration compared
with the vector group (Fig. 4E).
Furthermore, restoration of RUNX3 expression led to a significant
decrease in the expression of MMP-9 (Fig. 4F and G; P<0.01), which can
promote invasion and LNM through ECM degradation (28).
Upregulation of RUNX3 may reverse EMT
and decrease Smad2/3 phosphorylation in ESCC cells
To evaluate the mechanism of RUNX3 in ESCC
metastasis, the levels of EMT markers (E-cadherin, N-cadherin and
Snail) were detected by western blotting and it was revealed that
N-cadherin and Snail expression levels were downregulated in
RUNX3-overexpressing cells but that E-cadherin expression was
significantly upregulated (Fig.
5A). TGF-β/Smad-induced EMT has been suggested to be associated
with the development and progression of ESCC. To further assess
whether RUNX3 reverses ESCC EMT through the TGF-β/Smad pathway, the
levels of Smad2, Smad3 and their phosphorylated versions (pSmad2,
pSmad3) were evaluated. The results of the western blot analysis
revealed that levels of both pSmad2 and pSmad3 were decreased in
the RUNX3 group compared to the vector group; in contrast, the
levels of Smad2 and Smad3 were unaltered (Fig. 5B). Immunofluorescence assays were
also used to validate the effect of RUNX3 overexpression on pSmad2
and pSmad3 staining, and as revealed in Fig. 5C, pSmad2 and pSmad3 staining was
markedly decreased in the RUNX3 group.
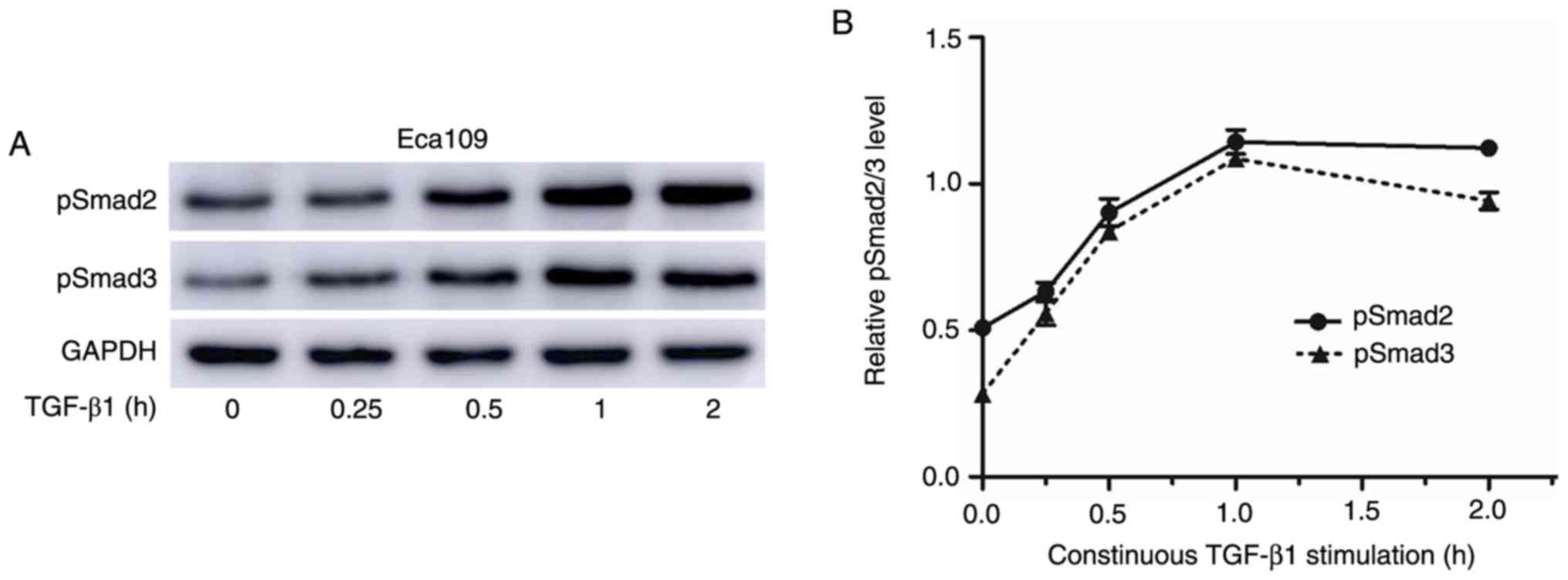
RUNX3-overexpressing cells display
diminished responsiveness to TGF-β1-induced EMT
A previous study indicated that Runx3-null gastric
epithelial lines are unexpectedly sensitive to TGF-β1-induced EMT
(29). The sensitivity of RUNX3 to
TGF-β1-induced EMT was next investigated and it was revealed that
the levels of pSmad2/3 peaked at 1 h after TGF-β treatment
(Fig. 6A and B). Thus, the temporal
pattern of Smad2/3 phosphorylation after treatment with 10 ng/ml
TGF-β1 for 1 h was investigated. Based on the results, the levels
of pSmad2/3 significantly increased in the vector group, whereas no
significant changes were evident in RUNX3-overexpressing cells
(Fig. 7A). The expression of
EMT-related markers after 72 h of stimulation with TGF-β1 was also
assessed. TGF-β1-induced increases in N-cadherin and Snail and
decreases in E-cadherin levels were significantly greater in the
vector control cells than in the RUNX3-overexpressing cells
(Fig. 7B). One hallmark of EMT is
phenotypic change in epithelial cell morphology. Eca109 and EC9706
cells displayed an elongated fibroblast-like morphology and reduced
cell-cell contact after 72 h of TGF-β1 stimulation, whereas
noticeable morphological changes and cell-cell contact were not
detected in RUNX3-overexpressing cells (Fig. 7C and D).
Discussion
It is widely accepted that surgery is the standard
treatment for patients with localized ESCC (30). Although surgical treatment can cure
90% of patients with early ESCC, the prognosis is suboptimal for
advanced-stage ESCC due to a lack of early diagnosis methods, local
relapse, distant metastasis and resistance to traditional
treatments such as chemotherapy and radiotherapy. The esophagus has
a unique histological structure involving a rich
lymphatic-capillary network in the submucosa, which contributes to
variable lymphatic spread and skip metastasis (3,31). In
fact, Li et al reported that approximately 33.1% of ESCC
patients experience LNM when the submucosa (T1b) is invaded and
that 4.3% of ESCC patients still experience LNM even when the tumor
is confined to the mucosa (T1a) (31). We previously demonstrated that RUNX3
expression was significantly related to LNM and that RUNX3
inactivation was predictive of poor survival (32). In the present study, we focused on
exploring the underlying mechanisms of RUNX3-mediated EMT and
metastasis.
According to the present results, RUNX3 expression
was markedly lower in tumor tissues than in adjacent normal
tissues, and this decreased expression of RUNX3 was correlated with
T status and LNM in ESCC patients. In an attempt to validate the
biological function of RUNX3 in ESCC metastasis, Eca109 and EC9706
cells were transfected with a RUNX3-expressing lentivirus and it
was revealed that restoration of RUNX3 expression attenuated
invasion and migration abilities. Moreover, RUNX3 overexpression
led to a significant decrease MMP-9 expression. All of these
results indicated that RUNX3 markedly inhibited ESCC cell
metastasis. Since EMT plays an important role in tumor invasion and
metastasis, it was surmised that RUNX3 may prevent ESCC cell
metastasis by modulating EMT. The expression levels of E-cadherin
were determined to be increased and those of Snail and N-cadherin
to be decreased in the RUNX3-overexpressing group compared with the
vector control group. IHC results also revealed that RUNX3
expression was positively correlated with E-cadherin expression and
inversely with Snail and N-cadherin expression in ESCC tissues,
which strongly supports our hypothesis.
Recently, an accumulating number of studies have
demonstrated that EMT can be induced by numerous molecules, such as
inflammatory cytokines, growth factors and numerous transcription
factors (20). Among these factors,
TGF-β is a key driver of EMT (19).
Cumulative research has revealed that TGF-β signaling is tightly
controlled by the phosphorylation of R-Smads and that
dephosphorylation of R-Smads disrupts signal relay (33). Lin et al reported that PPM1A
dephosphorylates TGF-β-activated Smad2/3, dissociates the Smad
complex, and promotes the nuclear export of Smad2/3 (34). RUNX3, as a critical downstream
effector in the TGF-β signaling pathway, physically interacts with
R-Smad through its C-terminal region (35–38).
In this study, it was revealed that RUNX3 overexpression resulted
in significant dephosphorylation of pSmad2/3, indicating that the
effect of RUNX3 on reversing EMT may be attributable, at least in
part, to TGF-β/Smad signaling.
A previous study has indicated that the gastric
mucosa of Runx3-null mice is resistant to TGF-β1-induced growth
suppression (8). However, in
another study, Voon et al reported that Runx3-null gastric
epithelial lines were unexpectedly sensitive to TGF-β1-induced EMT
(29). Therefore, the sensitivity
of RUNX3 to TGF-β1-induced EMT was further assessed. Upon TGF-β1
stimulation, significantly decreased levels of E-cadherin and
increased levels of N-cadherin, Snail and pSmad2/3 were observed in
the vector group. However, no effect on these proteins was observed
in the RUNX3 group. Morphological studies also revealed that TGF-β1
treatment induced phenotypic changes, resulting in the cells in the
vector group adopting a more spindle-like morphology than the cells
in the RUNX3 group, which indicates that RUNX3-overexpressing ESCC
cells were resistant to TGF-β1-induced EMT.
RUNX3 is thought to exist in a basal, inactive state
in the cytoplasm and cannot act as a transcription factor (9). Increasing evidence indicates that
RUNX3 can be reactivated and is therefore considered to be a good
drug target (39). Our previous
studies demonstrated that reactivation of RUNX3 by 5-azacytidine
(an anticancer drug) inhibited the malignant behavior of ESCC cells
(40). Moreover, heterologous RUNX3
expression was able to reverse cisplatin resistance in ESCC cell
lines (41). In the present study,
evidence is provided that restoration of RUNX3 expression decreased
the invasion and migration of ESCC cells by inhibiting EMT.
Collectively, the present results revealed that novel drugs
targeting RUNX3 are a promising treatment strategy for ESCC.
There are several limitations to the present study.
Firstly, the number of samples was relatively small, which may
affect the reliability of our findings. Therefore, larger sample
sizes and multicenter randomized studies are required. Secondly,
the cells were not serum-starved in the wound-healing assay. In our
future studies, we will attempt to knock down RUNX3 and develop a
lung metastasis model to further validate our results in
vivo. Immunoprecipitation assays will also be performed to
verify the binding of RUNX3 to R-Smad.
In summary, in the present study it was revealed
that RUNX3 inhibited EMT by abrogating TGF-β1-mediated Smad2/3
phosphorylation and decreasing the expression of the transcription
factor Snail, thereby inhibiting the invasion and migration
abilities of ESCC cells. This study is the first to elucidate the
detailed mechanisms of RUNX3-mediated regulation of EMT and
metastasis in ESCC. The findings herein support the hypothesis that
targeted therapies for RUNX3 may serve as complementary treatment
approaches to control postoperative LNM and improve the survival of
ESCC patients.
Acknowledgements
The authors appreciate the help from the Departments
of Thoracic Surgery and Pathology of Shandong Provincial Hospital
affiliated to Shandong University and the Department of Thoracic
Surgery of The Second Hospital of Shandong University.
Funding
The present study was funded by the Natural Science
Foundation of Shandong Province (grant no. ZR2017MH089), and the
Major Program of Shandong Province Natural Science Foundation
(grant no. ZR2018ZC0232).
Availability of data and materials
The data used during the study are available from
the corresponding author upon reasonable request.
Authors' contributions
YT and QS collected the samples and acquired the
experimental data. WJ, GC and MS performed the analysis of the
data. ZX wrote the manuscript. ZX, YT, YJ and BS performed the
experiments. ZW and XGZ designed, supervised and funded the
experiments. All authors read and approved the manuscript and agree
to be accountable for all aspects of the research in ensuring that
the accuracy or integrity of any part of the work are appropriately
investigated and resolved.
Ethics approval and consent to
participate
The present study was approved by the Research
Ethics Committee of Shandong Provincial Hospital Affiliated to
Shandong University (protocol no. 2017550). Written informed
consent was obtained from each patient or their relatives for use
of the tissues and data analysis.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
RUNX3
|
runt-related transcription factor
3
|
|
ESCC
|
esophageal squamous cell carcinoma
|
|
LNM
|
lymph node metastasis
|
|
EMT
|
epithelial-mesenchymal transition
|
References
|
1
|
Fitzmaurice C, Dicker D, Pain A, Hamavid
H, Moradi-Lakeh M, MacIntyre MF, Allen C, Hansen G, Woodbrook R,
Wolfe C, et al: The global burden of cancer 2013. JAMA Oncol.
1:505–527. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ji X, Cai J, Chen Y and Chen LQ: Lymphatic
spreading and lymphadenectomy for esophageal carcinoma. World J
Gastrointest Surg. 8:90–94. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pennathur A, Gibson MK, Jobe BA and
Luketich JD: Oesophageal carcinoma. Lancet. 381:400–412. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Rutegard M, Charonis K, Lu Y, Lagergren P,
Lagergren J and Rouvelas I: Population-based esophageal cancer
survival after resection without neoadjuvant therapy: An update.
Surgery. 152:903–910. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liu Q, Cai XW, Wu B, Zhu ZF, Chen HQ and
Fu XL: Patterns of failure after radical surgery among patients
with thoracic esophageal squamous cell carcinoma: Implications for
the clinical target volume design of postoperative radiotherapy.
PLoS One. 9:e972252014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Peyre CG, Hagen JA, DeMeester SR, Van
Lanschot JJ, Holscher A, Law S, Ruol A, Ancona E, Griffin SM,
Altorki NK, et al: Predicting systemic disease in patients with
esophageal cancer after esophagectomy: A multinational study on the
significance of the number of involved lymph nodes. Ann Surg.
248:979–985. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chi XZ, Yang JO, Lee KY, Ito K, Sakakura
C, Li QL, Kim HR, Cha EJ, Lee YH, Kaneda A, et al: RUNX3 suppresses
gastric epithelial cell growth by inducing p21(WAF1/Cip1)
expression in cooperation with transforming growth factor
{beta}-activated SMAD. Mol Cell Biol. 25:8097–8107. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ito K, Liu Q, Salto-Tellez M, Yano T, Tada
K, Ida H, Huang C, Shah N, Inoue M, Rajnakova A, et al: RUNX3, a
novel tumor suppressor, is frequently inactivated in gastric cancer
by protein mislocalization. Cancer Res. 65:7743–7750. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hiramatsu T, Osaki M, Ito Y, Tanji Y,
Tokuyasu N and Ito H: Expression of RUNX3 protein in human
esophageal mucosa and squamous cell carcinoma. Pathobiology.
72:316–324. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Shiraha H, Nishina S and Yamamoto K: Loss
of runt-related transcription factor 3 causes development and
progression of hepatocellular carcinoma. J Cell Biochem.
112:745–749. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Soong R, Shah N, Peh BK, Chong PY, Ng SS,
Zeps N, Joseph D, Salto-Tellez M, Iacopetta B and Ito Y: The
expression of RUNX3 in colorectal cancer is associated with disease
stage and patient outcome. Br J Cancer. 100:676–679. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sugiura H, Ishiguro H, Kuwabara Y, Kimura
M, Mitsui A, Mori Y, Ogawa R, Katada T, Harata K and Fujii Y:
Decreased expression of RUNX3 is correlated with tumor progression
and poor prognosis in patients with esophageal squamous cell
carcinoma. Oncol Rep. 19:713–719. 2008.PubMed/NCBI
|
|
14
|
Chen F, Liu X, Bai J, Pei D and Zheng J:
The emerging role of RUNX3 in cancer metastasis (Review). Oncol
Rep. 35:1227–1236. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Torquati A, O'Rear L, Longobardi L,
Spagnoli A, Richards WO and Daniel Beauchamp R: RUNX3 inhibits cell
proliferation and induces apoptosis by reinstating transforming
growth factor beta responsiveness in esophageal adenocarcinoma
cells. Surgery. 136:310–316. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Guan X: Cancer metastases: Challenges and
opportunities. Acta Pharm Sin B. 5:402–418. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yilmaz M, Christofori G and Lehembre F:
Distinct mechanisms of tumor invasion and metastasis. Trends Mol
Med. 13:535–541. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chang TL, Ito K, Ko TK, Liu Q,
Salto-Tellez M, Yeoh KG, Fukamachi H and Ito Y: Claudin-1 has tumor
suppressive activity and is a direct target of RUNX3 in gastric
epithelial cells. Gastroenterology. 138:255–265. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Xu J, Lamouille S and Derynck R:
TGF-Beta-Induced epithelial to mesenchymal transition. Cell Res.
19:156–172. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mittal V: Epithelial mesenchymal
transition in tumor metastasis. Annu Rev Pathol. 13:395–412. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Foroutan M, Cursons J, Hediyeh-Zadeh S,
Thompson EW and Davis MJ: A transcriptional program for detecting
TGFβ-induced EMT in cancer. Mol Cancer Res. 15:619–631. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ikushima H and Miyazono K: TGFbeta
signalling: A complex web in cancer progression. Nat Rev Cancer.
10:415–424. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
23
|
Derynck R, Akhurst RJ and Balmain A:
TGF-Beta signaling in tumor suppression and cancer progression. Nat
Genet. 29:117–129. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Miyazono K, Suzuki H and Imamura T:
Regulation of TGF-beta signaling and its roles in progression of
tumors. Cancer Sci. 94:230–234. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wrighton KH, Lin X and Feng XH:
Phospho-control of TGF-beta superfamily signaling. Cell Res.
19:8–20. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Pastushenko I and Blanpain C: EMT
transition states during tumor progression and metastasis. Trends
Cell Biol. 29:212–226. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Xia T, Tong S, Fan K, Zhai W, Fang B, Wang
SH and Wang JJ: XBP1 induces MMP-9 expression to promote
proliferation and invasion in human esophageal squamous cell
carcinoma. Am J Cancer Res. 6:2031–2040. 2016.PubMed/NCBI
|
|
29
|
Voon DC, Wang H, Koo JK, Nguyen TA, Hor
YT, Chu YS, Ito K, Fukamachi H, Chan SL, Thiery JP and Ito Y: Runx3
protects gastric epithelial cells against epithelial-mesenchymal
transition-induced cellular plasticity and tumorigenicity. Stem
Cells. 30:2088–2099. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Swisher SG, Moughan J, Komaki RU, Ajani
JA, Wu TT, Hofstetter WL, Konski AA and Willett CG: Final results
of NRG oncology RTOG 0246: An organ-preserving selective resection
strategy in esophageal cancer patients treated with definitive
chemoradiation. J Thorac Oncol. 12:368–374. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li B, Chen H, Xiang J, Zhang Y, Kong Y,
Garfield DH and Li H: Prevalence of lymph node metastases in
superficial esophageal squamous cell carcinoma. J Thorac Cardiovasc
Surg. 146:1198–1203. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Shi M, Wang Z, Liu XY and Chen D:
Inactivation of RUNX3 predicts poor prognosis in esophageal
squamous cell carcinoma after ivor-lewis esophagectomy. Med Oncol.
31:3092014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Goto K, Tong KI, Ikura J and Okada H:
HLA-B-associated transcript 3 (Bat3/Scythe) negatively regulates
Smad phosphorylation in BMP signaling. Cell Death Dis. 2:e2362011.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lin X, Duan X, Liang YY, Su Y, Wrighton
KH, Long J, Hu M, Davis CM, Wang J, Brunicardi FC, et al: PPM1A
functions as a smad phosphatase to terminate TGFbeta signaling.
Cell. 125:915–928. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hanai J, Chen LF, Kanno T, Ohtani-Fujita
N, Kim WY, Guo WH, Imamura T, Ishidou Y, Fukuchi M, Shi MJ, et al:
Interaction and functional cooperation of PEBP2/CBF with Smads.
Synergistic induction of the immunoglobulin germline Calpha
promoter. J Biol Chem. 274:31577–31582. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ito Y and Miyazono K: RUNX transcription
factors as key targets of TGF-beta superfamily signaling. Curr Opin
Genet Dev. 13:43–47. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Miyazono K, Maeda S and Imamura T:
Coordinate regulation of cell growth and differentiation by
TGF-beta superfamily and Runx proteins. Oncogene. 23:4232–4237.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zaidi SK, Sullivan AJ, van Wijnen AJ,
Stein JL, Stein GS and Lian JB: Integration of runx and smad
regulatory signals at transcriptionally active subnuclear sites.
Proc Natl Acad Sci USA. 99:8048–8053. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Balmain A: Cancer: New-age tumour
suppressors. Nature. 417:235–237. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wang S, Liu H, Wang Z and Chen HX: Effects
of 5-azacytidine on RUNX3 gene expression and the biological
behavior of esophageal carcinoma cells. Mol Med Rep. 9:1259–1265.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Li DJ, Shi M and Wang Z: RUNX3 reverses
cisplatin resistance in esophageal squamous cell carcinoma via
suppression of the protein kinase B pathway. Thorac Cancer.
7:570–580. 2016. View Article : Google Scholar : PubMed/NCBI
|