Introduction
Liver cancer was the sixth most commonly diagnosed
cancer and the fourth leading cause of cancer deaths worldwide in
2018, with ~841,000 new cases and 782,000 deaths annually (1). Hepatocellular carcinoma (HCC) is the
most frequent primary liver cancer, accounting for 75–85% of all
cases (2). Despite substantial
improvements in diagnostic and therapeutic techniques, the overall
survival (OS) and recurrence-free survival (RFS) rates of HCC
remain comparatively low, mainly because HCC is a highly
heterogeneous malignancy (3,4).
Furthermore, no effective prognostic biomarkers have yet been
described for HCC. Such biomarkers might help to guide individual
treatment and improve the prediction of prognosis.
Long non-coding RNAs (lncRNAs), located in the
nucleus and cytoplasm of eukaryotic cells, are non-coding
transcripts >200 nucleotides in length (5). Studies suggest that lncRNAs serve
crucial roles in the occurrence and progression of malignant tumors
(6–8). For example, one study found that
lncRNA-KRTAP5-AS1 and lncRNA-TUBB2A acted as competing endogenous
RNAs to influence the function of claudin-4 and thereby affect the
prognosis of patients with gastric cancer (9). Specifically, in the case of hepatitis
B virus (HBV)-associated HCC, lncRNA HULC can activate HBV by
modulating STAT3-related signaling (10). Another study showed that
lncRNAlnc-EGFR stimulated the differentiation of T-regulatory
cells, thus promoting HCC immune evasion (11).
Hepatofibrosis is a type of liver tissue scar
reaction involved in chronic liver injury, which can progress to
cirrhosis and HCC. Numerous studies have suggested that
hepatofibrosis is an important risk factor in HCC (12–14).
The recurrence rates and OS of HCC patients are lower in the
presence of no or minimal fibrosis (15–17).
Therefore, the present study aimed to examine whether lncRNAs may
be useful in predicting the survival of HCC patients with or
without fibrosis. This possibility was tested using lncRNA
expression data from The Cancer Genome Atlas (TCGA).
Material and methods
Selection of patients with HCC
Expression profiles for lncRNAs and mRNAs, as well
as the corresponding clinical information for patients with HCC,
were downloaded from TCGA (version 09-14-2017 for HCC) via UCSC
Xena (https://xenabrowser.net/datapages/). Patients were
included in the present study if i) their HCC was confirmed
histologically, ii) complete RNA-Seq data for lncRNAs and mRNAs
were available, iii) data on presence or absence of fibrosis were
available and iv) survival outcomes were known. Based on the Ishak
fibrosis score (18), patients in
the TCGA were assigned as having no fibrosis, portal fibrosis,
fibrous septum, nodular formation and incomplete cirrhosis, or
established cirrhosis. In the present study, patients with ‘no
fibrosis’ were referred to as ‘without fibrosis’, while all others
were referred to as ‘with fibrosis’. Finally, 135 HCC patients with
fibrosis and 72 without fibrosis were included in the study
(Table I). This study complies with
TCGA publication guidelines (https://cancergenome.nih.gov/publications/publicationguidelines).
Since the data were obtained from TCGA, Guangxi Medical University
Cancer Hospital Ethics Committee waived the need for approval.
 | Table I.Clinicopathological characteristics
of 207 hepatocellular carcinoma patients with or without
fibrosis. |
Table I.
Clinicopathological characteristics
of 207 hepatocellular carcinoma patients with or without
fibrosis.
| Clinicopathological
characteristics | N (%) |
|---|
| Fibrosis |
|
| With
fibrosis | 135 (65.22) |
| Without
fibrosis | 72 (34.78) |
| Sex |
|
|
Female | 66 (31.88) |
|
Male | 141 (68.12) |
| Age (years) |
|
|
≤60 | 96 (46.38) |
|
>60 | 111 (53.62) |
| Ethnicity |
|
|
Nonasian | 121 (58.45) |
|
Asian | 80 (38.65) |
| Not
reported | 6 (2.90) |
| BMI |
|
|
<25 | 94 (45.41) |
|
≥25 | 103 (49.76) |
| Not
reported | 10 (4.83) |
| AFP (ng/ml) |
|
|
≤20 | 109 (52.66) |
|
>20 | 76 (36.71) |
| Not
reported | 22 (10.63) |
| Alcohol
consumption |
|
| No | 148 (71.50) |
|
Yes | 50 (24.15) |
| Not
reported | 9 (4.35) |
| Hepatitis B or
C |
|
| No | 94 (45.41) |
|
Yes | 104 (50.24) |
| Not
reported | 9 (4.35) |
| Histology
grade |
|
|
G1-2 | 133 (64.25) |
|
G3-4 | 72 (34.78) |
| Not
reported | 2 (0.97) |
| Pathologic
stage |
|
| Stage
I+II | 155 (74.88) |
| Stage
III+IV | 41 (19.81) |
| Not
reported | 11 (5.31) |
| New tumor
event |
|
| No | 97 (46.86) |
|
Yes | 101 (48.79) |
| Not
reported | 9 (4.35) |
| Cancer status |
|
| Tumor
free | 116 (56.04) |
| With
tumor | 84 (40.58) |
| Not
reported | 7 (3.38) |
| Residual tumor |
|
| R0 | 192 (92.75) |
|
Non-R0 | 13 (6.28) |
| Not
reported | 2 (0.97) |
| Vascular
invasion |
|
|
Negative | 138 (66.67) |
|
Positive | 60 (28.98) |
| Not
reported | 9 (4.35) |
| Family cancer
history |
|
| No | 111 (53.62) |
|
Yes | 68 (32.85) |
| Not
reported | 28 (13.53) |
Expression profile of lncRNAs in
HCC
First, lncRNAs with expression levels of 0 in
>50% of patients were removed, then the remaining lncRNAs were
analyzed using the edgeR algorithm within R software
(version 3.4.4; www.r-project.org) (19) in order to identify lncRNAs
differentially expressed (DElncRNAs) between HCC patients with or
without fibrosis. DElncRNAs were defined as those showing
|log2fold change (logFC)|>1 with a false discovery
rate (FDR)<0.05. Cluster heat maps and volcano maps were
generated using gplots and heatmap packages in R
software.
Construction of lncRNA
expression-based risk scoring systems and prognostic
assessment
A univariate Cox model was employed to identify the
relationships of DElncRNAs with OS or RFS. In this analysis,
lncRNAs in relationships associated with P<0.05 were regarded as
statistically significant. Multivariate Cox regression analysis was
subsequently used to assess the contribution of a lncRNA and to
select the best model via a backward stepwise method. A risk
scoring system was constructed based on a linear combination of the
lncRNA expression level and a multiplied regression coefficient
(β): Risk score = (β1 × expression level of lncRNA1) + (β2 ×
expression level of lncRNA2) + (β3 × expression level of lncRNA3) +
(β4 × expression level of …).
This formula was used to calculate the risk score of
each patient with HCC. Prognostic performance was assessed based on
the sensitivity and specificity of time-dependent receiver
operating characteristic (ROC) curves within 3 years. Based on the
cut-off of the median risk score, patients with HCC were divided
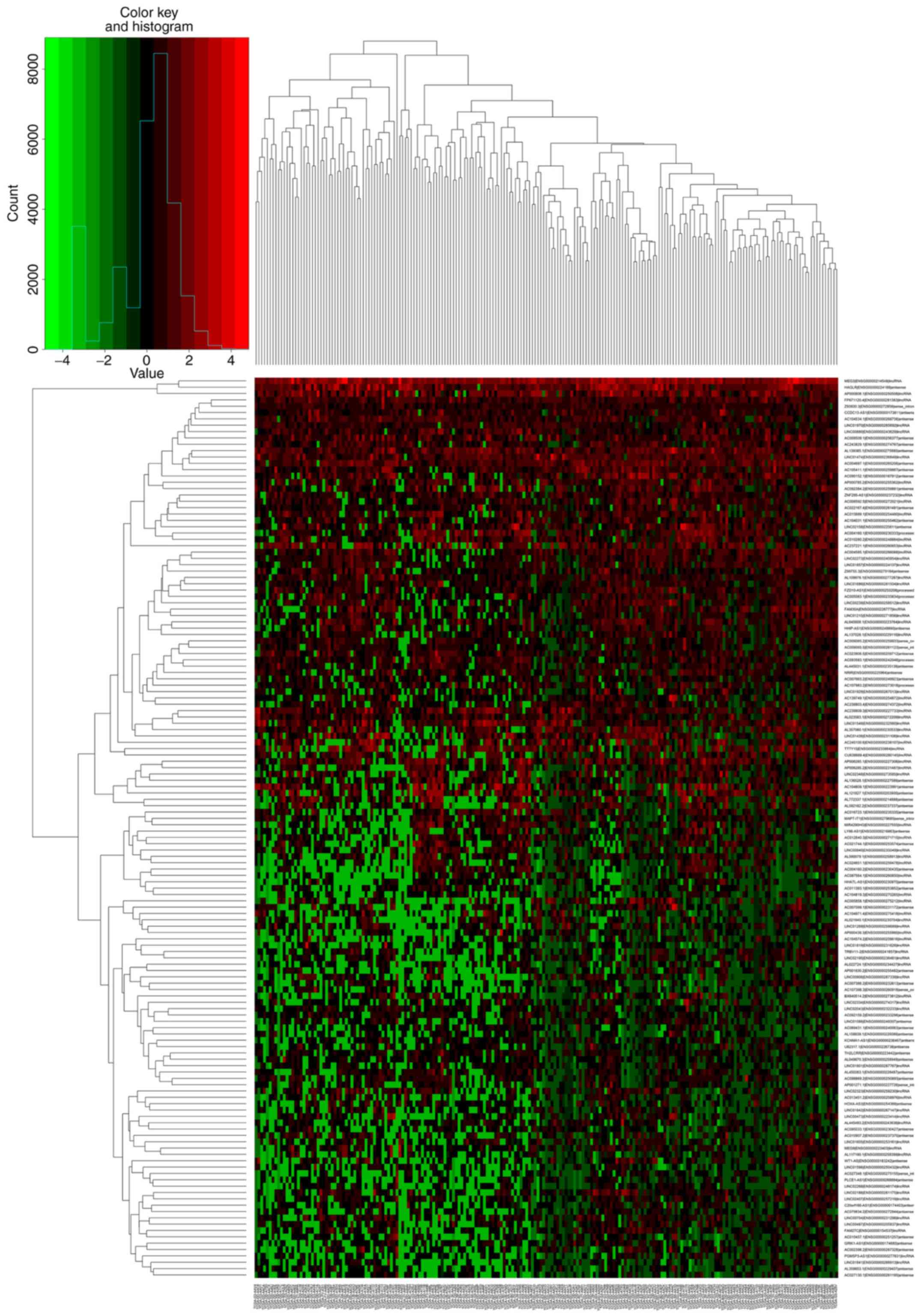
into high- or low-risk groups, as shown by a non-cluster heat map.
Kaplan-Meier survival curves predicted to be low or high risk were
created for patients with or without fibrosis. All analyses were
conducted using R/Bioconductor.
Prognostic significance of the risk
scoring system
To confirm the prognostic significance of the risk
scoring systems after adjusting for other clinical variables, uni-
and multivariate Cox regression analyses were performed. If the
results of these analyses were not significant, stratified analyses
were performed to identify potential impact factors using the
Chi-square test. All these analyses were carried out using SPSS
16.0 (SPSS, Inc.). All reported P-values were two-sided, and
P<0.05 was defined as significant. Hepatitis B and C were
considered together to avoid too many groups influencing the
results of the uni- and multivariate analysis.
Co-expression and functional analysis
of mRNAs related to lncRNAs in the risk scoring systems
To identify pairs of co-expressed lncRNA and mRNA,
Pearson correlation coefficients and the P-value of the z-test were
calculated based on the expression value between prognostic lncRNAs
in the risk scoring systems and mRNAs in the dataset of 207
patients with HCC. Protein-coding mRNAs showing a |Pearson
correlation coefficient|>0.30 and P<0.01 with a given lncRNA
were considered to be highly related to that lncRNA. These mRNAs
were subjected to Kyoto Encyclopedia of Genes and Genomes (KEGG)
pathway enrichment analysis using the clusterProfiler
package in R (20). P<0.05 was
considered to indicate a statistically significant difference in
the enrichment analyses.
Results
DElncRNAs associated with fibrosis in
HCC
The present study investigated the expression levels
of RNAs in 135 HCC patients with fibrosis and 72 HCC patients
without fibrosis. A total of 142 DElncRNAs were identified,
including 41 (28.87%) that were upregulated and 101 (71.13%) that
were downregulated. The first 20 up- and downregulated lncRNAs, and
their corresponding logFC, P-value and FDR values, sorted by
P-value, are shown in Table II.
The distribution of all DElncRNAs according to the two dimensions
of -log10 (FDR) and logFC is shown as a volcano map in
Fig. 1. The specificity of
DElncRNAs was evaluated using a heat map as shown in Fig. 2.
 | Table II.Differentially expressed lncRNAs in
hepatocellular carcinoma patients with or without fibrosis. |
Table II.
Differentially expressed lncRNAs in
hepatocellular carcinoma patients with or without fibrosis.
| A, Top 20
upregulated lncRNAs |
|---|
|
|---|
| lncRNA | logFC | P-value | FDR |
|---|
| AP000439.3 | 5.4751654 |
4.24×10−13 |
3.03×10−10 |
| LINC01819 | 5.0170809 |
1.22×10−12 |
7.60×10−10 |
| MEG9 | 2.5802684 |
2.10×10−8 |
4.78×10−6 |
| FAM30A | 1.9258543 |
3.12×10−7 |
5.24×10−5 |
| AC245100.6 | 2.6549937 |
3.52×10−7 |
5.58×10−5 |
| LINC02273 | 1.2146603 |
1.68×10−6 |
2.34×10−4 |
| AC104971.4 | 2.4240637 |
2.42×10−6 |
3.21×10−4 |
| HOXA-AS3 | 1.7517143 |
5.23×10−6 |
5.85×10−4 |
| TRBV11-2 | 1.5240872 |
9.87×10−6 |
9.70×10−4 |
| NRIR | 1.1734476 |
1.15×10−5 |
1.09×10−3 |
| AC004687.1 | 1.0076407 |
3.72×10−5 |
2.33×10−3 |
| LINC01857 | 1.0254128 |
3.98×10−5 |
2.44×10−3 |
| AC243829.1 | 1.0263797 |
4.79×10−5 |
2.79×10−3 |
| LINC00487 | 1.3755637 |
5.39×10−5 |
3.01×10−3 |
| LINC01215 | 1.0637294 |
1.15×10−4 |
5.57×10−3 |
| MEG3 | 1.4208994 |
1.21×10−4 |
5.71×10−3 |
| LINC01549 | 1.5173721 |
1.30×10−4 |
5.94×10−3 |
| AC004585.1 | 1.0120319 |
1.45×10−4 |
6.32×10−3 |
| LINC01269 | 1.595745 |
1.50×10−4 |
6.44×10−3 |
| Z99755.3 | 1.318727 |
1.85×10−4 |
7.37×10−3 |
|
| B, Top 20
downregulated lncRNAs |
|
| lncRNA | logFC | P-value | FDR |
|
| AC139749.1 | −4.63412 |
1.19×10−27 |
6.80×10−24 |
| AC069431.1 | −2.66384 |
7.24×10−19 |
2.07×10−15 |
| AC007099.1 | −4.01939 |
2.56×10−18 |
4.87×10−15 |
| AC022167.4 | −2.49441 |
5.22×10−17 |
7.45×10−14 |
| LINC02334 | −3.13188 |
8.87×10−15 |
1.01×10−11 |
| LINC01970 | −1.80509 |
1.39×10−13 |
1.33×10−10 |
| AL359853.1 | −2.81772 |
1.83×10−13 |
1.49×10−10 |
| AL445493.2 | −2.5131 |
1.33×10−12 |
7.60×10−10 |
| AL158839.1 | −2.44346 |
1.52×10−12 |
7.86×10−10 |
| AC010457.1 | −2.44059 |
9.53×10−12 |
4.53×10−9 |
| TH2LCRR | −1.77579 |
3.01×10−11 |
1.32×10−8 |
| CCDC13-AS1 | −1.75267 |
7.28×10−11 |
2.96×10−8 |
| PLCE1-AS1 | −2.39471 |
1.72×10−10 |
6.53×10−8 |
| LINC01686 | −1.62237 |
1.84×10−10 |
6.57×10−8 |
| HHATL-AS1 | −2.11296 |
9.54×10−10 |
3.20×10−7 |
| AC092159.2 | −1.52149 |
1.44×10−9 |
4.56×10−7 |
| AL645608.1 | −1.72757 |
4.16×10−9 |
1.25×10−6 |
| BX640514.2 | −2.39457 |
4.87×10−9 |
1.39×10−6 |
| AC021744.1 | −2.20589 |
9.84×10−9 |
2.67×10−6 |
| AC104031.1 | −1.82197 |
1.12×10−8 |
2.91×10−6 |
Using DElncRNAs to predict the OS of
HCC patients with fibrosis
Univariate Cox analysis was conducted to explore the
associations between DElncRNAs and OS in HCC patients with
fibrosis. Seven lncRNAs exhibited a significant association with
OS: AC012640.3, CU638689.4, AL772337.1, AL359853.1, HOXA-AS3,
AL022724.1 and Z93930.3. Multivariate Cox regression confirmed five
of these lncRNAs (AL359853.1, Z93930.3, HOXA-AS3, AL772337.1 and
AC012640.3) to be independent predictors of OS (Table III). The final risk scoring system
was as follows:
 | Table III.Five lncRNAs associated with the
overall survival of hepatocellular carcinoma patients with fibrosis
in the best statistical model. |
Table III.
Five lncRNAs associated with the
overall survival of hepatocellular carcinoma patients with fibrosis
in the best statistical model.
| lncRNA | β | HR | z | P-value |
|---|
| AL359853.1 | 0.2751 | 1.3167 | 1.66 | 0.09761 |
| Z93930.3 | 0.5679 | 1.7646 | 2.61 | 0.00913 |
| HOXA-AS3 | 0.1861 | 1.2045 | 1.92 | 0.05485 |
| AL772337.1 | 0.2186 | 1.2444 | 2.81 | 0.00499 |
| AC012640.3 | 0.4639 | 1.5902 | 3.65 | 0.00026 |
Risk score = (0.2751 × AL359853.1) + (0.5679 ×
Z93930.3) + (0.1861 × HOXA-AS3) + (0.2186 × AL772337.1) + (0.4639 ×
AC012640.3).
In this prognostic formula, higher expression levels
of the five lncRNAs were associated with higher risk of death
(β>0).
Based on the risk scores for OS, HCC patients with
fibrosis were divided into high- or low-risk groups using the
median score as a cut-off (Fig.
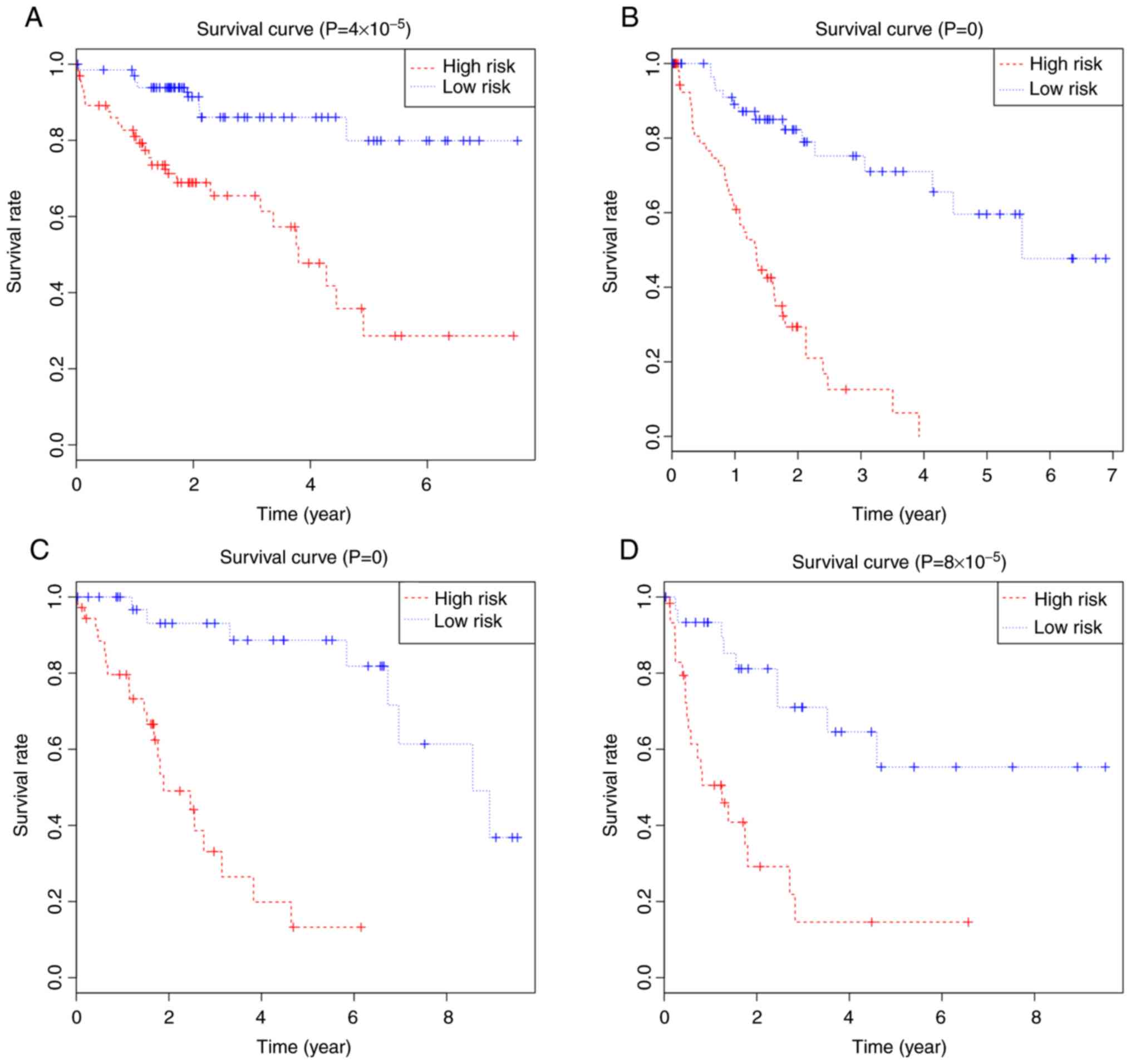
3A), and Kaplan-Meier curves were calculated for the two groups
Fig. 4A. Patients with a high-risk
score showed poorer OS than patients with a low-risk score at 3
years (65.4 vs. 86.1%) and 5 years (28.6 vs. 79.9%). The area under
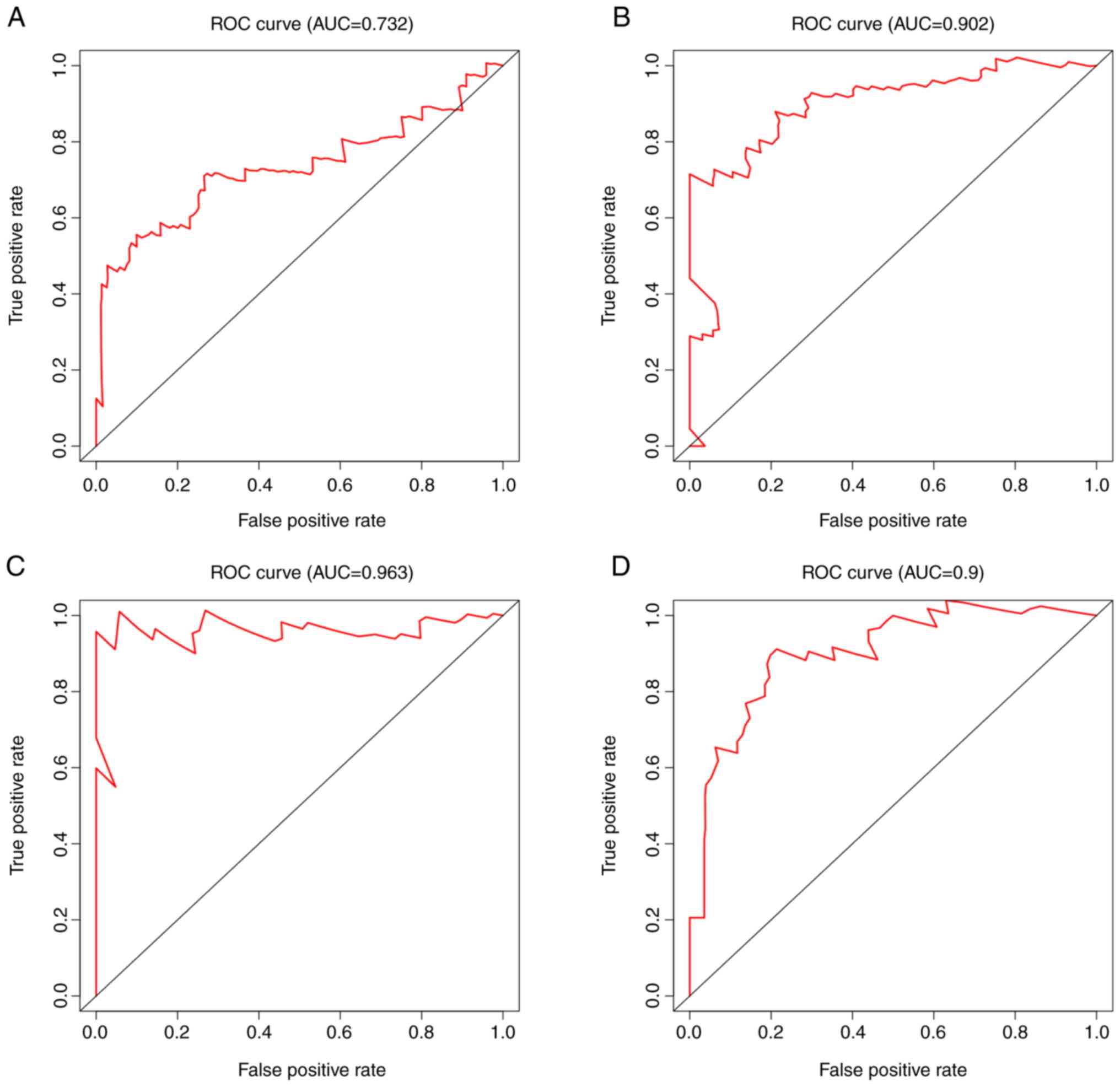
the ROC curve for the risk scores was 0.732 (Fig. 5A).
Using DElncRNAs to predict the RFS of
HCC patients with fibrosis
Uni- and multivariate Cox regression analyses were
also performed between the DElncRNAs and RFS of HCC patients with
fibrosis. Univariate analysis identified 27 lncRNAs that were
significantly associated with RFS: LINC01857, AC090152.1,
LINC02407, LINC01970, TRBV11-2, CU638689.4, C20orf166-AS1,
AC027348.1, Z93930.3, MEG3, LINC02273, HHIP-AS1, AC004585.1,
FZD10-AS1, LINC01215, LINC00239, AP000808.1, PLCE1-AS1, Z99755.3,
AL357060.1, AC005083.1, MEG9, LINC00473, AC004687.1, AL359853.1,
LINC02195 and LINC01842. Multivariate analysis confirmed the
following 12 as independent prognostic indicators of RFS:
PLCE1-AS1, Z93930.3, LINC02273, TRBV11-2, HHIP-AS1, AC004687.1,
LINC01857, AC004585.1, AP000808.1, CU638689.4, AC090152.1 and
AL357060.1 (Table IV). The risk
scoring system was as follows:
 | Table IV.Twelve lncRNAs associated with the
recurrence-free survival of hepatocellular carcinoma patients with
fibrosis in the best statistical model. |
Table IV.
Twelve lncRNAs associated with the
recurrence-free survival of hepatocellular carcinoma patients with
fibrosis in the best statistical model.
| lncRNA | β | HR | z | P-value |
|---|
| PLCE1-AS1 | −0.4792 | 0.6193 | −2.76 | 0.00587 |
| Z93930.3 | 0.4315 | 1.5396 | 2.53 | 0.01131 |
| LINC02273 | 0.4505 | 1.5690 | 2.19 | 0.02819 |
| TRBV11-2 | −0.2680 | 0.7649 | −1.94 | 0.05275 |
| HHIP-AS1 | −0.1816 | 0.8339 | −1.49 | 0.13637 |
| AC004687.1 | −0.2211 | 0.8016 | −1.66 | 0.09697 |
| LINC01857 | −0.3274 | 0.7208 | −2.08 | 0.03709 |
| AC004585.1 | 0.2398 | 1.2709 | 1.62 | 0.10467 |
| AP000808.1 | −0.1150 | 0.8914 | −1.97 | 0.04861 |
| CU638689.4 | −0.2929 | 0.7461 | −3.40 | 0.00066 |
| AC090152.1 | −0.2303 | 0.7943 | −2.48 | 0.01317 |
| AL357060.1 | −0.1530 | 0.8581 | −2.01 | 0.04431 |
Risk score = (−0.4792 × PLCE1-AS1) + (0.4315 ×
Z93930.3) + (0.4505 × LINC02273) + (−0.2680 × TRBV11-2) + (−0.1816
× HHIP-AS1) + (−0.2211 × AC004687.1) + (−0.3274 × LINC01857) +
(0.2398 × AC004585.1) + (−0.1150 × AP000808.1) + (−0.2929 ×
CU638689.4) + (−0.2303 × AC090152.1) + (−0.1530 × AL357060.1).
In this formula, higher expression of Z93930.3,
LINC02273 and AC004585.1 was associated with higher risk of
recurrence (β>0), while higher expression of the other lncRNAs
was associated with improved RFS (β<0).
HCC patients with fibrosis were stratified into
high- or low-risk groups using the median risk score as a cut-off
(Fig. 3B), and Kaplan-Meier curves
were calculated for both groups (Fig.
4B). Patients with a high-risk score had poorer RFS than
patients with a low-risk score at 3 years (12.59 vs. 75.20%) and 5
years (0.00 vs. 59.60%). The area under the ROC curve was 0.902
(Fig. 5B).
Using DElncRNAs to predict the OS of
HCC patients without fibrosis
Univariate analysis showed 27 lncRNAs to be
significantly associated with OS in HCC patients without fibrosis:
FAM27C, AC007099.1, AC005858.1, LINC00239, AC010280.2, LINC02323,
AC011383.1, NRIR, AC104971.4, AC004160.1, AL139385.1, AP001271.1,
AC237221.1, AC079834.2, AC093583.1, AL049870.3, AP006285.2,
AC098869.2, AC004160.2, AL445931.1, AC239803.4, AC009065.2,
LINC01269, AP006285.1, HAGLR, HOXA-AS3 and LINC01929. Multivariate
analysis confirmed seven of these as independent predictors of OS:
LINC00239, AC104971.4, AP006285.2, HOXA-AS3, AC079834.2, NRIR and
LINC01929 (Table V). The risk
scoring system was as follows:
 | Table V.Seven lncRNAs associated with the
overall survival of hepatocellular carcinoma patients without
fibrosis in the best statistical model. |
Table V.
Seven lncRNAs associated with the
overall survival of hepatocellular carcinoma patients without
fibrosis in the best statistical model.
| lncRNA | β | HR | z | P-value |
|---|
| LINC00239 | 0.280 | 1.324 | 2.62 | 0.0088 |
| AC104971.4 | 0.774 | 2.169 | 4.09 |
4.2×10−5 |
| AP006285.2 | −0.266 | 0.766 | −2.06 | 0.0395 |
| HOXA-AS3 | 0.362 | 1.437 | 1.86 | 0.0633 |
| AC079834.2 | −0.586 | 0.557 | −2.64 | 0.0083 |
| NRIR | −0.514 | 0.598 | −2.95 | 0.0032 |
| LINC01929 | −0.358 | 0.699 | −1.89 | 0.0585 |
Risk score = (0.280 × LINC00239) + (0.774 ×
AC104971.4) + (−0.266 × AP006285.2) + (0.362 × HOXA-AS3) + (−0.586
× AC079834.2) + (−0.514 × NRIR) + (−0.358 × LINC01929).
In the formula, lower expression of LINC00239,
AC104971.4 and HOXA-AS3 was associated with worse OS (β>0),
while higher expression of the remaining lncRNAs was associated
with improved OS (β<0).
Patients were stratified into low- or high-risk
groups using the median risk score groups (Fig. 6A), and Kaplan-Meier curves were
calculated (Fig. 4C). The high-risk
group had a poorer OS than the low-risk group at 3 years (33.10 vs.
93.10%) and 5 years (13.20 vs. 88.70%). The area under the ROC
curves was 0.963 (Fig. 5C).
Using DElncRNAs to predict the RFS of
HCC patients without fibrosis
Univariate analysis showed six lncRNAs (NRIR,
AC005858.1, LINC00487, AC107398.3, AC021744.1 and BX640514.2) to
have a significant association with the RFS of HCC patients without
fibrosis. Multivariate analysis found five lncRNAs to be
independent prognostic indicators of RFS: AC021744.1, NRIR,
LINC00487, AC005858.1 and AC107398.3 (Table VI). The risk scoring system was as
follows:
 | Table VI.Five lncRNAs associated with the
recurrence-free survival of hepatocellular carcinoma patients
without fibrosis in the best statistical model. |
Table VI.
Five lncRNAs associated with the
recurrence-free survival of hepatocellular carcinoma patients
without fibrosis in the best statistical model.
| lncRNA | β | HR | z | P-value |
|---|
| AC021744.1 | 0.1430 | 1.1537 | 1.95 | 0.0514 |
| NRIR | −0.4181 | 0.6583 | −2.14 | 0.0324 |
| LINC00487 | −0.5324 | 0.5872 | −2.10 | 0.0361 |
| AC005858.1 | 0.2145 | 1.2392 | 2.71 | 0.0068 |
| AC107398.3 | 0.2217 | 1.2482 | 2.03 | 0.0421 |
Risk score = (0.1430 × AC021744.1) + (−0.4181 ×
NRIR) + (−0.5324 × LINC00487) + (0.2145 × AC005858.1) + (0.2217 ×
AC107398.3).
Higher expression of AC021744.1, AC005858.1 and
AC107398.3 was associated with higher risk of recurrence (β>0),
while higher expression of the other lncRNAs was associated with
improved RFS (β<0).
Patients were stratified into high- or low-risk
groups using the median risk score as cut-off (Fig. 6B), and Kaplan-Meier curves showed
that high-risk patients had poorer RFS than low-risk patients at 3
years (14.6 vs. 71.0%) and 5 years (14.6 vs. 55.3%; Fig. 4D). The area under the ROC curves was
0.90 (Fig. 5D).
Prognostic significance of the risk
scoring system after adjustment for other clinical
characteristics
Each of the four risk scoring systems was validated
after adjusting for other clinical characteristics that can
influence survival. First, univariate Cox regression analysis was
conducted between clinical features and OS for HCC patients with
fibrosis. The risk scoring system, age, body mass index (BMI) and
ethnicity were significantly associated with OS. The following
characteristics were not associated with OS: α-fetoprotein (AFP),
sex, hepatitis, alcohol consumption, histology grade, new tumor
event, pathologic stage, cancer status, family cancer history,
residual tumor and vascular invasion. Subsequently, multivariate
Cox regression was performed using the covariates that were
significant in the univariate analysis. The hazard ratio (HR) of
the risk scoring system was 3.92 [95% confidence interval (CI)
1.32–11.66] in the univariate Cox regression, and 2.65 (95% CI
1.12–6.26) in the multivariate Cox regression after adjusting for
the other clinical covariates. These results confirm that the risk
scoring system is a significant independent predictor of OS for HCC
patients with fibrosis. Multivariate Cox regression identified
another two independent predictors: BMI (HR 0.38, 95% CI 0.16–0.88)
and ethnicity (HR 0.20, 95% CI 0.08–0.51) (Table VII).
 | Table VII.Uni- and multivariate Cox regression
analysis of factors affecting overall survival in hepatocellular
carcinoma patients with fibrosis. |
Table VII.
Uni- and multivariate Cox regression
analysis of factors affecting overall survival in hepatocellular
carcinoma patients with fibrosis.
|
| Univariate Cox
regression | Multivariate Cox
regression |
|---|
|
|
|
|
|---|
| Variables | P-value | HR | 95% CI | P-value | HR | 95% CI |
|---|
| Risk score
(high/low) | 0.01 | 3.92 | 1.32 | 11.66 | 0.03 | 2.65 | 1.12 | 6.26 |
| Age (>60/≤60
years) | 0.01 | 3.41 | 1.31 | 8.89 |
|
|
|
|
| BMI | 0.01 |
|
|
|
|
|
|
|
|
<25 |
|
| Ref. |
|
|
| Ref. |
|
|
≥25 |
| 0.26 | 0.07 | 0.89 | 0.02 | 0.38 | 0.16 | 0.88 |
| Not
reported |
| 1.71 | 0.25 | 11.75 | 0.51 | 1.41 | 0.51 | 3.88 |
| Ethnicity | 0.01 |
|
|
|
|
|
|
|
|
Non-Asian |
|
| Ref. |
|
|
| Ref. |
|
|
Asian |
| 0.16 | 0.05 | 0.51 | <0.01 | 0.20 | 0.08 | 0.51 |
| AFP (ng/ml) | 0.64 |
|
|
|
|
|
|
|
|
≤20 |
|
| Ref. |
|
|
|
|
|
|
>20 |
| 0.99 | 0.33 | 2.99 |
|
|
|
|
| Not
reported |
| 0.32 | 0.03 | 3.69 |
|
|
|
|
| Sex
(male/female) | 0.87 | 0.90 | 0.26 | 3.11 |
|
|
|
|
| Hepatitis B or
C | 0.44 |
|
|
|
|
|
|
|
| No |
|
| Ref. |
|
|
|
|
|
|
Yes |
| 1.90 | 0.58 | 6.24 |
|
|
|
|
| Not
reported |
| 0.30 | 0.01 | 11.80 |
|
|
|
|
| Alcohol consumption
(yes/no) | 0.45 | 0.61 | 0.17 | 2.20 |
|
|
|
|
| Histology
grade | 0.26 |
|
|
|
|
|
|
|
|
G1-2 |
|
| Ref. |
|
|
|
|
|
|
G3-4 |
| 2.11 | 0.86 | 5.17 |
|
|
|
|
| New tumor
event | 0.46 |
|
|
|
|
|
|
|
| No |
|
| Ref. |
|
|
|
|
|
|
Yes |
| 0.76 | 0.18 | 3.15 |
|
|
|
|
| Not
reported |
| 3.88 | 0.20 | 73.85 |
|
|
|
|
| Pathologic
stage | 0.37 |
|
|
|
|
|
|
|
| Stage
I+II |
|
| Ref. |
|
|
|
|
|
| Stage
III+IV |
| 0.39 | 0.11 | 1.46 |
|
|
|
|
| Not
reported |
| 0.84 | 0.08 | 8.49 |
|
|
|
|
| Cancer status | 0.14 |
|
|
|
|
|
|
|
| Tumor
free |
|
| Ref. |
|
|
|
|
|
| With
tumor |
| 3.04 | 0.84 | 10.98 |
|
|
|
|
| Not
reported |
| 7.77 | 0.26 | 236.32 |
|
|
|
|
| Family cancer
history | 0.45 |
|
|
|
|
|
|
|
| No |
|
| Ref. |
|
|
|
|
|
|
Yes |
| 2.06 | 0.64 | 6.56 |
|
|
|
|
| Not
reported |
| 1.05 | 0.18 | 5.96 |
|
|
|
|
| Residual tumor | 0.67 |
|
|
|
|
|
|
|
| R0 |
|
| Ref. |
|
|
|
|
|
|
Non-R0 |
| 2.39 | 0.35 | 16.25 |
|
|
|
|
| Vascular
invasion | 0.16 |
|
|
|
|
|
|
|
|
Negative |
|
| Ref. |
|
|
|
|
|
|
Positive |
| 2.48 | 0.92 | 6.68 |
|
|
|
|
| Not
reported |
| 2.39 | 0.53 | 10.73 |
|
|
|
|
Second, univariate Cox regression analysis was
conducted between the clinical features and RFS of patients with
HCC and fibrosis. None of the clinical covariates, with the
exception of cancer status, were associated with RFS (Table VIII). Stratified analyses were
conducted to identify factors affecting the risk scoring system.
These factors included age, ethnicity, alcohol consumption, new
tumor event, pathology stage and cancer status (P<0.05; Table IX).
 | Table VIII.Univariate Cox regression analysis of
factors affecting recurrence-free survival in hepatocellular
carcinoma patients with fibrosis. |
Table VIII.
Univariate Cox regression analysis of
factors affecting recurrence-free survival in hepatocellular
carcinoma patients with fibrosis.
|
| Univariate Cox
regression |
|---|
|
|
|
|---|
| Variables | P-value | HR | 95% CI |
|---|
| Risk score
(high/low) | 0.15 | 2.25 | 0.75 | 6.81 |
| Age (>60/≤60
years) | 0.17 | 0.58 | 0.26 | 1.27 |
| BMI | 0.08 |
|
|
|
|
<25 |
|
|
|
|
|
≥25 |
| 0.30 | 0.10 | 0.87 |
| Not
reported |
| 0.17 | 0.01 | 2.66 |
| Ethnicity | 0.87 |
|
|
|
|
Non-Asian |
|
|
|
|
|
Asian |
| 0.76 | 0.27 | 2.15 |
| AFP (ng/ml) | 0.38 |
|
|
|
|
≤20 |
|
|
|
|
|
>20 |
| 1.52 | 0.65 | 3.55 |
| Not
reported |
| 2.94 | 0.52 | 16.58 |
| Sex
(male/female) | 0.33 | 0.56 | 0.17 | 1.80 |
| Hepatitis B or
C | 0.37 |
|
|
|
| No |
|
|
|
|
|
Yes |
| 0.78 | 0.20 | 3.07 |
| Not
reported |
| 0.28 | 0.04 | 2.10 |
| Alcohol consumption
(yes/no) | 0.36 | 1.84 | 0.51 | 6.65 |
| Histology
grade | 0.30 |
|
|
|
|
G1-2 |
|
|
|
|
|
G3-4 |
| 0.75 | 0.30 | 1.86 |
| Not
reported |
| 12.31 | 0.41 | 371.37 |
| New tumor event
(yes/no) | 0.84 |
3.51×105 | 0.00 |
2.15×1059 |
| Pathologic
stage | 0.55 |
|
|
|
| Stage
I+II |
|
|
|
|
| Stage
III+IV |
| 1.63 | 0.50 | 5.30 |
| Not
reported |
| 0.50 | 0.04 | 6.68 |
| Cancer status | <0.05 |
|
|
|
| Tumor
free |
|
|
|
|
| With
tumor |
| 3.82 | 1.32 | 11.07 |
| Not
reported |
| 4.60 | 0.00 | . |
| Family cancer
history | 0.36 |
|
|
|
| No |
|
|
|
|
|
Yes |
| 0.56 | 0.19 | 1.66 |
| Not
reported |
| 0.41 | 0.12 | 1.50 |
| Residual tumor | 0.85 |
|
|
|
| R0 |
|
|
|
|
|
Non-R0 |
| 1.70 | 0.26 | 11.08 |
| Not
reported |
| 1.39 | 0.08 | 23.58 |
| Vascular
invasion | 0.26 |
|
|
|
|
Negative |
|
|
|
|
|
Positive |
| 2.08 | 0.75 | 5.77 |
| Not
reported |
| 0.94 | 0.14 | 6.18 |
 | Table IX.Stratified analyses to explore
factors influencing the relationship between the risk scoring
system and recurrence-free survival in hepatocellular carcinoma
patients with fibrosis. |
Table IX.
Stratified analyses to explore
factors influencing the relationship between the risk scoring
system and recurrence-free survival in hepatocellular carcinoma
patients with fibrosis.
|
| Risk score |
|
|---|
|
|
|
|
|---|
| Variables | Low-risk (n) | High-risk (n) | P-value |
|---|
| Age (years) |
|
| <0.01 |
|
≤60 | 40 | 23 |
|
|
>60 | 20 | 36 |
|
| BMI |
|
| 0.19 |
|
<25 | 32 | 23 |
|
|
≥25 | 27 | 32 |
|
| Ethnicity |
|
| 0.01 |
|
Non-Asian | 20 | 34 |
|
|
Asian | 38 | 25 |
|
| AFP (ng/ml) |
|
| 0.33 |
|
≤20 | 31 | 36 |
|
|
>20 | 24 | 19 |
|
| Sex |
|
| 0.54 |
|
Female | 15 | 12 |
|
|
Male | 45 | 47 |
|
| Hepatitis B or
C |
|
| 0.08 |
| No | 12 | 20 |
|
|
Yes | 47 | 37 |
|
| Alcohol
consumption |
|
| 0.03 |
| No | 47 | 35 |
|
|
Yes | 12 | 22 |
|
| Histology
grade |
|
| 0.20 |
|
G1-2 | 42 | 34 |
|
|
G3-4 | 18 | 24 |
|
| New tumor
event |
|
| <0.01 |
| No | 45 | 19 |
|
|
Yes | 15 | 40 |
|
| Pathologic
stage |
|
| 0.03 |
| Stage
I+II | 52 | 42 |
|
| Stage
III+IV | 5 | 13 |
|
| Cancer status |
|
| <0.01 |
| Tumor
free | 48 | 23 |
|
| With
tumor | 11 | 36 |
|
| Family cancer
history |
|
| 0.78 |
| No | 37 | 35 |
|
|
Yes | 15 | 16 |
|
| Residual tumor |
|
| 0.13 |
| R0 | 58 | 52 |
|
|
Non-R0 | 2 | 6 |
|
| Vascular
invasion |
|
| 0.65 |
|
Negative | 41 | 36 |
|
|
Positive | 17 | 18 |
|
Third, univariate Cox regression analysis was
conducted between the clinical features and OS of HCC patients
without fibrosis. The risk scoring system, BMI, new tumor event and
pathology stage were significantly associated with the OS of HCC
patients without fibrosis. Multivariate analysis of these
covariates identified the risk scoring system (HR 23.15, 95% CI
5.65–94.91) and pathology stage (HR 3.82, 95% CI 1.42–10.25) as
significant independent predictors of OS (Table X).
 | Table X.Uni- and multivariate Cox regression
analysis of factors affecting overall survival in hepatocellular
carcinoma patients without fibrosis. |
Table X.
Uni- and multivariate Cox regression
analysis of factors affecting overall survival in hepatocellular
carcinoma patients without fibrosis.
|
| Univariate Cox
regression | Multivariate Cox
regression |
|---|
|
|
|
|
|---|
| Variables | P-value | HR | 95.0% CI | P-value | HR | 95.0% CI |
|---|
| Risk score
(high/low) | <0.01 | 56.17 | 8.42 | 374.46 | <0.01 | 23.15 | 5.65 | 94.91 |
| Age (>60/≤60
years) | 0.37 | 0.45 | 0.08 | 2.62 |
|
|
|
|
| BMI | 0.01 |
|
|
|
|
|
|
|
|
<25 |
|
| Ref. |
|
|
| Ref. |
|
|
≥25 |
| 2.33 | 0.42 | 12.98 | 0.99 | 1.00 | 0.41 | 2.44 |
| Not
reported |
| 46.08 | 3.34 | 635.97 | 0.03 | 8.16 | 1.29 | 51.68 |
| Ethnicity | 0.10 |
|
|
|
|
|
|
|
|
Non-Asian |
|
| Ref. |
|
|
|
|
|
|
Asian |
| 49.26 | 0.68 | 3,591.00 |
|
|
|
|
| Not
reported |
| 0.20 | 0.00 | 16.50 |
|
|
|
|
| AFP (ng/ml) | 0.51 |
|
|
|
|
|
|
|
|
≤20 |
|
| Ref. |
|
|
|
|
|
|
>20 |
| 0.88 | 0.15 | 5.21 |
|
|
|
|
| Not
reported |
| 1.92 | 0.47 | 7.87 |
|
|
|
|
| Sex
(male/female) | 0.37 | 0.48 | 0.10 | 2.40 |
|
|
|
|
| Hepatitis B or
C | 0.66 |
|
|
|
|
|
|
|
| No |
|
| Ref. |
|
|
|
|
|
|
Yes |
| 3.81 | 0.21 | 68.88 |
|
|
|
|
| Not
reported |
| 0.01 | 0.00 |
4.95×1080 |
|
|
|
|
| Alcohol consumption
(yes/no) | 0.06 | 6.99 | 0.90 | 54.44 |
|
|
|
|
| Histology
grade | 0.12 |
|
|
|
|
|
|
|
|
G1-2 |
|
| Ref. |
|
|
|
|
|
|
G3-4 |
| 2.29 | 0.50 | 10.51 |
|
|
|
|
| Not
reported |
| 0.00 | 0.00 | 1.44 |
|
|
|
|
| New tumor
event | <0.01 |
|
|
|
|
|
|
|
| No |
|
| Ref. |
|
|
| Ref. |
|
|
Yes |
| 0 | 0 |
5.76×1083 | 0.90 | 1.07 | 0.38 | 3.05 |
| Not
reported |
| 1,014.00 | 18.59 | 55,300.00 | <0.01 | 8.53 | 2.46 | 29.63 |
| Pathologic
stage | <0.01 |
|
|
|
|
|
|
|
| Stage
I+II |
|
| Ref. |
|
|
| Ref. |
|
| Stage
III+IV |
| 2.19 | 0.51 | 9.47 | 0.01 | 3.82 | 1.42 | 10.25 |
| Not
reported |
| 1,093.00 | 17.61 | 67,880.00 | 0.15 | 3.37 | 0.65 | 17.56 |
| Cancer status | 0.13 |
|
|
|
|
|
|
|
| Tumor
free |
|
| Ref. |
|
|
|
|
|
| With
tumor |
| 131,600.00 | 0.00 |
3.82×1092 |
|
|
|
|
| Not
reported |
| 0.00 | 0.00 | 0.80 |
|
|
|
|
| Family cancer
history | 0.30 |
|
|
|
|
|
|
|
| No |
|
| Ref. |
|
|
|
|
|
|
Yes |
| 4.90 | 0.67 | 36.05 |
|
|
|
|
| Not
reported |
| 2.08 | 0.14 | 31.22 |
|
|
|
|
| Residual tumor | 0.84 |
|
|
|
|
|
|
|
| R0 |
|
| Ref. |
|
|
|
|
|
|
Non-R0 |
| 0.13 | 0.00 | 113.04 |
|
|
|
|
| Vascular
invasion | 0.08 |
|
|
|
|
|
|
|
|
Negative |
|
| Ref. |
|
|
|
|
|
|
Positive |
| 6.24 | 0.80 | 48.34 |
|
|
|
|
Finally, univariate Cox regression analysis was
conducted between the clinical features and RFS of HCC patients
without fibrosis. The risk scoring system, age, BMI and pathology
stage exhibited a significant association with RFS. The remaining
factors did not: Ethnicity, AFP, sex, hepatitis, alcohol
consumption, histology grade, new tumor event, cancer status,
family cancer history, residual tumor and vascular invasion.
Multivariate analysis identified the risk scoring system (HR 6.42,
95% CI 2.62–15.70) and age (HR 0.36, 95% CI 0.16–0.80) as
significant independent predictors of RFS (Table XI).
 | Table XI.Uni- and multivariate Cox regression
analysis of factors affecting recurrence-free survival in
hepatocellular carcinoma patients without fibrosis. |
Table XI.
Uni- and multivariate Cox regression
analysis of factors affecting recurrence-free survival in
hepatocellular carcinoma patients without fibrosis.
|
| Univariate Cox
regression | Multivariate Cox
regression |
|---|
|
|
|
|
|---|
| Variables | P-value | HR | 95% CI | P-value | HR | 95% CI |
|---|
| Risk score
(high/low) | 0.01 | 11.52 | 1.70 | 78.15 | <0.01 | 6.42 | 2.62 | 15.70 |
| Age (>60/≤60
years) | 0.01 | 0.06 | 0.01 | 0.56 | 0.01 | 0.36 | 0.16 |
0.80 |
| BMI | 0.01 |
|
|
|
|
|
|
|
|
<25 |
|
| Ref. |
|
|
|
|
|
|
≥25 |
| 1.53 | 0.31 | 7.48 |
|
|
|
|
| Not
reported |
| 221.65 | 6.47 | 7,597.00 |
|
|
|
|
| Ethnicity | 0.69 |
|
|
|
|
|
|
|
|
Non-Asian |
|
| Ref. |
|
|
|
|
|
|
Asian |
| 1.54 | 0.00 |
7.83×1051 |
|
|
|
|
| Not
reported |
| 6.48 | 0.09 | 471.12 |
|
|
|
|
| AFP (ng/ml) | 0.34 |
|
|
|
|
|
|
|
|
≤20 |
|
| Ref. |
|
|
|
|
|
|
>20 |
| 2.41 | 0.37 | 15.84 |
|
|
|
|
| Not
reported |
| 12.05 | 0.35 | 415.85 |
|
|
|
|
| Sex
(male/female) | 0.12 | 0.11 | 0.01 | 1.70 |
|
|
|
|
| Hepatitis B or
C | 0.88 |
|
|
|
|
|
|
|
| No |
|
| Ref. |
|
|
|
|
|
|
Yes |
| 0.49 | 0.02 | 11.15 |
|
|
|
|
| Not
reported |
| 0.47 | 0.01 | 27.36 |
|
|
|
|
| Alcohol consumption
(yes/no) | 0.66 | 1.86 | 0.12 | 28.92 |
|
|
|
|
| Histology
grade | 0.54 |
|
|
|
|
|
|
|
|
G1-2 |
|
| Ref. |
|
|
|
|
|
|
G3-4 |
| 2.65 | 0.47 | 14.89 |
|
|
|
|
| Not
reported |
| 0.45 | 0.00 |
2.64×10171 |
|
|
|
|
| New tumor event
(yes/no) | 0.55 | 242,100.00 | 0.00 |
6.504×1022 |
|
|
|
|
| Pathologic
stage | 0.01 |
|
|
|
|
|
|
|
| Stage
I+II |
|
| Ref. |
|
|
|
|
|
| Stage
III+IV |
| 23.11 | 3.14 | 169.93 |
|
|
|
|
| Not
reported |
| 2.47 | 0.00 |
1.40×10172 |
|
|
|
|
| Cancer status | 0.91 |
|
|
|
|
|
|
|
| Tumor
free |
|
| Ref. |
|
|
|
|
|
| With
tumor |
| 1.84 | 0.10 | 35.34 |
|
|
|
|
| Not
reported |
| 1.84 | 0.03 | 125.06 |
|
|
|
|
| Family cancer
history | 0.33 |
|
|
|
|
|
|
|
| No |
|
| Ref. |
|
|
|
|
|
|
Yes |
| 1.07 | 0.12 | 9.75 |
|
|
|
|
| Not
reported |
| 0.01 | 0.00 | 4.99 |
|
|
|
|
| Residual tumor | 0.14 |
|
|
|
|
|
|
|
| R0 |
|
| Ref. |
|
|
|
|
|
|
Non-R0 |
| 255.36 | 1.10 | 59,270.00 |
|
|
|
|
| Vascular
invasion | 0.82 |
|
|
|
|
|
|
|
|
Negative |
|
| Ref. |
|
|
|
|
|
|
Positive |
| 1.80 | 0.01 | 273.16 |
|
|
|
|
Co-expression analysis of DElncRNAs
and mRNAs, and functional analysis of the mRNAs
Potential co-expression of the lncRNAs in the risk
scoring systems and mRNAs in RNA-seq data was explored using
Pearson's correlation (Tables
SI–SIV). The mRNAs found to be
strongly associated with these lncRNAs were then analyzed using
KEGG signal pathway databases. The top 10 significantly enriched
KEGG signal pathways are shown in Fig.
7. Functional enrichment analysis showed that mRNAs strongly
associated with the risk scoring systems are involved mainly in
cell cycle-related pathways (in the case of OS of patients with HCC
and fibrosis), chemokine-related pathways (RFS of patients with HCC
and fibrosis), Th17 cell differentiation-related pathways (OS of
HCC patients without fibrosis) and thermogenesis-related pathways
(RFS of HCC patients without fibrosis).
Discussion
HCC has a high morbidity and mortality (1,2), the
risk of which differs depending on whether fibrosis is present or
not (15–17). Thus, prognostic biomarkers specific
for each situation are required in order to improve patient
management. Toward this end, the present study explored lncRNAs in
patients with HCC that are differentially expressed in the presence
or absence of fibrosis, and then identified which of the DElncRNAs
used to construct risk scoring systems may be useful for predicting
survival. The risk scoring systems were validated using uni- and
multivariate Cox analyses following adjustment for several clinical
characteristics that can also influence survival.
It was possible to predict the risk of OS for HCC
patients with or without fibrosis using 5 or 7 lncRNAs. The areas
under the ROC curves were 0.732 or 0.963, respectively, suggesting
reasonable predictive power. Furthermore, multivariate Cox analysis
identified additional significant predictors of OS: BMI and
ethnicity among patients with fibrosis, or pathology stage among
patients without fibrosis. Other studies have also associated these
factors with OS in patients with HCC (21,22).
The present study predicted the RFS of HCC patients
without fibrosis using 5 lncRNAs. The area under the ROC curve was
0.90, suggesting good predictive ability. Multivariate Cox analysis
further identified age as a significant independent predictor. The
DElncRNA-based risk scoring approach used in the present study was
less successful in predicting the RFS of patients with fibrosis.
Univariate Cox analysis failed to show that the risk scoring system
could significantly predict prognosis after adjusting for other
clinical factors. Stratified analyses based on risk scoring
identified the following factors as influencing risk: age,
ethnicity, alcohol consumption, new tumor event, pathology stage
and cancer status. Therefore, these factors need to be taken into
account in the prediction of RFS for HCC patients with
fibrosis.
The present study identified several DElncRNAs that
may be useful targets in efforts to understand why prognosis of
patients with HCC is worse in the presence of fibrosis. Numerous
studies have shown improved survival outcomes among patients with
no or mild fibrosis than among those with severe fibrosis (15–17),
including one analysis of 11,783 patients with HCC (23). It is even possible that fibrosis
promotes genetic mutations in patients with HCC (14).
As a first step towards using the DElncRNAs
identified in the present study to understand the prognosis more
clearly, the molecular functions of protein-coding genes highly
associated with the lncRNAs included in the present study's risk
scoring systems were analyzed. KEGG pathway analysis showed these
genes to be involved mainly in the cell cycle, chemokine-related
pathways, Th17 cell differentiation or thermogenesis, depending on
whether fibrosis was present and depending on whether OS or RFS was
the target outcome. These differential results for HCC
subpopulations may help guide future research in understanding,
predicting and managing recurrence and fibrosis.
Several previous studies have also constructed risk
scoring systems to predict the prognosis of patients with HCC
(24–31). However, those risk scoring systems
were based on DElncRNAs between HCC and normal samples, while the
risk scoring systems in the present study are based on the
DElncRNAs between HCC patients with or without fibrosis, and have
greater specificity in HCC patients with fibrosis. Moreover, the
DElncRNAs of risk scoring systems in the present study are
different from those in previous studies. To the best of our
knowledge, this is the first study to construct a risk scoring
system to predict survival in HCC patients with or without
fibrosis.
Despite its advantages, the present study has
several limitations. First, the prognostic value of the lncRNAs in
this study has not been validated in sample tissues or cells.
Second, the multivariate Cox regression analysis did not include
type of HCC treatment because these data were lacking from TCGA;
treatment history may affect OS and RFS (32). Indeed, the adjustment for potential
effects of other clinical characteristics on survival in the
present study may have been biased because relevant data for some
patients were not reported. Third, HCC patients were not stratified
based on early or advanced fibrosis, which may have biased the
results, such as the finding that the risk scoring system was not a
significant predictor of RFS in HCC patients with fibrosis. Fourth,
the hepatic fibrosis of patients in the TCGA database was evaluated
using only the Ishak fibrosis score, which may be inaccurate.
Despite these limitations, the results describe
novel risk scoring systems based on the expression of 5–7 lncRNAs
for predicting the OS of HCC patients with or without fibrosis, and
for predicting the RFS of patients without fibrosis. Further
studies are required to explore the possibility of using lncRNA
expression to predict the RFS of HCC patients with fibrosis.
Supplementary Material
Supporting Data
Supporting Data
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
This study was supported by The Young and
Middle-aged Teacher Basic Ability Enhancement Project of Guangxi
University (grant no. 2018KY0111), and Innovation Project of
Guangxi Graduate Education (no. YCBZ2019036).
Availability of data and materials
All the datasets generated and/or analyzed during
the present study are included in this published article.
Authors' contributions
LG and JY conceived the study. JY and SW conducted
the data curation and formal analysis. JY and SP were responsible
for methodology. SP and JH were responsible for data collection. LG
supervised the study. JY and JH were responsible for data
visualization. JY drafted the manuscript and LG revised the
manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
lncRNA
|
long non-coding RNA
|
|
HCC
|
hepatocellular carcinoma
|
|
OS
|
overall survival
|
|
RFS
|
recurrence-free survival
|
|
TCGA
|
The Cancer Genome Atlas
|
|
FC
|
fold change
|
|
FDR
|
false discovery rate
|
|
ROC
|
receiver operating characteristic
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
|
BMI
|
body mass index
|
|
AFP
|
α-fetoprotein
|
|
HR
|
hazard ratio
|
|
AUC
|
area under ROC curve
|
References
|
1
|
Forner A, Reig M and Bruix J:
Hepatocellular carcinoma. Lancet. 391:1301–1314. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Fong ZV and Tanabe KK: The clinical
management of hepatocellular carcinoma in the United States,
Europe, and Asia: A comprehensive and evidence-based comparison and
review. Cancer. 120:2824–2838. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bruix J, Gores GJ and Mazzaferro V:
Hepatocellular carcinoma: Clinical frontiers and perspectives. Gut.
63:844–855. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wu L, Pan C, Wei X, Shi Y, Zheng J, Lin X
and Shi L: lncRNA KRAL reverses 5-fluorouracil resistance in
hepatocellular carcinoma cells by acting as a ceRNA against
miR-141. Cell Commun Signal. 16:472018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Huang Y, Xiang B, Liu Y, Wang Y and Kan H:
LncRNA CDKN2B-AS1 promotes tumor growth and metastasis of human
hepatocellular carcinoma by targeting let-7c-5p/NAP1L1 axis. Cancer
Lett. 437:56–66. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Schmitt AM and Chang HY: Long noncoding
RNAs in cancer pathways. Cancer Cell. 29:452–463. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Song YX, Sun JX, Zhao JH, Yang YC, Shi JX,
Wu ZH, Chen XW, Gao P, Miao ZF and Wang ZN: Non-coding RNAs
participate in the regulatory network of CLDN4 via ceRNA mediated
miRNA evasion. Nat Commun. 8:2892017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu Y, Feng J, Sun M, Yang G, Yuan H, Wang
Y, Bu Y, Zhao M, Zhang S and Zhang X: Long non-coding RNA HULC
activates HBV by modulating HBx/STAT3/miR-539/APOBEC3B signaling in
HBV-related hepatocellular carcinoma. Cancer Lett. 454:158–170.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jiang R, Tang J, Chen Y, Deng L, Ji J, Xie
Y, Wang K, Jia W, Chu WM and Sun B: The long noncoding RNA lnc-EGFR
stimulates T-regulatory cells differentiation thus promoting
hepatocellular carcinoma immune evasion. Nat Commun. 8:151292017.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Friedman SL: Evolving challenges in
hepatic fibrosis. Nat Rev Gastroenterol Hepatol. 7:425–436. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Takaya H, Kawaratani H, Tsuji Y, Nakanishi
K, Saikawa S, Sato S, Sawada Y, Kaji K, Okura Y, Shimozato N, et
al: von Willebrand factor is a useful biomarker for liver fibrosis
and prediction of hepatocellular carcinoma development in patients
with hepatitis B and C. United European Gastroenterol J.
6:1401–1409. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
O'Rourke JM, Sagar VM, Shah T and Shetty
S: Carcinogenesis on the background of liver fibrosis: Implications
for the management of hepatocellular cancer. World J Gastroenterol.
24:4436–4447. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ko CJ, Lin PY, Lin KH, Lin CC and Chen YL:
Presence of fibrosis is predictive of postoperative survival in
patients with small hepatocellular carcinoma.
Hepatogastroenterology. 61:2295–2300. 2014.PubMed/NCBI
|
|
16
|
Kadri HS, Blank S, Wang Q, Kim KW, Fiel
MI, Luan W and Hiotis SP: Outcomes following liver resection and
clinical pathologic characteristics of hepatocellular carcinoma
occurring in patients with chronic hepatitis B and minimally
fibrotic liver. Eur J Surg Oncol. 39:1371–1376. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hung HH, Su CW, Lai CR, Chau GY, Chan CC,
Huang YH, Huo TI, Lee PC, Kao WY, Lee SD and Wu JC: Fibrosis and
AST to platelet ratio index predict post-operative prognosis for
solitary small hepatitis B-related hepatocellular carcinoma.
Hepatol Int. 4:691–699. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Abdalla AF, Zalata KR, Ismail AF, Shiha G,
Attiya M and Abo-Alyazeed A: Regression of fibrosis in paediatric
autoimmune hepatitis: Morphometric assessment of fibrosis versus
semiquantiatative methods. Fibrogenesis Tissue Repair. 2:22009.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Robinson MD, McCarthy DJ and Smyth GK:
edgeR: A Bioconductor package for differential expression analysis
of digital gene expression data. Bioinformatics. 26:139–140. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. Omics. 16:284–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Artinyan A, Mailey B, Sanchez-Luege N,
Khalili J, Sun CL, Bhatia S, Wagman LD, Nissen N, Colquhoun SD and
Kim J: Race, ethnicity, and socioeconomic status influence the
survival of patients with hepatocellular carcinoma in the United
States. Cancer. 116:1367–1377. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shebl FM, Capo-Ramos DE, Graubard BI,
McGlynn KA and Altekruse SF: Socioeconomic status and
hepatocellular carcinoma in the United States. Cancer Epidemiol
Biomarkers Prev. 21:1330–1335. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liu H, Cen D, Yu Y, Wang Y, Liang X, Lin H
and Cai X: Does fibrosis have an impact on survival of patients
with hepatocellular carcinoma: Evidence from the SEER database? BMC
Cancer. 18:11252018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yan J, Zhou C, Guo K, Li Q and Wang Z: A
novel seven-lncRNA signature for prognosis prediction in
hepatocellular carcinoma. J Cell Biochem. 120:213–223. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhao QJ, Zhang J, Xu L and Liu FF:
Identification of a five-long non-coding RNA signature to improve
the prognosis prediction for patients with hepatocellular
carcinoma. World J Gastroenterol. 24:3426–3439. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wu Y, Wang PS, Wang BG, Xu L, Fang WX, Che
XF, Qu XJ, Liu YP and Li Z: Genomewide identification of a novel
six-LncRNA signature to improve prognosis prediction in resectable
hepatocellular carcinoma. Cancer Med. 7:6219–6233. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sui J, Miao Y, Han J, Nan H, Shen B, Zhang
X, Zhang Y, Wu Y, Wu W, Liu T, et al: Systematic analyses of a
novel lncRNA-associated signature as the prognostic biomarker for
hepatocellular carcinoma. Cancer Med. May 15–2018.(Epub ahead of
print). doi: 10.1002/cam4.1541. View Article : Google Scholar
|
|
28
|
Shi YM, Li YY, Lin JY, Zheng L, Zhu YM and
Huang J: The discovery of a novel eight-mRNA-lncRNA signature
predicting survival of hepatocellular carcinoma patients. J Cell
Biochem. Nov 28–2018.(Epub ahead of print). doi:
10.1002/jcb.28028.
|
|
29
|
Ma Y, Luo T, Dong D, Wu X and Wang Y:
Characterization of long non-coding RNAs to reveal potential
prognostic biomarkers in hepatocellular carcinoma. Gene.
663:148–156. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liao X, Yang C, Huang R, Han C, Yu T,
Huang K, Liu X, Yu L, Zhu G, Su H, et al: Identification of
potential prognostic long non-coding RNA biomarkers for predicting
survival in patients with hepatocellular carcinoma. Cell Physiol
Biochem. 48:1854–1869. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Gu J, Zhang X, Miao R, Ma X, Xiang X, Fu
Y, Liu C, Niu W and Qu K: A three-long non-coding
RNA-expression-based risk score system can better predict both
overall and recurrence-free survival in patients with small
hepatocellular carcinoma. Aging (Albany NY). 10:1627–1639. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
El-Serag HB, Siegel AB, Davila JA, Shaib
YH, Cayton-Woody M, McBride R and McGlynn KA: Treatment and
outcomes of treating of hepatocellular carcinoma among medicare
recipients in the United States: A population-based study. J
Hepatol. 44:158–166. 2006. View Article : Google Scholar : PubMed/NCBI
|