Introduction
Renal cell carcinoma (RCC) with sarcomatoid
differentiation is a relatively rare renal tumor, and clear cell
renal cell carcinoma (CCRCC) with sarcomatoid differentiation
accounts for approximately 79% (1).
According to the World Health Organization (WHO) 2016
classification system, RCC with sarcomatoid differentiation was not
classified into a distinct subtype, however, it is considered to be
a specific histological characteristic of RCC (2,3). CCRCC
with sarcomatoid differentiation (CCRCCS) is of Fuhrman IV grade,
however, its prognosis is worse in comparison to other high-grade
RCCs. Approximately 80% of patients have been reported to be prone
to distant metastasis with a median overall survival of only 5.8
months, and 38% of patients had a survival of one year. The
mortality rate of CCRCCS has been reported to be 3.2 times higher
than that of CCRCC due to its poor response to systemic therapy and
owing to lack of any ideal therapeutic strategy (4–6).
Although great progress has been made in
understanding RCC, molecular mechanisms of sarcomatoid
differentiation have not yet been fully elucidated (7) According to a current prevailing
theory, sarcomatoid elements represent a subclonal
dedifferentiation or transformation from carcinomatous elements
based on their shared genetic patterns, such as X chromosome
inactivation and sarcomatoid element specific mutation, including
high frequent TP53, ARID1A, and BAP1 mutations (6,8,9).
Furthermore, frequently reduced expression of epithelial adhesion
molecules, such as E-cadherin and aberrant expression of N-cadherin
suggested that epithelial-mesenchymal transformation (EMT) may be
involved in the development of sarcomatoid elements (9,10).
To further expound the underlying molecular basis of
sarcomatoid differentiation in CCRCC, whole exome sequencing of
matched carcinomatous-sarcomatoid specimens of the same tumor from
five patients with CCRCCS was performed. Numerous candidate genes
were screened from the results of whole exome sequencing, and of
the identified genes, a cadherin gene, CDH23, that displayed a
highly frequent mutation, drew our attention. A single-nucleotide
polymorphism (SNP) site of CDH23 (rs3802711) was identified
concurrently in the exon areas of three groups of sarcomatoid
elements and two groups of carcinomatous elements. CDH23 is an
atypical cadherin that lacks the β-catenin binding motif, and
anchors to the actin cytoskeleton to mediate inter- and
intracellular adhesion (11).
Studies on CDH23 have confirmed its association with autosomal
recessive non-syndromic hearing loss (ARNSHL), sensorineural
deafness (DFNB12) and Usher syndrome type 1D (USH1D), age-related
HL, and noise induced HL (12,13).
However, the role CDH23 in cancer has not been
elucidated yet. It has been reported that cadherins play key roles
in the EMT of tumors which is a key factor in the progression and
metastasis of malignant tumors (14). Therefore, our study was further
expanded to another 40 specimens with CCRCCS and 50 specimens with
CCRCC by conducting Sanger sequencing to identify the SNP status of
CDH23. Concomitantly, the gene and protein expressions of CDH23
were detected, and the association between its mutation and a
series of clinicopathological features was also investigated.
Therefore, the aim of the present study, was to provide new
insights into the molecular mechanisms of sarcomatoid
transformation of CCRCC and identify novel gene targets for
prognostic predication and therapy of CCRCCS.
Materials and methods
Sample collection for whole exome
sequencing
Whole exome sequencing of five matched
carcinomatous-sarcomatoid paraffin-embedded specimens with CCRCCS,
was performed. The age range of the 5 patients was 53–70 years old
(with a median age of 62 years), including 3 males and 2 females.
Tissues were fixed with 4% neutral formaldehyde and embedded in
paraffin, and all sections were 4-µm-thick for H&E staining.
After deparaffinization and hydration, sections were stained with
hematoxylin for 5 min and stained with eosin for 3 min. After
dehydration and transparency, the sections were sealed with neutral
gum. Carcinomatous and sarcomatoid areas were labeled under a light
microscope (×4; OLYMPUS BX41 Olympus Corporation) after H&E
staining and separately dissected into 10-µm thick sections for DNA
extraction and whole exome sequencing. All sections were confirmed
by two experienced pathologists. All patients provided informed
consent and the study was approved by the Ethics Committee of The
Affiliated Hospital of Qingdao University, and was conducted in
full compliance with all the principles of the Helsinki
Declaration.
DNA extraction, exome capture, and
whole exome sequencing
DNA was extracted from formalin-fixed,
paraffin-embedded (FFPE) blocks of five matched
carcinoma-sarcomatoid tissues using GenElute FFPE DNA Purification
Kit (Sigma-Aldrich; Merck KGaA). Exome capture was performed using
Roche NimbleGen SeqCap EZ Exome V3 based on the manufacturer's
instructions. Thereafter, paired-end sequencing was performed using
Illumina HiSeq 4000 (Illumina, Inc.). Sequences were aligned to the
human reference genome (UCSC hg19) using Burrows-Wheeler Aligner
(BWA) software (15,16) SSNV calling was performed using
Mutect (http://www.broadinstitute.org/cancer/cga/mutect)
(17). Single-nucleotide variant
(SNV) and SNP were detected using Genome Analysis Tool Kit (GATK
4.1.0.0) (https://gatk.broadinstitute.org/hc/en-us) (18) and annotated through ANNOVAR
(20180416) (19,20). Kyoto Encyclopedia of Genes and
Genomes (KEGG) (https://www.genome.jp/kegg/) annotation and Gene
Ontology (GO) (http://geneontology.org/) annotation were applied to
annotate the function of the variant genes.
Candidate gene selection
Among numerous mutant genes, we focused on the
common mutant genes that occurred in more than three groups of
carcinomatous elements or three groups of sarcomatoid elements to
identify the most possible pathogenic genes of CCRCCS. A
non-synonymous SNP site in the exon 40 of CDH23 (rs3802711), a
cadherin gene, that may be related to EMT in tumors, was selected
for further study. Evolutionary conserved sequences and structures
of the protein and nucleic acid of CDH23 were assessed through the
National Center for Biology Information (NCBI). The effect of the
identified novel non-synonymous mutation was assessed using
PolyPhen-2 tool (http://genetics.bwh.harvard.edu/pph2/) (21) for prediction of the possible impact
of an amino acid substitution on the structure and function of a
human protein. The 3D molecular structure of the extracellular
domain (EC) of CDH23 was modeled using SWISS-MODEL (https://swissmodel.expasy.org/interactive) (22).
Expanded specimen acquisition and
Sanger sequencing for CDH23
A total of 40 specimens with CCRCCS (the median age:
63 years old, including 30 males and 10 females) and 50 specimens
with CCRCC (the median age: 59 years old, including 32 males and 18
females) were collected from The Affiliated Hospital of Qingdao
University from January 2008 to October 2018, and reviewed by two
genitourinary pathologists. The clinicopathological characteristics
and survival data of the patients were collected. Matched
carcinomatous, sarcomatoid tissues and normal renal tissues were
labeled under a light microscope after H&E staining as
aforementioned, and separately dissected into 10-µm thick sections.
Thereafter, DNA extraction and Sanger sequencing were performed to
assess the genotype of the CDH23 gene at a base position of 5411
from FFPE samples. In accordance with the results of whole-exome
sequencing, the specific PCR primers were designed and custom
synthesized (Shenggong Techonology). The PCR primers were designed
as follows: Forward, 5′-GGGCACAGATGGTCAGGGTTG-3′ and reverse,
3′-ACCTGTGACGAGTGAGGCTT-5′. The amplified PCR products were used
for Sanger sequencing (Shenggong Techonology). Subsequently, the
sequencing results were screened using Chromas program (2.6.5)
(Technelysium Pty Ltd.) and analyzed manually.
Quantitative real-time PCR (RT-qPCR)
to detect the expression of the CDH23 gene
CDH23 gene expression was successfully detected in
21 cases with CCRCCS and 34 cases with CCRCC through RT-qPCR using
SuperScript™ IV First-Strand Synthesis System (cat. no. 18091050,
Thermo Fisher, Scientific, Inc.) and Power SYBR® Green
Master Mix (cat. no. 4367659, Thermo Fisher, Scientific, Inc.)
after the total RNA extraction from paraffin-embedded tissues using
the Total RNA Extraction Kit (cat. no. DP439; Tiangen Biotech Co.,
Ltd.). The PCR primers for CDH23 gene were designed as follows:
Forward, 5′-CCGGCTGCCCTTCTTCACCAACCA-3′ and reverse,
5′-GGCCTCCTCCCCAGACACGCC-3′. The GAPDH gene was used as the
control. The PCR primers for the GAPDH gene were: Forward,
5′-GGATTTGGTCGTATTGGG-3′ and reverse, 5′-GGAAGATGGTGATGGGATT-3′.
The thermal cycling protocol was: 95°C for 5 min, 1 cycle; 95°C for
1 min, 60°C for 30 sec and 72°C for 30 sec, 38 cycles; 72°C, 10
min, 1 cycle. PCR was performed on Applied Biosystems 7500
Real-Time PCR System (Thermo Fisher, Scientific, Inc.). Relative
expression was calculated using the 2−ΔΔCq method
(23).
Immunohistochemistry
Immunohistochemistry was performed on 4-µm thick
sections of 90 FFPE samples. The primary antibody used in the
present study was CDH23 rabbit polyclonal antibody (cat. no.
PA5-53564, Thermo Fisher, Scientific, Inc.). All
immunohistochemistry sections were analyzed using Roche BenchMark
XT fully automatic IHC/ISH instrument following the optimized
protocols. The known positive sections were used as a positive
control, and phosphate-buffered saline (PBS) was used as a negative
control instead of a primary antibody.
The results were determined using a double-blind
method. Concurrently, each section was observed independently by
two senior pathologists. CDH23 was positively expressed at the
cytomembrane and/or cytoplasm. The positive staining was initially
scored as 0, 1, 2, and 3 based on the staining intensity (no
staining, faint, mild, and strong, respectively) and the percentage
of positively-stained cells (0%, ≤25%, ~26-75%, and >75%),
respectively. Subsequently, the two scores were multiplied, and a
positive score indicated that the product of the two fractions was
>4.
Clinical follow-up and survival
analysis
Follow-up data were collected for 34 cases with
CCRCCS and 41 cases with CCRCC. Survival was calculated from the
date of surgery to the death or the last follow-up visit. For the
analysis, only deaths for RCCs were considered as events. Survival
curves were derived from Kaplan-Meier analysis and log-rank test to
compare overall survival between CCRCCS and CCRCC based on
different clinicopathological features. Furthermore, to investigate
the factors that may affect survival patterns, the P-values for
prognostic factors analysis were adjusted for multiple analysis
using Cox regression model.
Statistical analysis
SPSS 20.0 software (IBM Corp.) was used for
statistical analysis. The differences in clinicopathological
characteristics, CDH23 genotypes, CDH23 gene and protein expression
between CCRCCS and CCRCC were evaluated by Chi-square test,
Fisher's exact test or T-test. The associations between CDH23
genotype and clinicopathological characteristics were evaluated by
Chi-square test or Fisher's exact test. Furthermore, the
correlation between CDH23 genotype and protein expression was
analyzed by Pearson correlation test. Two-tailed tests were used
for all comparisons, and P<0.05 was considered to indicate a
statistically significant difference.
Results
Whole exome sequencing
The median of the cleaned bases of ten samples was
observed to be 10.74 Gbp. The average contrast efficiency between
the samples and the reference genome UCSC hg19 was observed to be
99.74%. The median sequencing depth of the target area was
76.89X.
Somatic single nucleotide variants (SSNVs) in CCRCCS
were first screened in the five groups through whole exome
sequencing. The number of common SSNVs in more than one group, each
belonging to carcinomatous and sarcomatoid elements, were observed
to be 1544 and 1653, respectively. Carcinomatous and sarcomatoid
elements were found to share most SSNVs (1198). Sarcomatoid
elements had a higher overall SSNV burden than carcinomatous
elements (1653 vs. 1544) (Fig.
1A).
While investigating the element-specific mutant
genes, a total of 679 candidate genes, including 319 common mutant
genes occurring in more than three groups of carcinomatous
elements, and 360 common mutant genes emerging in more than three
groups of sarcomatoid elements, were obtained. The 679 candidate
genes comprised 12 genes in the downstream area, 61 genes in the
exonic region, 167 genes in the intergenic region, 316 genes in the
intronic region, 18 genes in ncRNA_exonic region, 45 genes in the
ncRNA_intronic region, 12 genes in the upstream area, 37 genes in
the UTR3, and 11 genes in the UTR5 (Fig. 1B). The results of KEGG pathway
enrichment revealed that most mutant genes in carcinomatous
elements were involved in the same functions as those in
sarcomatoid elements, such as gap junction, endocytosis, phagosome,
calcium signaling pathway, Rap1 signaling pathway, and neuroactive
ligand-receptor interaction (Fig. 2A
and B). However, some mutant genes played specific roles in
carcinomatous elements, including Ras signaling pathway, cell
adhesion molecules, cytokine-cytokine receptor interaction, and
central carbon metabolism in cancer (Fig. 2A). Furthermore, in sarcomatoid
element, some distinct genes were involved in specific functions,
such as, regulation of actin cytoskeleton, local adhesion, FOXO
signaling pathway, MAPK signaling pathway, PI3K/Akt signaling
pathway, and vascular smooth muscle contraction (Fig. 2B). Moreover, GO enrichment analysis
revealed the mutant genes in carcinomatous elements (Fig. S1) and sarcinomatoid elements
(Fig. S2), including the genes
involved in biological processes, cellular components and molecular
functions.
Among them, there were 25 common mutant genes in
more than three groups of carcinomatous elements in the exon
region, including 12 synonymous mutant genes and 13 non-synonymous
mutant genes. The 13 non-synonymous mutant genes were KIF17,
MROH2B, UBQLN3, HLA-DPB1, RP1L1, SBSPON, IQCE, FMN1, ZNF592, ALPK2,
FGCBP, C19orf54, and ZFHX3 (Table
I). A total of 21 common mutant genes were identified in more
than three groups of sarcomatoid elements in the exon region,
including 13 synonymous mutant genes and 8 non-synonymous mutant
genes. The 13 non-synonymous mutant genes were RNF207, FAM107B,
PALD1, CDH23, SPON2, SLC37A1, PRDM10, and FPR1 (Table II).
 | Table I.SNP sites existing in more than 3
groups of carcinomatous elements in the exon area. |
Table I.
SNP sites existing in more than 3
groups of carcinomatous elements in the exon area.
| Gene | Chr | Start | End | Ref | Mut | Mutation type |
|---|
| KIF17 | 1 | 20704549 | 20704549 | T | A | Non-synonymous |
| MROH2B | 5 | 41008678 | 41008678 | A | G | Non-synonymous |
| UBQLN3 | 11 | 5508690 | 5508690 | G | C | Non-synonymous |
| HLA-DPB1 | 6 | 33080761 | 33080761 | T | C | Non-synonymous |
| RP1L1 | 8 | 10616532 | 10616532 | T | G | Non-synonymous |
| SBSPON | 8 | 73092896 | 73092896 | A | G | Non-synonymous |
| IQCE | 7 | 2604885 | 2604885 | C | T | Non-synonymous |
| FMN1 | 15 | 33067373 | 33067373 | C | A | Non-synonymous |
| ZNF592 | 15 | 84798628 | 84798628 | G | A | Non-synonymous |
| ALPK2 | 18 | 59537018 | 59537018 | A | C | Non-synonymous |
| FCGBP | 19 | 39902034 | 39902034 | G | T | Non-synonymous |
| C19orf54 | 19 | 40749595 | 40749595 | C | G | Non-synonymous |
| ZFHX3 | 16 | 72957816 | 72957816 | A | G | Non-synonymous |
 | Table II.SNP sites existing in more than 3
groups of sarcomatoid elements in the exon region. |
Table II.
SNP sites existing in more than 3
groups of sarcomatoid elements in the exon region.
| Gene | Chr | Start | End | Ref | Mut | Mutation type |
|---|
| RNF207 | 1 | 6218354 | 6218354 | A | G | Non-synonymous |
| FAM107B | 10 | 14774449 | 14774449 | G | A | Non-synonymous |
| PALD1 | 10 | 70530022 | 70530022 | C | T | Non-synonymous |
| CDH23 | 10 | 71784329 | 71784329 | G | A | Non-synonymous |
| SPON2 | 4 | 1171342 | 1171342 | G | T | Non-synonymous |
| SLC37A1 | 21 | 42565845 | 42565845 | G | A | Non-synonymous |
| PRDM10 | 11 | 129925055 | 129925055 | T | C | Non-synonymous |
| FPR1 | 19 | 51746419 | 51746419 | A | C | Non-synonymous |
Target gene selection
Notably, among the 21 common mutant genes in more
than three groups of sarcomatoid elements in the exon area, there
was a high frequent point mutation in exon 40 of CDH23 gene (c.
G5411A) occurring concurrently in the three groups of sarcomatoid
elements and two groups of carcinomatous elements. This point
mutation is an SNP site of CDH23 gene in exon 40 namely, rs3802711
in the NCBI database, and the identified amino acid substitution
(p.Arg1804Gln) is in the highly conserved calcium-binding sites of
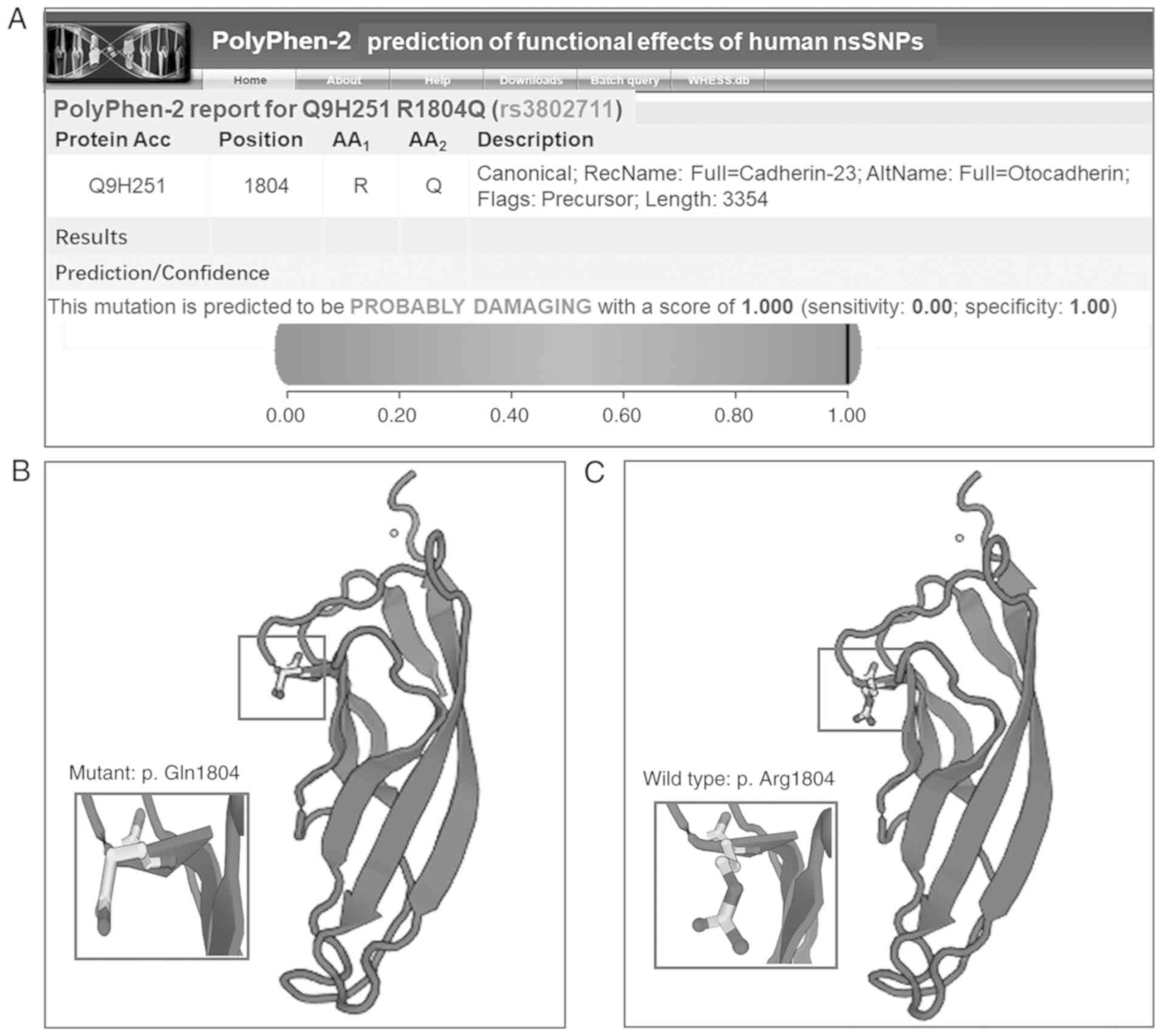
the extracellular cadherin 17 (EC17) domain. PolyPhen-2 analysis
predicated that the mutation (p.Arg1804Gln) may be involved in
damaging the structure and function of CDH23 with a score of 1.000
(Fig. 3A). The three-dimensional
(3D) structure of models predicted by SWISS-MODEL revealed a
typical folding pattern with several β-strands. The p.Arg1804Gln
was observed to be in the highly conserved EC calcium-binding sites
in the cadherin repeat domain. The 3D structure of the mutation
region of CDH23 (p.Gln1804; Fig.
3B) was different from that of the wild-type protein (p.
Arg1804; Fig. 3C), which could
disturb the protein function and interaction with other
proteins.
Clinicopathological characteristics of
40 patients with CCRCCS and 50 patients with CCRCC
The clinicopathological characteristics of 40
patients with CCRCCS and 50 patients with CCRCC are presented in
Table III. Compared with CCRCC,
CCRCCS were larger in diameter (7.7 cm vs. 2.6 cm; P<0.001), and
more frequently associated with metastasis (18 vs. 2; P<0.001)
and local invasion (stage T3/T4, 21 vs. 3; P<0.001). The
follow-up data were available in 34 patients with CCRCCS and 42
patients with CCRCC. Cancer-specific survival of CCRCCS was poorer
than that of CCRCC, and 17 patients with CCRCCS succumbed 3–24
months after the surgery, while only three patients with CCRCC
succumbed 17–22 months after the surgery.
 | Table III.Comparison of the clinicopathological
features between CCRCCS and CCRCC |
Table III.
Comparison of the clinicopathological
features between CCRCCS and CCRCC
|
| Groups |
|
|
|---|
|
|
|
|
|
|---|
|
Characteristics | CCRCCS (n=40) | CCRCC (n=50) |
χ2/t-test | P-value |
|---|
| Sex |
|
Male | 30 | 32 | 1.255 | 0.263 |
|
Female | 10 | 18 |
|
|
| Age |
|
Median | 63 | 59 | 1.607 | 0.112 |
| Diameter (cm) |
|
Median | 7.7 | 2.64 | 12.692 |
<0.001a |
| Metastasis |
| No | 22 | 48 | 21.613 |
<0.001a |
|
Yes | 18 | 2 |
|
|
| TNM stage |
|
I–II | 19 | 47 | 24.571 |
<0.001a |
|
III–IV | 21 | 3 |
|
|
Frequencies of CDH23 genotype in
CCRCCS and CCRCC
Sanger sequencing results revealed that among the 40
specimens with CCRCC, 22 specimens exhibited A genotype (rs3802711)
at the 5411th base of the CDH23 gene, and only 18 presented G
genotype (wild-type, WT). Among the 22 mutant specimens, three were
observed to be heterozygous mutations and the others were observed
to be homozygous mutations (Fig.
4A-D). Carcinomatous and sarcomatoid elements in the same
CCRCCS showed similar genotype at the 5411th base of the CDH23
gene. Whereas, among the 50 specimens with CCRCCS, 40 uniformly
displayed G genotype (WT) rather than A genotype (rs3802711). Among
the 10 mutant specimens, only one was a heterozygous mutation
(Fig. 4E and F). There was a
significant difference in the genotypic frequency (rs3802711) of
the CDH23 gene between CCRCCS and CCRCC (55 vs. 20%,
χ2=11.88, P=0.001; Table
IV). The genotypic frequency (rs3802711) of the CDH23 gene
between CCRCCS and high-grade CCRCC (Grade III–IV) revealed a
significant difference (55 vs. 18.2%, P=0.042; Table IV). Furthermore, the genotypic
frequency (rs3802711) of the CDH23 gene in CCRCCS is higher than
that in low-grade CCRCC (Grade I–II) (55 vs. 20.5%, P=0.002;
Table IV). The normal renal
tissues of the 90 specimens were all wild-types. There was no
significant association between CDH23 mutation and a series of
clinicopathological features of CCRCCS including the age, sex,
tumor diameter, metastasis, TNM stage, and survival of patients
(Table V).
 | Table IV.Comparison of the frequencies of
rs3802711 (A) and wild-type (G) of CDH23 in CCRCCS and CCRCC. |
Table IV.
Comparison of the frequencies of
rs3802711 (A) and wild-type (G) of CDH23 in CCRCCS and CCRCC.
|
|
| SNP of CDH23 |
|
|
|---|
|
|
|
|
|
|
|---|
| Groups | N | rs3802711 (A) | Wild-type (G) | χ2 | P-value |
|---|
| CCRCCS | 40 | 22 | 18 | 11.88 | 0.001a |
| CCRCC | 50 | 10 | 40 |
|
|
| CCRCCS | 40 | 22 | 18 | – | 0.042a |
| CCRCC (Grade
III–IV) | 11 | 2 | 9 |
|
|
| CCRCCS | 40 | 22 | 18 | 9.971 | 0.002a |
| CCRCC (Grade
I–II) | 39 | 8 | 31 |
|
|
 | Table V.The association between the genotype
frequency of the CDH23 gene with the clinicopathological features
of CCRCCS. |
Table V.
The association between the genotype
frequency of the CDH23 gene with the clinicopathological features
of CCRCCS.
|
|
| SNP of CDH23 |
|
|
|---|
|
|
|
|
|
|
|---|
| Groups | N | rs3802711 (A) | Wild-type (G) | χ2 | P-value |
|---|
| Sex |
|
Male | 30 | 16 | 14 | 0.135 | 0.714 |
|
Female | 10 | 6 | 4 |
|
|
| Age |
|
≤63 | 20 | 9 | 11 | 1.616 | 0.204 |
|
>63 | 20 | 13 | 7 |
|
|
| Diameter |
|
≤8.0 | 24 | 11 | 13 | 2.037 | 0.154 |
|
>8.0 | 16 | 11 | 5 |
|
|
| Metastasis |
| No | 22 | 10 | 12 | 1.800 | 0.180 |
|
Yes | 18 | 12 | 6 |
|
|
| TNM stage |
| I-
II | 19 | 8 | 11 | 2.431 | 0.119 |
|
III–IV | 21 | 14 | 7 |
|
|
|
Survival | – | – | – | – | >0.05 |
Kaplan-Meier analysis and Cox
multivariate analysis for the factors affecting the overall
survival of patients with RCC
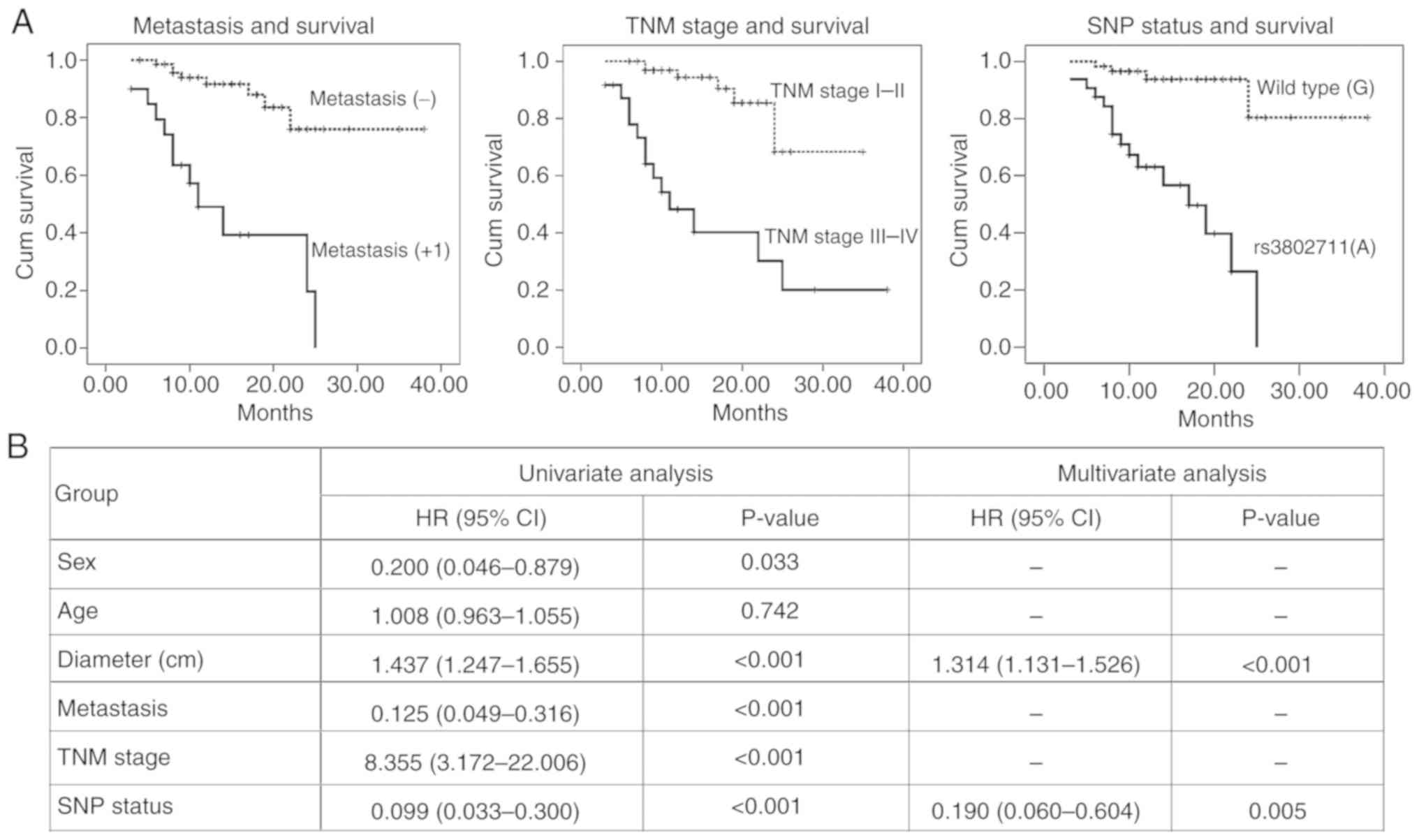
Kaplan-Meier analysis revealed that the
clinicopathological features, including, distant metastasis
(Fig. 5A and B), TNM stage
(Fig. 5A and B), CDH23 SNP type
(rs3802711) (Fig. 5A and B) and
tumor diameter (Fig. 5B) were
associated with the overall survival of a total of 90 RCCs. The Cox
multivariate analysis indicated that the tumor diameter and CDH23
SNP type were the independent prognostic factors affecting the
overall survival of the 90 RCCs (Fig.
5B).
Expression of the CDH23 gene in CCRCCS
and CCRCC
In RT-qPCR analysis, CDH23 gene expression was
observed to be lower in 21 cases with CCRCCS (Fig. S3) than that in 34 cases with CCRCC
(P<0.001; Fig. S4; Table VI).
 | Table VI.CDH23 gene expression in CCRCCS and
CCRCC. |
Table VI.
CDH23 gene expression in CCRCCS and
CCRCC.
|
|
| CDH23 gene
expression |
|
|
|---|
|
|
|
|
|
|
|---|
| Group | N | Negative | Positive | χ2 | P-value |
|---|
| CCRCCS | 21 | 16 | 5 | 17.429 |
<0.001a |
| CCRCC | 34 | 6 | 28 |
|
|
Expression of the CDH23 protein in
CCRCCS and CCRCC
Through immunohistochemistry, the positive
expression rates of the CDH23 protein in CCRCCS and CCRCC were
observed to be 42.5% (17/40) and 76% (38/50), respectively
(P=0.001), exhibiting a lower expression rate in CCRCCS compared
with CCRCC (Fig. 6; Table VII). CDH23 protein expression in
CCRCCS was not associated with clinicopathological characteristics,
such as the age, sex, tumor diameter, metastasis, TNM stage, and
survival of patients (Table
SI).
 | Figure 6.The morphology of (A) CCRCC (H&E,
×400), (B) strong expression and (C) weak expression of CDH23 in
CCRCC (immunohistochemistry, ×400). (D) The morphology of
sarcomatoid elements of CCRCCS (H&E, ×400), (E) strong
expression and (F) negative expression of CDH23 in sarcomatoid
elements of CCRCCS (immunohistochemistry, ×400). CCRCC, clear cell
renal cell carcinoma; H&E, hematoxylin and eosin; CDH23,
cadherin 23; CCRCCS, clear cell renal cell carcinoma with
sarcomatoid differentiation. |
 | Table VII.CDH23 protein expression in CCRCCS
and CCRCC. |
Table VII.
CDH23 protein expression in CCRCCS
and CCRCC.
|
|
| CDH23 protein
expression |
|
|
|---|
|
|
|
|
|
|
|---|
| Group | N | Negative | Positive | χ2 | P-value |
|---|
| CCRCCS | 40 | 23 | 17 | 10.494 | 0.001a |
| CCRCC | 12 | 38 |
|
|
|
Correlation between the genotype of
the CDH23 gene and expression of the CDH23 protein in a total of 90
studied cases
The genotype (rs3802711) of the CDH23 gene was
significantly positively correlated with the expression of the
CDH23 protein in 90 studied cases (P<0.001). The CDH23 protein
was negatively or weakly expressed in most CCRCCS specimens with
CDH23 mutation indicating that the mutation impaired the expression
of the CDH23 protein (Table
VIII).
 | Table VIII.The association between the genotype
of the CDH23 gene and the CDH23 protein expression in CCRCCS and
CCRCC. |
Table VIII.
The association between the genotype
of the CDH23 gene and the CDH23 protein expression in CCRCCS and
CCRCC.
|
|
| CDH23 protein
expression |
|
|
|---|
|
|
|
|
|
|
|---|
| Genotype | N | Negative | Positive | r | P-value |
|---|
| rs3802711 (A) | 25 | 7 | 5 | 0.598 |
<0.001a |
| Wild-type (G) | 10 | 48 |
|
|
|
Discussion
Given the characteristics of aggressive growth,
early metastasis, extremely poor prognosis, and short survival time
of CCRCCS, the pathogenesis of sarcomatoid elements is a key point
and has always been an obstacle in renal tumor research. With the
advent of next generation sequencing (NGS), few researchers have
applied this technology to the research of CCRCCS. In a previous
study, it was revealed that CCRCCS was more likely to lose 9q, 15q,
18p/q and 22q, and gain 1q and 8q than CCRCC, papillary renal cell
carcinoma and chromophobe renal cell carcinoma. Furthermore, CCRCCS
was revealed to have a higher CNV burden through SNP microarray
analysis (24). Malouf et al
(25) revealed that TP53, VHL,
CDKN2A, and NF2 were most susceptible to mutation through next
generation sequencing of 26 tumors with CCRCCS. CCRCCS and CCRCC
had some different tumor-driving genes, and CCRCCS exhibited higher
CNV burdens. Sircar et al (26) carried out cDNA microarray and
RNA-sequencing to reveal that CCRCCS was molecularly distinct from
non-sarcomatoid CCRCC, with its genetic programming mostly shared
by the epithelioid and sarcomatoid elements.
However, to date, the molecular mechanisms of
sarcomatoid transformation had not been well elucidated. In the
present study, the whole exome sequencing results revealed that
most SSNVs were shared between carcinomatous and sarcomatoid
elements providing reliable evidence that these elements originated
from a common ancestor. Bi et al (6) revealed that the two elements had the
same mutations with an average of 42% shared SSNVs through exome
sequencing. The data in the present study and the previous
literature (6,26) have provided strong evidence of a
carcinomatous origin of the two elements. However, the burden of
element-specific SSNVs in known cancer drivers was higher in
sarcomatoid elements than in carcinomatous elements suggesting that
the sarcomatoid element has arisen in a process of
de-differentiation from a preexisting carcinomatous element during
the progression of tumors, which was consistent with previous
studies (6,26).
To investigate the element-specific mutant genes,
319 common mutant genes occurring in more than three groups of
carcinomatous elements, and 360 common mutant genes emerging in
more than three groups of sarcomatoid elements were obtained. Most
mutant genes in carcinomatous elements were involved in the same
functions as those in sarcomatoid elements. However, some mutant
genes played specific roles in carcinomatous elements, including
Ras signaling pathway, cell adhesion molecules, cytokine-cytokine
receptor interaction, and central carbon metabolism in cancer.
Furthermore, in sarcomatoid elements, some distinct genes were
specifically involved in regulation of actin cytoskeleton, focal
adhesion, FOXO signaling pathway, MAPK signaling pathway, PI3K/Akt
signaling pathway, and vascular smooth muscle contraction, which
may be involved in the transformation of sarcomatoid elements in
CCRCCS. Among them, there were 25 common mutant genes in more than
three groups of carcinomatous elements in the exon region,
including 13 non-synonymous mutant genes KIF17, MROH2B, UBQLN3,
HLA-DPB1, RP1L1, SBSPON, IQCE, FMN1, ZNF592, ALPK2, FGCBP, C19orf54
and ZFHX3. Another 21 common mutant genes in more than three groups
of sarcomatoid elements in the exon region contained eight
non-synonymous mutant genes RNF207, FAM107B, PALD1, CDH23, SPON2,
SLC37A1, PRDM10, and FPR1. These genes may act as candidate genes,
further revealing the molecular mechanisms of sarcomatoid
transformation in CCRCCS.
Notably, simultaneous occurrence of non-synonymous
mutation of CDH23 c.G5411A (p.Arg1804 Gln), an SNP locus of CDH23
namely, rs3802711, in two groups of carcinomatous elements and
three groups of sarcomatoid elements drew our attention. It has
been reported that the CDH23 is a member of cadherin family that
plays crucial roles in epithelial and mesenchymal transformation of
tumors. Furthermore, Sanger sequencing in extended samples with
CCRCCS and CCRCC revealed that the frequency of CDH23 mutation
(rs3802711) was significantly higher in CCRCCS compared to CCRCC.
Moreover, the frequency of CDH23 mutation (rs3802711) was markedly
higher in CCRCCS than that in high-grade CCRCC (Fuhrman grade
III–IV). It was also revealed that the sarcomatoid and
carcinomatous elements in the same tumor shared similar CDH23
(rs3802711) mutation, which provided further evidence of a common
origin of the two elements in CCRCCS.
CDH23 (NM_022124) is located on chromosome
10q21-q22, encoding a 3354aa atypical cadherin with an elongated
extracellular region containing 27 EC domains, a single
transmembrane domain, and a cytoplasmic domain that is mostly
expressed in the cilia of neuroretina, cochlea, and vestibular hair
cells, but also in the brain, heart, lung, kidney, nose, eye, and
ear cells (27). The repeat EC
domains are highly conserved among human, mice and rats, which
responsibly mediate the functions of cadherin members. Thus far,
most research concerning CDH23 had been focused on its association
with hearing loss. Various mutations in CDH23 have resulted either
in the loss of tip links or hair cell death causing sensorineural
deafness (DFNB12), USH1D, age-related HL or noise-induced HL
(12,13). Certain deafness mutations in CDH23,
including p.Pro240Leu, p.Glu1595Lys, p.Asn342Ser, and p.Glu1595Lys
may have affected highly conserved calcium binding motifs in the EC
domain or in the linker region between the EC domain, both of which
are essential for calcium binding or dimerization of CDH23, and
then impaired cell-to-cell adhesion through calcium-dependent
interactions (28,29). Zhang et al (30) identified a new non-synonymous
heterozygous mutation c.G 4136T (p.Arg1379Leu) in CDH23 in
pituitary adenoma through whole exome sequencing, which caused an
amino acid substitution in the calcium-binding motif of the EC
domains of CDH23 and was predicted to impair cell-cell
adhesion.
In the present study, p.Arg1804Gln was revealed as a
new variant in CDH23 that was not shared with either hearing loss
or pituitary adenoma, and none of the patients in the present study
were linked to any symptoms of deafness or pituitary adenoma.
PolyPhen-2 analysis predicted the mutation (p.Arg1804Gln) that may
be involved in damaging the structure and function of CDH23.
Furthermore, the normal 3D structure of CDH23 was impaired in the
mutant protein (p.Arg1804Gln) predicted by SWISS-MODEL, which could
disturb the protein function and its interaction with other
proteins. Furthermore, the CDH23 gene and protein were negatively
or weakly expressed in most CCRCCS specimens with CDH23 mutation
indicating that the mutation of CDH23 impaired the expression of
CDH23 and then disturbed its normal function. Cox multivariate
analysis indicated that the CDH23 SNP type was an independent
prognostic factor affecting the overall survival of the total study
cohort, including CCRCC and CCRCCS, despite being independently
associated with the prognosis of patients with CCRCCS. The possible
interpretation was that a variety of factors may be involved in the
sarcomatoid transformation and prognosis of CCRCCS, of which the
CDH23 (rs3802711) genotype may be a high genetic risk factor of
sarcomatoid transformation and a predictive factor for the poor
prognosis of RCCs.
The role of CDH23 in cancer has not been fully
elucidated. Cadherins are well known to play crucial roles in EMT,
through which the epithelial cells are reprogrammed to become
mesenchymal cells, lose adhesion, and are prone to metastasis. The
altered cadherin expression is a known marker for EMT, mainly
including cadherin 1 (E-Cadherin), cadherin 2, cadherin 4, cadherin
13, cadherin 3, and cadherin 11 (31). CDH23 is a unique non-classic member
of the cadherin family that lacks the β-catenin binding motif
anchoring to the actin cytoskeleton to mediate intercellular
adhesion. The CDH23 interacting partners, such as USH1C and MAGI-1
are known to interact with the actin cytoskeleton to anchor the
proteins that may play important roles in the CDH23 anchorage
mechanism and cellular adhesion (32). However, studies have revealed its
possible role in complex diseases, such as cancer and Alzheimer's
disease (33,34). CDH23 has been recently reported to
mediate predominant heterotypic cell adhesion between the
epithelial cells and fibroblasts, thus playing an important role in
breast cancer cell metastasis through the TGF-β pathway. The TGF-β
signaling pathway could enhance cell invasion, migration, and an
immunosuppressive effect (7). CDH23
may be involved in EMT and tumor metastasis similarly to the other
cadherins. CDH23 mutations have always been studied with respect to
hearing loss, but exploring the somatic mutations in CDH23 and
protein expression levels in various carcinomas, especially in
carcinoma with sarcomatoid differentiation will be helpful in
further understanding its role in EMT and tumor metastasis
(35). However, the mechanisms
through which CDH23 is involved in EMT and tumor metastasis have
not been reported to date. As the future direction of our research,
we plan to perform RNA sequencing in cell lines to screen the
upstream and downstream factors related to CDH23 after the
overexpression or knockdown of CDH23, which may be helpful to
explain the molecular mechanisms of CDH23 in the CCRCCS
process.
The present study has identified, for the first
time, the CDH23 SNP (rs3802711) as a high genetic risk factor for
CCRCCS. CDH23 mutation (rs3802711) was associated with the
decreased expression of the CDH23 protein, resulting in the absence
of cadherin function of CDH23, which indicated that the CDH23
mutation may be involved in the sarcomatoid transformation in
CCRCCS. Furthermore, the present results provided a new prognostic
evaluation factor and potential target therapeutic site for CCRCCS.
New insight was gained into the molecular mechanisms of sarcomatoid
transformation in CCRCCS and a new prognosis evaluation factor for
CCRCCS was revealed. Furthermore, a novel and specific SNP of CDH23
was identified in CCRCCS and a new candidate cadherin involved in
EMT was revealed. These results have implications for the therapy
of patients with this type of tumor which has a poor prognosis.
However, a greater number of tumors with CCRCCS are required for
study and further functional studies of the CDH23 gene are required
to gain insights into the detailed pathogenic mechanisms involved
in CCRCCS.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the Natural
Science Foundation of Shandong Province (ZR2017MH009), and the
National Natural Science Foundation of China (81201654).
Availability of data and materials
The datasets analyzed during the present study are
publicly available from the following online databases: PolyPhen-2,
http://genetics.bwh.harvard.edu/pph2/; SWISS-MODEL,
https://swissmodel.expasy. org/interactive; https://www.ncbi.nlm.nih.gov/pubmed/.
Authors' contributions
WY designed the study, analyzed the data and wrote
the manuscript. XW performed Sanger sequencing and analyzed the
data. YW analyzed and interpreted the data of the study. WZ
analyzed the imaging features, clinical data and IHC results. YJ
and YL reviewed the pathologic diagnosis and contributed to the
conception and design of the study. HS performed the IHC experiment
and mRNA extraction. YL also revised the manuscript critically for
important intellectual content. All authors read and approved the
final version of the manuscript and agree to be accountable for all
aspects of the work in ensuring that questions related to the
accuracy or integrity of any part of the work are appropriately
investigated and resolved.
Ethics approval and consent to
participate
This study was performed in accordance with standard
guidelines and was approved by the Ethics Committee of the
Affiliated Hospital of Qingdao University. All patients provided
informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Nguyen DP, Vilaseca A, Vertosick EA,
Corradi RB, Touijer KA, Benfante NE, Sjoberg DD and Russo P:
Histologic subtype impacts cancer-specific survival in patients
with sarcomatoid-variant renal cell carcinoma treated surgically.
World J Urol. 34:539–544. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Farrow GM, Harrison EG Jr, Utz DC and
ReMine WH: Sarcomas and sarcomatoid and mixed malignant tumors of
the kidney in adults. Cancer. 22:556–563. 1968. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Moch H, Cubilla AL, Humphrey PA, Reuter VE
and Ulbright TM: The 2016 WHO Classification of tumours of the
urinary system and male genital organs-part a: Renal, penile, and
testicular tumours. Eur Urol. 70:93–105. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Trudeau V, Larcher A, Sun M, Boehm K,
Dell'Oglio P, Sosa J, Tian Z, Fossati N, Briganti A, Shariat SF and
Karakiewicz P: Comparison of oncologic outcomes between sarcomatoid
and clear cell renal cell carcinoma. World J Urol. 34:1429–1436.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Morra L, Rechsteiner M, Casagrande S, Duc
Luu V, Santimaria R, Diener PA, Sulser T, Kristiansen G, Schraml P,
Moch H and Soltermann A: Relevance of periostin splice variants in
renal cell carcinoma. Am J Pathol. 179:1513–1521. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bi M, Zhao S, Said JW, Merino MJ, Adeniran
AJ, Xie Z, Nawaf CB, Choi J, Belldegrun AS, Pantuck AJ, et al:
Genomic characterization of sarcomatoid transformation in clear
cell renal cell carcinoma. Proc Natl Acad Sci USA. 113:2170–2175.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Shuch B, Bratslavsky G, Linehan WM and
Srinivasan R: Sarcomatoid renal cell carcinoma: A comprehensive
review of the biology and current treatment strategies. Oncologist.
17:46–54. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jones TD, Eble JN, Wang M, Maclennan GT,
Jain S and Cheng L: Clonal divergence and genetic heterogeneity in
clear cell renal cell carcinomas with sarcomatoid transformation.
Cancer. 104:1195–1203. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Shuch B, Said J, LaRochelle JC, Zhou Y, Li
G, Klatte T, Pouliot F, Kabbinavar FF, Belldegrun AS and Pantuck
AJ: Histologic evaluation of metastases in renal cell carcinoma
with sarcomatoid transformation and its implications for systemic
therapy. Cancer. 116:616–624. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Conant JL, Peng Z, Evans MF, Naud S and
Cooper K: Sarcomatoid renal cell carcinoma is an example of
epithelial-mesenchymal transition. J Clin Pathol. 64:1088–1092.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Siemens J, Lillo C, Dumont RA, Reynolds A,
Williams DS, Gillespie PG and Müller U: Cadherin 23 is a component
of the tip link in hair-cell stereocilia. Nature. 428:950–955.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mizutari K, Mutai H, Namba K, Miyanaga Y,
Nakano A, Arimoto Y, Masuda S, Morimoto N, Sakamoto H, Kaga K and
Matsunaga T: High prevalence of CDH23 mutations in patients with
congenital high-frequency sporadic or recessively inherited hearing
loss. Orphanet J Rare Dis. 10:602015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Schultz JM, Bhatti R, Madeo AC, Turriff A,
Muskett JA, Zalewski CK, King KA, Ahmed ZM, Riazuddin S, Ahmad N,
et al: Allelic hierarchy of CDH23 mutations causing non-syndromic
deafness DFNB12 or Usher syndrome USH1D in compound heterozygotes.
J Med Genet. 48:767–775. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Taniuchi K, Nakagawa H, Hosokawa M,
Nakamura T, Eguchi H, Ohigashi H, Ishikawa O, Katagiri T and
Nakamura Y: Overexpressed P-cadherin/CDH3 promotes motility of
pancreatic cancer cells by interacting with p120ctn and activating
rho-family GTPases. Cancer Res. 65:3092–3099. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li H and Durbin R: Fast and accurate short
read alignment with Burrows-Wheeler transform. Bioinformatics.
25:1754–1760. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li H and Durbin R: Fast and accurate
long-read alignment with Burrows-Wheeler transform. Bioinformatics.
26:589–595. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cibulskis K, Lawrence MS, Carter SL,
Sivachenko A, Jaffe D, Sougnez C, Gabriel S, Meyerson M, Lander ES
and Getz G: Sensitive detection of somatic point mutations in
impure and heterogeneous cancer samples. Nat Biotechnol.
31:213–219. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
McKenna A, Hanna M, Banks E, Sivachenko A,
Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly
M and DePristo MA: The Genome Analysis Toolkit: A MapReduce
framework for analyzing next-generation DNA sequencing data. Genome
Res. 20:1297–1303. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yang H and Wang K: Genomic variant
annotation and prioritization with ANNOVAR and wANNOVAR. Nat
Protoc. 10:1556–1566. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chang X and Wang K: wANNOVAR: Annotating
genetic variants for personal genomes via the web. J Med Genet.
49:433–436. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Adzhubei I, Jordan DM and Sunyaev SR:
Predicting functional effect of human missense mutations using
polyphen-2. Curr Protoc Hum Genet. 7:Unit7.20. 2013.PubMed/NCBI
|
|
22
|
Waterhouse A, Bertoni M, Bienert S, Studer
G, Tauriello G, Gumienny R, Heer FT, de Beer TAP, Rempfer C,
Bordoli L, et al: SWISS-MODEL: Homology modelling of protein
structures and complexes. Nucleic Acids Res. 46:(W1):W296–W303.
2018. View Article : Google Scholar
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2 (Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ito T, Pei J, Dulaimi E, Menges C, Abbosh
PH, Smaldone MC, Chen DY, Greenberg RE, Kutikov A, Viterbo R, et
al: Genomic copy number alterations in renal cell carcinoma with
sarcomatoid features. J Urol. 195:852–858. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Malouf GG, Ali SM, Wang K, Balasubramanian
S, Ross JS, Miller VA, Stephens PJ, Khayat D, Pal SK, Su X, et al:
Genomic characterization of renal cell carcinoma with sarcomatoid
dedifferentiation pinpoints recurrent genomic alterations. Eur
Urol. 70:348–357. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sircar K, Yoo SY, Majewski T, Wani K,
Patel LR, Voicu H, Torres-Garcia W, Verhaak RG, Tannir N, Karam JA,
et al: Biphasic elements of sarcomatoid clear cell renal cell
carcinomas are molecularly similar to each other, but distinct
from, non-sarcomatoid renal carcinomas. J Biphasic Pathol Clin Res.
1:212–224. 2015. View
Article : Google Scholar
|
|
27
|
Diez-Roux G, Banfi S, Sultan M, Geffers L,
Anand S, Rozado D, Magen A, Canidio E, Pagani M, Peluso I, et al: A
high-resolution anatomical atlas of the transcriptome in the mouse
embryo. PLoS Biol. 9:e10005822011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Woo HM, Park HJ, Park MH, Kim BY, Shin JW,
Yoo WG and Koo SK: Identification of CDH23 mutations in Korean
families with hearing loss by whole-exome sequencing. BMC Med
Genet. 15:462014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kowalski TJ, Pawelczyk M, Rajkowska E,
Dudarewicz A and Sliwinska-Kowalska M: Genetic variants of CDH23
associated with noise-induced hearing loss. Otol Neurotol.
35:358–365. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang Q, Peng C, Song J, Zhang Y, Chen J,
Song Z, Shou X, Ma Z, Peng H, Jian X, et al: Germline mutations in
CDH23, encoding cadherin-related 23, are associated with both
familial and sporadic pituitary adenomas. Am J Hum Genet.
100:817–823. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Karlsson MC, Gonzalez SF, Welin J and Fuxe
J: Epithelial-mesenchymal transition in cancer metastasis through
the lymphatic system. Mol Oncol. 11:781–791. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zaric J, Joseph JM, Tercier S, Sengstag T,
Ponsonnet L, Delorenzi M and Rüegg C: Identification of MAGI1 as a
tumor-suppressor protein induced by cyclooxygenase-2 inhibitors in
colorectal cancer cells. Oncogene. 31:48–59. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
De Jager PL, Srivastava G, Lunnon K,
Burgess J, Schalkwyk LC, Yu L, Eaton ML, Keenan BT, Ernst J, McCabe
C, et al: Alzheimer's disease: Early alterationsin brain DNA
methylation at ANK1, BIN1, RHBDF2 and other loci. Nat Neurosci.
17:1156–63. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lunnon K, Smith R, Hannon E, De Jager PL,
Srivastava G, Volta M, Troakes C, Al-Sarraj S, Burrage J, Macdonald
R, et al: Methylomic profiling implicates cortical deregulation of
ANK1 in Alzheimer's disease. Nat Neurosci. 17:1164–1170. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Vanniya SP, Srisailapathy CRS and Kunka
Mohanram R: The tip link protein Cadherin-23: From hearing loss to
cancer. Pharmacol Res. 130:25–35. 2018. View Article : Google Scholar : PubMed/NCBI
|