Cancer immunotherapy, which aims to foster the host
immune response against cancer to obtain durable anticancer
responses, has achieved marked success in the past decade (1). The programmed death-1 (PD-1)-PD
ligand-1 (PD-L1) receptor-ligand pair is an important immune
checkpoint pathway exploited by tumor cells to evade immune attack
(2). Blockade of the PD-1/PD-L1
axis represents an effective form of cancer immunotherapy (3). To date, several anti-PD-1/PD-L1
antibodies have been designed and assessed in clinical trials for
cancer treatment. Nivolumab and pembrolizumab are humanized,
engineered anti-PD-1 monoantibodies that have demonstrated durable
objective response and improved overall survival in patients with
advanced melanoma or non-small cell lung cancer (NSCLC), supporting
their approved applications in these cancer types (4–10).
Nivolumab also exhibited marked therapeutic activity in patients
with metastatic renal cell carcinoma (RCC) or relapsed/refractory
Hodgkin's lymphoma (11,12). In addition to anti-PD-1 drugs, there
are numerous agents targeting PD-L1 in clinical development at
various phases. Atezolizumab, a monoclonal antibody against PD-L1,
was approved for the treatment of metastatic bladder carcinoma and
NSCLC, based on a prolonged overall survival compared with
chemotherapy (13,14). Another recent phase Ia study
revealed the potential antitumor activity of atezolizumab in
metastatic RCC (15). Other
anti-PD-L1 antibodies, such as durvalumab and avelumab, have been
launched for the treatment of advanced NSCLC, urothelial cancer and
Merkel cell carcinoma.
Despite significant progress of PD-1/PD-L1-directed
immunotherapy, the efficacy and safety profiles of these agents
varies greatly across individual patients and among different tumor
types. Not all patients respond to PD-1/PD-L1 blockade (5,8,9,15).
Moreover, immune checkpoint inhibitors (ICIs) may have
immune-associated adverse events and are usually costly (16,17).
Thus, it is of utmost value to define predictive biomarkers of
response, in order to optimize the application of anti-PD-1/PD-L1
ICIs. The detection of tumor-cell PD-L1 expression via
immunohistochemistry (IHC) has been thus far the most widely
studied biomarker for predicting response to anti-PD-1/PD-L1
immunotherapy. However, a variety of limitations have been found
with the PD-L1 testing, such as different antibodies, different
analysis systems and different cut-off criteria for positivity in
previous clinical trials, and the heterogeneity of PD-L1 expression
between serial sections of one tumor sample (18,19).
While tumor PD-L1 expression is predictive of response to
PD-1/PD-L1 inhibitors, a small fraction of PD-L1-negative patients
can also benefit from PD-1/PD-L1 blockade (6,9,20,21).
There is an urgent need to develop alternative biomarkers to
identify patients who are most likely to respond. This review
provides an overview of the mechanisms of action of the established
PD-L1 testing and other evolving biomarkers to predict the response
to anti-PD-1/PD-L1 therapies. The review also details the
challenges faced by the application of predictive biomarkers, and
proposes directions for future biomarker development and combined
biomarker strategies. This review represents the latest evidence
regarding biomarkers of response to PD-1/PD-L1 blockade for cancer
treatment.
Immune tolerance is considered one of the hallmarks
of cancer that is exploited by tumor cells to evade immune
surveillance and elimination (22)
(Fig. 1). In general, antigen
presenting cells (APCs) in lymphoid tissue can dispose and present
mutant or non-mutant neoantigens to CD4+ and
CD8+ T cells for priming via the interaction between
major histocompatibility complex (MHC) II and T-cell receptor
(TCR). The CD4+ T helper cells also contribute to the
priming of CD8+ T cytotoxic cells via various cytokines.
Both CD4+ and CD8+ T cells are subsequently
activated by the APCs through B7.1/B7.2/CD28 co-stimulatory
pathways, which trigger their proliferation, migration to tumor
sites, secretion of inflammatory cytokines and cytotoxic
activities, leading ultimately to tumor eradication (23). There are multiple mechanisms of
immune tolerance in tumors, including the well-established
B7.1/B7.2/cytotoxic T-lymphocyte-associated antigen-4 (CTLA-4) and
PD-L1/PD-L2/PD-1 checkpoint pathways (24). PD-1 is an inhibitory receptor found
on activated T and B cells, natural killer cells and monocytes,
while its main ligand, PD-L1, is expressed on tumor cells,
dendritic cells, macrophages, stromal cells and activated T cells
(25). Another ligand PD-L2 is
mostly restricted to APCs, such as dendritic cells and monocytes
(26). The mechanism of action is
that PD-L1 results in the tyrosine phosphorylation of PD-1
cytoplasmic immunoreceptor tyrosine-based inhibition motif (ITIM)
and immunoreceptor tyrosine-based switch motif (ITSM), which
recruit phosphatases, particularly Src homology region 2
domain-containing phosphatase-2 (SHP-2) (27). This leads to the dephosphorylation
of TCR proximal signaling molecules, such as ZAP70, PKCθ and CD3δ,
attenuating TCR and CD28 signals, which ultimately promotes T cell
apoptosis, anergy and functional exhaustion (28).
PD-L1 expression in tumors has been hypothesized to
be associated with response to PD-1/PD-L1 blockade. In a phase I
trial, Topalian et al firstly demonstrated an association
between PD-L1 expression in tumor cells and objective response to
nivolumab in multiple cancer types (16). They determined surface PD-L1
expression of pretreated tumor samples via IHC, with a cut-off
value of 5% defined to be PD-L1-positive, and found that 9 out of
25 PD-L1-positive patients had an objective response to nivolumab,
while none of the 17 PD-L1-negative patients had an objective
response. The KEYNOTE-024 study revealed superior progression-free
survival (PFS) and overall survival (OS) in a pembrolizumab
treatment group vs. a platinum-doublet chemotherapy group in
patients with advanced NSCLC and PD-L1 expression in at least 50%
of tumor cells (29). Thus far,
several clinical trials have been performed to compare the
treatment efficiency of anti-PD-1/PD-L1 antibodies between
PD-L1-positive and -negative tumors (6–11,17,21,30–43),
which are summarized in Table SI.
Despite different pretreatments and cut-off points to define PD-L1
positivity, these studies have largely supported a role for PD-L1
expression, either on tumor cells or on tumor-infiltrating immune
cells, as a predictive biomarker of response to PD-1/PD-L1 blockade
in a variety of tumors. Notably, by analyzing multiple tumor types,
Taube et al determined that membranous PD-L1 expression by
tumor cells and infiltrating immune cells was most abundant in
melanoma, NSCLC and RCC; tumors that exhibit objective response to
anti-PD-1 immunotherapy (44).
In addition to PD-L1 expression on tumor cells or
tumor-infiltrating immune cells, other forms of PD-L1 can also
predict response to anti-PD-1/PD-L1 therapy. A recent study by Chen
et al revealed the presence of PD-L1 on the surface of
exosomes released by melanoma cells (45). They found that a fold change in
circulating exosomal PD-L1 >2.43 at weeks 3–6 was associated
with an improved objective response rate (ORR), PFS and OS to
pembrolizumab. Another study of NSCLC suggested that the baseline
plasma soluble PD-L1 concentration, determined using the
enzyme-linked immunosorbent assay method, was significantly
associated with clinical benefit in nivolumab therapy (46). However, lower response rate and
shorter OS were detected in patients with NSCLC and high
plasma-soluble PD-L1 levels.
In numerous tumors, PD-L1 expression can be induced
either via oncogenic drivers and transcriptional factors, or via
cytokines produced by tumor-infiltrating immune cells (47). Thus, PD-L1 acts as a constitutive
and adaptive immune resistance against antitumor immune responses.
The predictive value of PD-L1 expression can be explained by the
fact that inhibiting the PD-1/PD-L1 axis with therapeutic
antibodies allows the host to overcome immune resistance and
thereby activate the antitumor immunity.
Although the results suggest PD-L1 expression as a
predictive biomarker, several clinical trials have repeatedly
demonstrated that there is a small but definite proportion of
PD-L1-negative patients who can also derive clinical benefit from
PD-1/PD-L1 blockade (6,9,20,21).
As summarized in Table SI, ORR to
PD-1/PD-L1 antibodies in PD-L1-negative groups was revealed to be
20–40% in melanomas, 10–20% in NSCLC, and 5–20% in urothelial
carcinomas. Brahmer et al even observed similar ORRs and
survival outcomes between patients with PD-L1-positive and
-negative squamous-cell NSCLC treated with second-line nivolumab,
collectively revealing that there should be predictive biomarkers
other than PD-L1 expression that can also determine the efficacy of
PD-1/PD-L1 inhibitors (9). PD-L1
testing alone is insufficient for the selection of patients for
anti-PD-1/PD-L1 immunotherapy. On the other hand, several studies
indicated that anti-PD-L1 is somewhat less effective than anti-PD-1
therapy, which may be associated with slightly lower toxicity in
cancer treatment (16,48). This discrepancy is potentially due
to the mode of action, targeting the ligand vs. the receptor,
between anti-PD-1 and anti-PD-L1 antibodies. Consistently, our data
also revealed that anti-PD-1 therapy, but not anti-PD-L1, was
effective against FXRhighPD-L1low mouse Lewis
lung carcinoma (LLC) tumors. It speculated that the absence of
targetable PD-L1 on tumor cells may be responsible for the
ineffectiveness of anti-PD-L1 antibody (49). To date, no clinical trials have
directly compared the treatment efficiency and toxicity between
anti-PD-1 and anti-PD-L1 antibodies, particularly in
PD-L1-low/negative patients.
Notably, the application of PD-L1 testing via IHC as
a predictive biomarker is associated with several issues.
Technically, different PD-L1 IHC antibodies with different analysis
systems and different cut-off values for PD-L1 positivity were
employed in early clinical trials (Table SI). The anti-PD-L1 28-2 clone and
22C3 clone were, thus far, the most prevalent antibodies for IHC.
The common thresholds for PD-L1 positivity were 1, 5 and 10% in
multiple cancer types (Table SI).
Encouragingly, recent studies have compared three PD-L1 diagnostic
assays (Dako 28-8, 22C3 and Ventana SP263), revealing preferable
concordance rates at various cut-offs in resected NSCLC samples
(50,51), and great efforts have been paid to
develop a consensus for the use of PD-L1 IHC testing as a
predictive biomarker for anti-PD-1/PD-L1 immunotherapy (52). Essentially, the expression of PD-L1
on tumor cells and infiltrating immune cells is dynamic. It has
been discovered that PD-L1 expression levels can be influenced by
various mechanisms, including the change in intracellular
transcriptional factors or extracellular inflammatory cytokines, as
well as antitumor therapies, such as radiation therapy,
chemotherapy and targeted therapy (53–62).
Recently, we detected an inverse correlation between FXR and PD-L1
expression in a cohort of NSCLC specimens (49). Our data demonstrated that FXR could
directly bind to an FXR-responsive element in PD-L1 promoter
and repress its transcription, suggesting FXR as a novel
transcriptional factor for the regulation of PD-L1. In general,
contemporaneous tumor samples, obtained at the beginning of immune
checkpoint blockade (ICB), should represent the PD-L1 expression
status better compared with archival tumor samples. Another
ineluctable variable is the heterogeneity of PD-L1 expression,
which exists both within the same tumor lesion, and between primary
and metastatic lesions in the same patient. It has been reported
that PD-L1 expression is widely heterogeneous within the tumor,
which often accumulates at the tumor-immune interface (44). Takamori et al compared PD-L1
expression between lung metastases and corresponding primary
tumors, including tumors in the rectum, colon, liver and bile duct.
Although the proportion of PD-L1-positive tumor cells was not
significantly different between lung metastases and primary tumors,
PD-L1 expression on immune cells was significantly higher in lung
metastases compared with the corresponding primary tumors (63). In this regard, tumor sampling aimed
at a particular proportion of one tumor site may not accurately
reflect on the PD-L1 status of that patient. Finally, factors
enabling the prediction of response, such as tumor mutational
burden (TMB) and the tumor microenvironment (TME), should be
incorporated with PD-L1 IHC staining, in order to achieve a
paradigm of precise anti-PD-1/PD-L1 immunotherapy.
Owing to advances in DNA sequencing techniques, a
large amount of information on cancer genetics and genomics has
been gained in the past few decades. There is increasing evidence
that the TMB can predict response to ICIs, including
anti-PD-1/PD-L1 drugs. The first indication was from the combined
result, revealing that tumors with a high TMB (melanoma and NSCLC)
often have a higher response rate to anti-PD-1 or anti-PD-L1
therapy across multiple tumor types (16,48,64–66).
Moreover, within certain tumors, Rizvi et al revealed that
patients with NSCLC and a high non-synonymous TMB exhibited durable
clinical benefit to pembrolizumab, defined as a partial or stable
response rate, for >6 months, compared with patients with less
frequent non-synonymous mutations (67). Consistently, a pilot study of
nivolumab in early-stage NSCLC revealed a significantly higher mean
TMB in tumors with a major pathological response compared with
tumors without a major pathological response (68). Recently, Wang et al reported
that a higher blood TMB (bTMB), established by a 150-cancer gene
panel of circulating tumor DNA, was significantly associated with
superior PFS and ORR in patients with NSCLC treated with anti-PD-1
and anti-PD-L1 drugs (69),
collectively suggesting the potential predictive value of TMB in
antitumor PD-1/PD-L1 blockade.
Notably, recent evidence suggests that mismatch
repair deficiency (MMRD) may be associated with an increased
response to PD-1/PD-L1 blockade (70). Mismatch repair (MMR) is an intrinsic
mechanism that can identify and correct errors in DNA replication
and recombination, such as miss-incorporation deletions and base
insertions (71). Mutations in MMR
genes can produce log-fold increase of TMB, leading to the
detectable DNA microsatellite instability (MSI) (23). It was estimated that tumors with a
MMRD genotype possess 10 to 100-fold the mutational load of their
MMR-proficient counterparts (23).
Le et al have formally evaluated the predictive value of
MMRD in patients with colorectal and non-colorectal cancer treated
with pembrolizumab, and revealed that patients with MMRD colorectal
cancer had a significantly improved ORR and PFS rate compared with
those with MMR-proficient colorectal cancer (72). Moreover, it was demonstrated that
the immune-associated ORR and PFS rate of pembrolizumab treatment
were similar between MMRD colorectal cancers and MMRD
non-colorectal cancers. Whole-exome sequencing revealed a mean of
1782 somatic mutations per tumor in MMRD tumors compared with 73 in
MMR-proficient tumors; however, there were in total 41 cases in
this phase II trial. Subsequently, the study was expanded to 86
patients of 12 tumor types with MMRD, which displayed an objective
radiographic response rate of 53% and a complete response rate of
21% to anti-PD-1 therapy, although the median PFS and OS were not
reached (73). Large clinical
studies are required to verify the potential of MMRD or MSI in
predicting the response to different PD-1/PD-L1 antibodies within
different tumor types.
The association between TMB and response to
anti-PD-1/PD-L1 drugs is considered to be primarily due to the
generation of neoantigens, as a result of somatic mutations in
tumor cells. The theory is that tumors with a high mutational load
often possess more neoantigens, which can be recognized as non-self
epitopes by the immune system, thereby enhancing T cell responses
against tumors, as well as killing tumor cells when the PD-1/PD-L1
axis is blocked (74). It has been
documented that the sensitivity to PD-1 blockade in advanced NSCLC
and melanoma was increased in tumors enriched for clonal
neoantigens (75). In parallel,
active T cells against clonal neoantigens were detected in tumors
with durable clinical benefits. Another study revealed a
significant correlation between higher neoantigen burden and
improved treatment efficacy in patients with NSCLC treated with
pembrolizumab (67). In addition,
the increased PD-L1 expression in the context of certain mutations
is proposed as another determinant. A recent study demonstrated
that the activating mutations in Janus kinase 3 (Jak3) promoted
PD-L1 expression in lung cancer cells and the tumor immune
microenvironment, thus contributing to the durable clinical benefit
from anti-PD-L1 therapy (76). To
date, a variety of oncogenic mutations, such as EGFR, PTEN
and ALK, have been reported to be associated with PD-L1
upregulation in tumor cells, although further investigation is
required to better define the role of PD-L1 in TMB-associated
response to anti-PD-1/PD-L1 immunotherapy (77–79).
In this respect, it is also reasonable to speculate that the
somatic mutations in tumor cells would have a broad effect on the
tumor immune microenvironment; specifically, those affecting other
immune checkpoints, cytokines and chemokines that may also
determine anti-PD-1/PD-L1 therapeutic response. Further studies are
warranted for a comprehensive examination of host immune make-up in
tumors with somatic mutations in comparison with others.
TMB is a promising predictive biomarker, albeit with
its own limitations. Although a number of studies correlate TMB
with tumor response to anti-PD-1/PD-L1 drugs, there has been thus
far, seldom numerical cut-off value of TMB that was formally
defined (16,39,48,66,68).
Rizvi et al established a cut-off point of 178
non-synonymous mutations per sample to predict durable clinical
benefit in a discovery cohort of NSCLC treated with pembrolizumab
(67). In the validation set, this
cut-off point yielded a clinical benefit rate of 75% in patients
harboring at least 178 mutations, compared with 14% in patients
with <178 mutations. Another clinical trial defined ≥10
mutations per megabase in DNA sequences of tumor cells as high TMB
(34). Moreover, while somatic
mutations in tumor cells may produce neoantigens that are prone to
immune attack, not all neoantigens can elicit a T-cell response. A
previous study has revealed that only 10% of non-synonymous point
mutations of an MC38 mouse tumor model could generate peptides
capable of binding to MHC-I with high affinity, and only a
proportion of these peptides, rather than the overall peptide load,
were necessary to elicit immunogenicity (80). Despite recent technological
advances, for example, whole exome sequencing or computational
algorithm, it is still challenging to identify meaningful
neoantigens that are responsible for T-cell responses. In addition,
intratumor mutation heterogeneity should be another obstacle for
predicting response to PD-1/PD-L1 blockade. McGranahan et al
detected considerable intratumor neoantigen heterogeneity within
NSCLC tumors (75). It was
demonstrated in the same study that decreased intratumor neoantigen
heterogeneity was correlated with improved OS of patients with lung
adenocarcinoma. Therefore, mutations that arise early and are
shared by most cancer cells in an individual should elicit more
potent antitumor T-cell responses compared with mutations that
arise later or are limited to a fraction of cancer cells. Lastly,
several studies revealed primary resistance to anti-PD-1/PD-L1 in
tumors with specific mutations, for example LKB1 loss, which
may be ascribed to the impaired antitumor immune responses
(81,82).
The TME consists of non-malignant stromal cells,
such as cancer-associated fibroblasts (CAFs), bone marrow-derived
cells and tumor-infiltrating immune cells, extracellular matrix,
and the blood and lymphatic vascular networks (83). The tumor-infiltrating immune cells
include macrophages, dendritic cells, natural killer cells, B
cells, effector T helper cells, regulatory T cells (Treg) and
cytotoxic T cells. The stromal cells aforementioned can release
growth factors, matrix-degrading enzymes, cytokines and chemokines,
responsible for either antitumor immune response or
immunosuppressive response (84,85).
In addition, both activating markers, such as MHC-II and CD86, and
exhausting markers, such as PD-1, CTLA-4, LAG-3 and TIM-3, can be
expressed in tumor-infiltrating immune cells, which collectively
form a complex host of factors to either combat or promote tumor
progression (86). Previous studies
have revealed that the response of tumor cells to anti-PD-1/PD-L1
therapy is determined not only by intrinsic properties of tumor
cells but also by cellular and molecular components of TME
(87,88).
Firstly, the presence of tumor-infiltrating T cells
was demonstrated to be associated with clinical benefit from
anti-PD-1/PD-L1 therapy. Tumeh et al analyzed tumor samples
from 46 patients with metastatic melanoma treated with
pembrolizumab (89). Higher numbers
of CD8-, PD-1- and PD-L1-expressing T cells were found at the
invasive tumor margin and inside tumors in responding patients
compared with non-responders. Ultimately, it was validated that the
presence of CD8+ T cells at the invasive tumor margin
serves as a potential predictive biomarker to anti-PD-1 therapy in
melanoma. Chen et al revealed a modest association between
CD8+, CD3+ and CD45RO+ T cells in
pretreated tumor samples and responsiveness to PD-1 blockade in
patients with advanced melanoma (90). Notably, this association became more
significant after anti-PD-1 therapy. Another study on melanoma
found that an increasing proportion of
PD-1highCTLA-4high subset within
tumor-infiltrating CD8+ T cells strongly correlated with
objective response to anti-PD-1 therapy (91). In contrast, PD-1+
tumor-infiltrating T cells were significantly decreased in brain
metastatic lesions compared with primary lung cancer, which was
associated with a lower likelihood of objective response to
anti-PD-1 in brain metastases (92). Consistently, our study revealed that
aside from the downregulated PD-L1 expression, enforced FXR
expression generated an immunosuppressive microenvironment in mouse
LLC tumors, characterized by the inactivated and exhausted
CD8+ T cells (shown as decreased
TNFα+CD8+ T cells, as well as increased LAG-3
expression on CD8+ T cells), which was correlated with
significant tumor growth inhibition in FXR-overexpressed LLC tumors
treated with anti-PD-1 antibody (49). These findings collectively suggested
that pretreated tumor-infiltrating T cells, particularly for the
exhausted CD8+ T cells, can act as a promising
predictive biomarker for PD-1/PD-L1 blockade (Fig. 2). This phenomenon can be logically
ascribed to the pre-existing T cell-mediated antitumor immunity,
which is restrained by the PD-1/PD-L1-induced suppression, but can
be reactivated via PD-1/PD-L1 blockade. However, it was also
proposed that intratumor T cells expressing multiple exhaustion
markers may be irresponsive to anti-PD-1/PD-L1 drugs (93). Kim et al reported that VEGF-A
could induce transcription factor TOX expression in T cells to
drive exhaustion-specific transcription program, accounting for the
resistance to PD-1 blockade in microsatellite-stable colorectal
cancer (94). Another study even
defined a threshold for PD-1 downregulation on tumor-infiltrating
CD8+ T cells, for which the release of adaptive immune
resistance could be achieved via PD-1 blockade (95). It was revealed that the
functionality of PD-1high T cells in resistant tumors
failed to be rescued by anti-PD-1 therapy. All these lend credence
to the theory that the less exhausted tumor-infiltrating
CD8+ T cells, rather than the over-exhausted
populations, are probably the determinants of response to
anti-PD-1/PD-L1 immunotherapy. A future area of research should be
to evaluate the predictive value of exhausted CD8+ T
cells, of various phenotypes within TME, in PD-1/PD-L1-based
immunotherapy.
Secondly, the immunosuppressive cell populations in
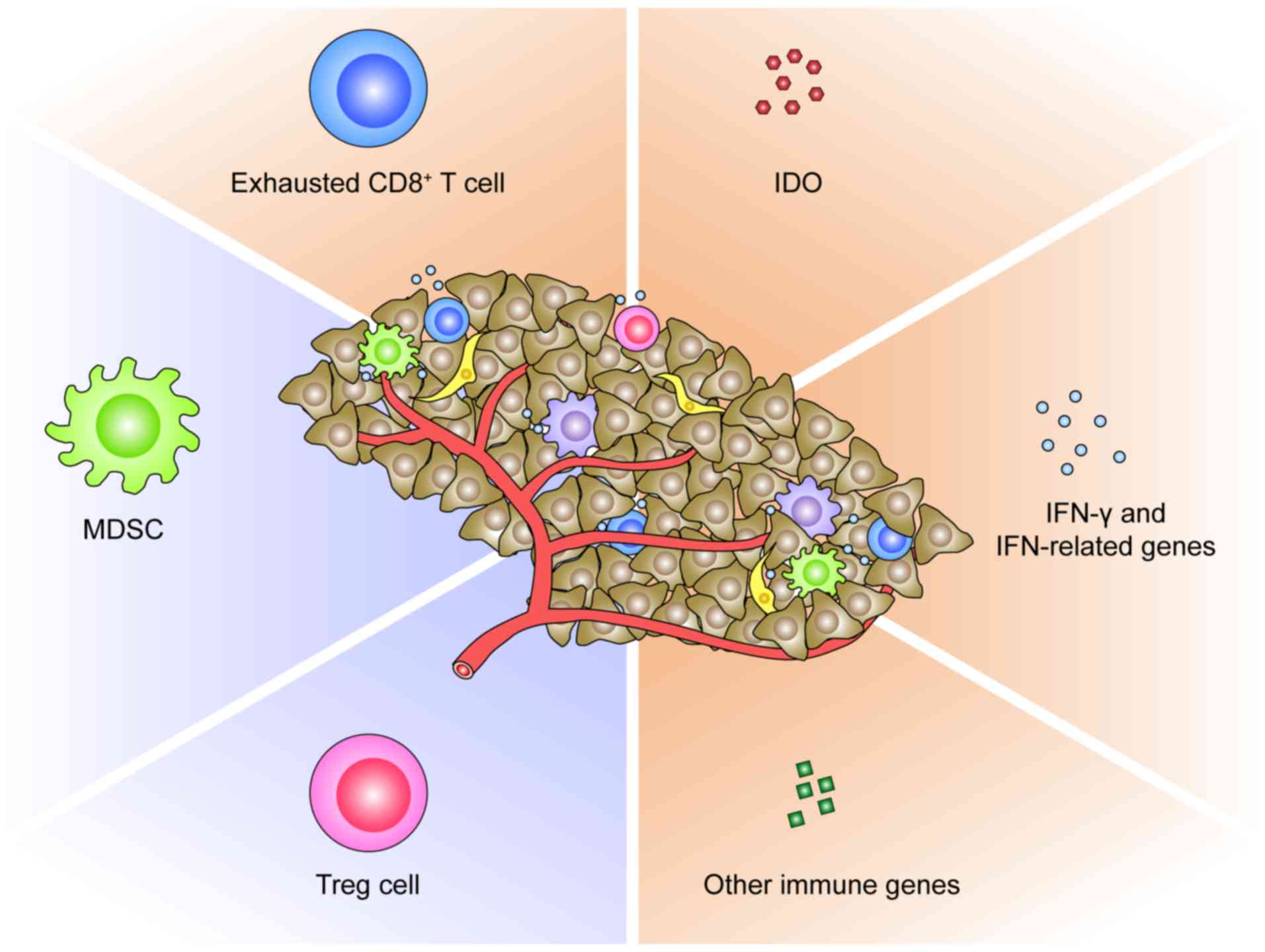
TME could restrain the response to PD-1/PD-L1 blockade (Fig. 2). It has been documented that
tumor-infiltrating myeloid cells and Treg cells are partially
responsible for the development of anti-PD-1 resistance in mouse
colorectal and mammary cancer (95). Davis et al revealed that the
functional inhibition of myeloid-derived suppressor cells (MDSCs)
could enhance the objective response to anti-PD-L1 treatment in T
cell-inflamed mouse tumor models of head and neck cancers (96). In a transgenic mouse model of
neuroblastoma, Mao et al revealed that while checkpoint
inhibitors were insufficient in controlling mouse neuroblastoma
growth, combining suppressive myeloid cell inhibitor with
anti-PD-1/PD-L1 antibodies resulted in superior tumor control
(97). In this regard, the
inhibition of immunosuppressive cell subsets within TME represents
a potential predictive biomarker or rational approach to enhance
antitumor PD-1/PD-L1 blockade.
Thirdly, the molecular profiles of TME pre- or
post-anti- PD-1/PD-L1 immunotherapy represent alternative
biomarkers of response (Fig. 2).
Indoleamine 2,3-dioxygenase (IDO), a rate-limiting enzyme in the
degradation of tryptophan via the kynurenine pathway, plays a
critical role in suppressing T-cell immunity within tumors
(98). A recent study revealed that
MSI colorectal cancer overexpressing IDO were more responsive to
anti-PD-1 treatment compared with microsatellite-stable cancer
(99). The IFN-related genes are
also relevant in patient selection for anti-PD-1/PD-L1 therapies.
In a phase I/II study of durvalumab, an ORR of 33% was revealed in
NSCLC patients positive for IFN-γ mRNA and 8% in those negative for
IFN-γ mRNA (100). Consistently,
in another study on atezolizumab, pretreatment melanoma samples
from responding patients had increased expression of IFN-γ and
IFN-related genes compared with non-responding samples (66). In contrast, Zaretsky et al
revealed that defects in IFN receptor signaling pathways resulted
in acquired resistance to anti-PD-1 therapy in melanoma (81). In addition, in a study
characterizing the gene expression profiles of RCC treated with
anti-PD-1 antibodies, immune genes such as BACH2, a
regulator of CD4+ T cell differentiation, and
CCL3 involved in leukocyte migration were revealed to be
overexpressed in responding patients as compared with
non-responders (101). Chen et
al analyzed immune signatures in longitudinal tumor samples
obtained at multiple time-points during anti-PD-1 therapies, and
identified numerous genes to be differentially expressed between
responders and non-responders (90). However, there is thus far no
conclusive data on the predictive power of either IDO, or
IFN-related genes, or other immune genes in TME for anti-PD-1/PD-L1
immunotherapy.
The close association between the aforementioned TME
components and treatment efficacy in PD-1/PD-L1 blockade can be
explained by the following theory. Immune recognition of tumors
results in a host-immune response, which promotes tumor eradication
through immune mechanisms, including antigen presentation, T cell
priming and trafficking, cytokine production, cytotoxic activity
and the expression of other immune genes. However, the antitumor
Th1 and CD8+ T cell responses are negatively regulated
by PD-1/PD-L1-mediated adaptive immune resistance (89). Other immunosuppressive constituents
also contribute to the adaptive immune resistance. This is
supported by the combined result, revealing that the upregulation
of PD-L1 and IDO in response to IFN-γ promotes the development of
tumor-infiltrating myeloid cells and Treg cells, thereby
facilitating tumor immune evasion (102–104). Response to anti-PD-1/PD-L1
immunotherapy occurs primarily in cancer patients with a T
cell-inflamed but adaptive immune-resistant TME (105). In addition, the potential
interactions between TME components and PD-L1 expression have
partially been disclosed before. PD-L1 expression in tumors
represents a negative feedback to IFN-γ released by
tumor-infiltrating immune cells, including macrophages, dendritic
cells, natural killer cells, B cells and effector T helper cells
(56,106). Other immune factors within TME,
such as tumor necrosis factor-α (TNF-α), interleukin-6 (IL-6),
IL-12, IL-4, IL-10 and transforming growth factor-β (TGF-β), have
also been revealed to induce the expression of PD-L1 (57–59,107–109). In return, PD-L1 was documented to
compromise the effector T-cell responses, promote the
differentiation of induced Treg cells, as well as mediate the
immunosuppressive activity of myeloid cells in tumors (89,102,103).
Traditionally, the immune profiles of tumors can be
classified into three main phenotypes: The immune-inflamed
phenotype, the immune-excluded phenotype and the immune-desert
phenotype; based on whether tumors harbor an inflammatory TME or
not (93). In this scenario, the
immune-inflamed phenotype is postulated to correlate with a higher
response rate to anti-PD-1/PD-L1 therapy. Recently, a new theory
arose that according to both tumor-infiltrating lymphocytes (TILs)
and PD-L1 expression in tumors, the TME can be stratified into four
groups: TILs−PD-L1−,
TILs−PD-L1+,
TILs+PD-L1− and
TILs+PD-L1+ (88). While the
TILs−PD-L1− group may exhibit lack of
response to anti-PD-1/PD-L1 antibodies, and the
TILs+PD-L1− group is irresponsive to
anti-PD-L1, the TILs+PD-L1+ group would
expect the best response to anti-PD-1/PD-L1 treatments. An
alternative classification of the TME immune types provided by
Kondou et al was determined by the expression level of the
PD-L1 and CD8b genes (110). The
aforementioned stratifications should enable the selection of
optimal treatment strategies for patients with cancer. More
recently, the effects of TME on PD-L1 expression have attracted
much attention, particularly for those with a pre-existing T
cell-inflamed phenotype. The adaptive CD8+ T cells,
CAFs, M2-like macrophages, and corresponding cytokines were
revealed to engage in the dynamic change of PD-L1 within local
tumors (106,108,111). These findings support the
predictive value of TME-derived cellular and molecular elements as
a whole population, rather than in individuals, for anti-PD-1/PD-L1
immunotherapy. With the discovery of more determinants for ICB in
the past decade, more attention is expected to be devoted to
evaluating a spectrum of ‘immunome’ before anti-PD-1/PD-L1
therapies.
Despite the known predictive biomarkers, such as
PD-L1 expression, TMB and TME profiles, additional research is
still necessary to explore other reliable candidate biomarkers for
predicting the response of patients to anti-PD-1/PD-L1 therapies.
In fact, emerging data have indicated future directions for
biomarker development.
Immunogenic cell death (ICD) in tumors has been
implicated in the response to PD-1/PD-L1 blockade. Recently, Zhao
et al reported that irreversible electroporation could
induce ICD in pancreatic ductal adenocarcinomas, which activated
dendritic cells and alleviated stroma-induced immunosuppression
(112). The combination of
irreversible electroporation and anti-PD-1 resulted in a
significantly longer median survival in mouse orthotopic pancreatic
ductal adenocarcinoma models compared with mice administered either
treatment alone. Another study revealed that the combination of
cisplatin and high-dose crizotinib induced ICD in NSCLC cells,
which synergized with anti-PD-1 to induce a superior cure rate and
long-term survival in mouse orthotopic NSCLC models (113). It is considered that the dying
tumor cells may function as tumor vaccines to stimulate antitumor
immune responses during ICD.
The diverse T-cell repertoire, corresponding to
different antigenic peptides bounding to class I or II MHC, is
generated by random recombination of discrete TCR-αβ gene segments
(114). Tumeh et al
analyzed tumor samples from 46 patients with metastatic melanoma
obtained before and during anti-PD-1 therapy (89). It was revealed that patients who met
the criteria for radiographic response had more than 10 times as
many TCR clones expansion after pembrolizumab treatment, suggesting
that the TCR clonality may represent a promising on-treatment
predictive biomarker for anti-PD-1 therapies. The predictive value
of TCR clonality during anti-PD-1 can be interpreted as an
enrichment of a diverse T-cell repertoire, which will eventually be
tailored as antigenic peptides on MHC interaction with TCR,
reflecting an activated immune response against tumor cells
(115).
Recently, intensive studies have been conducted to
dissect the impact of the gut microbiome on response to
anti-PD-1/PD-L1 immunotherapy in human malignancies, including
melanoma, hepatocellular carcinoma, gastric cancer and NSCLC
(116–119). It has been acknowledged that
patients responding to PD-1/PD-L1 blockade often harbor higher
diversity of gut microbiome compared with non-responders (116,118,119). This effect can be partially
attributed to the ability of the gut microbiome to activate the
host innate immune responses (120). In addition, subsequent studies
revealed a clearer association between an intact commensal
bacterial community and robust antitumor T-cell responses (121,122). Other studies revealed that
interventions to modulate gut bacterial profiles exhibited great
promise for improving the therapeutic responses (121,123). Dong et al reported that
diosgenin, a natural steroidal saponin, could modulate the
composition of the intestinal microbiome in melanoma-bearing mice,
thereby enhancing the growth-inhibitory effect of anti-PD-1
antibody (123). Another study
revealed that oral administration of Bifidobacterium
collaborated with anti-PD-L1 therapy to control tumor growth in
mouse melanoma models (121).
Although promising, the favorable bacterial species that improve
antitumor PD-1/PD-L1 blockade remain to be determined.
The evaluation of peripheral blood could be another
interesting approach. It has been reported by Weide et al
that relative eosinophil count (≥1.5%), relative lymphocyte count
(≥17.5%), LDH level (≤2.5-fold elevation), and the absence of
metastases other than soft-tissue/lung were confirmed as
independent baseline characteristics associated with favorable OS
in patients with melanoma treated with pembrolizumab (124). Another study reported that low
neutrophil to lymphocyte ratio and absolute neutrophil count during
treatment was correlated with superior objective response and
treatment duration in patients with advanced NSCLC cured by
PD-1/PD-L1 inhibitors (125).
Similarly, data from a study on patients with advanced/metastatic
melanoma treated with nivolumab or pembrolizumab revealed that
patients with an increased baseline LDH had a significantly shorter
OS compared with those with normal LDH. Furthermore, patients with
a relative increase of >10% from elevated baseline LDH during
treatment had a significantly shorter OS compared with those with
≤10% change (126). Other
parameters, such as peripheral Treg cells, antigen-specific
CD8+ T cells and MDSCs, were also revealed to be
associated with response to nivolumab in patients with melanoma
(21,127). These factors may be associated
with adaptive immune resistance, which can be overcome when the
PD-1/PD-L1 axis is blocked, although the detailed mechanisms need
to be elucidated in future. In clinical practice, these peripheral
blood biomarkers have the advantage of being readily assessable,
and are suitable for serial sampling during treatment.
Owing to advances in technology, medical imaging
can not only assess macroscopic features, but also characterize the
cellular and molecular properties of malignant lesions, which may
serve as a novel approach to select patients for PD-1/PD-L1
blockade. It has recently been revealed that PD-L1 and PD-1
expression in NSCLC could be quantified non-invasively with PET-CT
imaging using the radiotracers 18F-BMS-986192 and
89Zr-nivolumab, respectively (128). Bensch et al conducted
another first-in-human study characterizing the biodistribution of
zirconium-89-labeled atezolizumab via PET within 22 patients across
three tumor types (129). Notably,
they found that zirconium-89-labeled atezolizumab tumor uptake was
positively correlated with ORR, PFS and OS of enrolled patients
treated with atezolizumab. TILs can also be traced by medical
imaging. In a retrospective study by Sun et al a radiomic
signature that included eight variables was established for
assessing tumor-infiltrating CD8+ T cells in solid
cancer types. Their data revealed that a high baseline radiomic
score was associated with a higher proportion of patients who had
an objective response at 3 or 6 months, as well as associated with
improved OS either in univariate or in multivariate analysis
(130). These imaging biomarkers
were designed to detect and monitor antitumor immune activities
within tumors, thereby predicting response to PD-1/PD-L1 blockade.
Although just the beginning, image-driven biomarkers have the
unique advantage of being non-invasive, which should make them
promising candidates to aid future anti-PD-1/PD-L1
immunotherapy.
The clinical use, applicable tumors, as well as
predicted clinical outcomes of each biomarker discussed in this
review are summarized in Table I.
ICIs, particularly PD-1/PD-L1-targeted antibodies, are proven to be
effective in a variety of cancer types. The establishment of
reliable predictive biomarkers to ensure the rational use of these
agents is crucial, given the reality that only a subset of patients
can benefit from PD-1/PD-L1 blockade, and that treatment with these
agents carries a risk of immune-associated toxicities and
substantial financial burden (5,8,9,15).
Conversely, based on the result of multiple validated biomarker
assays, even patients who are stratified as non-responders for
anti-PD-1/PD-L1 monotherapy may be treated with other antitumor
regimens or combined treatment strategies to maximize clinical
outcomes (131).
Combined biomarker strategies may enrich responders
to anti-PD-1/PD-L1 immunotherapy in future. Factors enabling
response prediction, such as PD-L1 IHC testing, TMB and TME
profiles, should be incorporated together, since it has been
indicated that high tumor PD-L1 expression does not equate to a T
cell-inflamed content, and that high TMB does not always indicate
pre-existing antitumor immune activities (132,133). In fact, evidence has been
increasingly showing that combining data from tumor immune
profiling, including CD3, CD8, FoxP3, CD163, PD-L1, PD-1 and
CTLA-4, could be more ideal approaches to predicting response to
PD-1/PD-L1 blockade, although the optimal model has yet to be
determined (91,134).
Overall, anti-PD-1/PD-L1 immunotherapy is one of
the predominant methods for cancer treatment. The improvement in
the understanding of the interactions among multiple factors, as
well as the dynamic changes of certain variables within different
tumor types will certainly help identify more reliable and
effective predictors for PD-1/PD-L1 blockade, thereby paving the
way for a framework that allows treatment decisions to be made on a
personalized basis.
Not applicable.
The present study was supported in part by grant
nos. 81902325 and 81903633 from the National Natural Science
Foundation of China, grant no. 2017WS026 from the Health and Family
Planning Commission of Shandong Province (China), and grant nos.
2019GSF107042 and 2019GSF107051 from the Key Research and
Development Plan of Shandong Province (China).
Not applicable.
LS and SJ devised the conceptual idea. WY, BS and
JS performed the literature search. WY prepared the figures. WY,
XL, LS and SJ wrote, reviewed and revised the manuscript. All
authors read and approved the final manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Mellman I, Coukos G and Dranoff G: Cancer
immunotherapy comes of age. Nature. 480:480–489. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Butte MJ, Keir ME, Phamduy TB, Sharpe AH
and Freeman GJ: Programmed death-1 ligand 1 interacts specifically
with the B7-1 costimulatory molecule to inhibit T cell responses.
Immunity. 27:111–122. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zou W, Wolchok JD and Chen L: PD-L1
(B7-H1) and PD-1 pathway blockade for cancer therapy: Mechanisms,
response biomarkers, and combinations. Sci Transl Med.
8:328rv42016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Topalian SL, Sznol M, McDermott DF, Kluger
HM, Carvajal RD, Sharfman WH, Brahmer JR, Lawrence DP, Atkins MB,
Powderly JD, et al: Survival, durable tumor remission, and
long-term safety in patients with advanced melanoma receiving
nivolumab. J Clin Oncol. 32:1020–1030. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Robert C, Schachter J, Long GV, Arance A,
Grob JJ, Mortier L, Daud A, Carlino MS, McNeil C, Lotem M, et al:
Pembrolizumab versus Ipilimumab in advanced melanoma. N Engl J Med.
372:2521–2532. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Larkin J, Chiarion-Sileni V, Gonzalez R,
Grob JJ, Cowey CL, Lao CD, Schadendorf D, Dummer R, Smylie M,
Rutkowski P, et al: Combined nivolumab and Ipilimumab or
Monotherapy in untreated melanoma. N Engl J Med. 373:23–34. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Robert C, Long GV, Brady B, Dutriaux C,
Maio M, Mortier L, Hassel JC, Rutkowski P, McNeil C,
Kalinka-Warzocha E, et al: Nivolumab in previously untreated
melanoma without BRAF mutation. N Engl J Med. 372:320–330. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Borghaei H, Paz-Ares L, Horn L, Spigel DR,
Steins M, Ready NE, Chow LQ, Vokes EE, Felip E, Holgado E, et al:
Nivolumab versus Docetaxel in advanced nonsquamous non-small-cell
lung cancer. N Engl J Med. 373:1627–1639. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Brahmer J, Reckamp KL, Baas P, Crino L,
Eberhardt WE, Poddubskaya E, Antonia S, Pluzanski A, Vokes EE,
Holgado E, et al: Nivolumab versus Docetaxel in advanced
squamous-cell non-small-cell lung cancer. N Engl J Med.
373:123–135. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Garon EB, Rizvi NA, Hui R, Leighl N,
Balmanoukian AS, Eder JP, Patnaik A, Aggarwal C, Gubens M, Horn L,
et al: Pembrolizumab for the treatment of non-small-cell lung
cancer. N Engl J Med. 372:2018–2028. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Motzer RJ, Escudier B, McDermott DF,
George S, Hammers HJ, Srinivas S, Tykodi SS, Sosman JA, Procopio G,
Plimack ER, et al: Nivolumab versus Everolimus in advanced
renal-cell carcinoma. N Engl J Med. 373:1803–1813. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ansell SM, Lesokhin AM, Borrello I,
Halwani A, Scott EC, Gutierrez M, Schuster SJ, Millenson MM, Cattry
D, Freeman GJ, et al: PD-1 blockade with nivolumab in relapsed or
refractory Hodgkin's lymphoma. N Engl J Med. 372:311–319. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Powles T, Eder JP, Fine GD, Braiteh FS,
Loriot Y, Cruz C, Bellmunt J, Burris HA, Petrylak DP, Teng SL, et
al: MPDL3280A (anti-PD-L1) treatment leads to clinical activity in
metastatic bladder cancer. Nature. 515:558–562. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Rittmeyer A, Barlesi F, Waterkamp D, Park
K, Ciardiello F, von Pawel J, Gadgeel SM, Hida T, Kowalski DM, Dols
MC, et al: Atezolizumab versus docetaxel in patients with
previously treated non-small-cell lung cancer (OAK): A phase 3,
open-label, multicentre randomised controlled trial. Lancet.
389:255–265. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
McDermott DF, Sosman JA, Sznol M, Massard
C, Gordon MS, Hamid O, Powderly JD, Infante JR, Fasso M, Wang YV,
et al: Atezolizumab, an Anti-programmed Death-Ligand 1 antibody, in
metastatic renal cell carcinoma: Long-term safety, clinical
activity, and immune correlates from a phase Ia study. J Clin
Oncol. 34:833–842. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Topalian SL, Hodi FS, Brahmer JR,
Gettinger SN, Smith DC, McDermott DF, Powderly JD, Carvajal RD,
Sosman JA, Atkins MB, et al: Safety, activity, and immune
correlates of anti-PD-1 antibody in cancer. N Engl J Med.
366:2443–2454. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Fehrenbacher L, Spira A, Ballinger M,
Kowanetz M, Vansteenkiste J, Mazieres J, Park K, Smith D,
Artal-Cortes A, Lewanski C, et al: Atezolizumab versus docetaxel
for patients with previously treated non-small-cell lung cancer
(POPLAR): A multicentre, open-label, phase 2 randomised controlled
trial. Lancet. 387:1837–1846. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Patel SP and Kurzrock R: PD-L1 expression
as a predictive biomarker in cancer immunotherapy. Mol Cancer Ther.
14:847–856. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
McLaughlin J, Han G, Schalper KA,
Carvajal-Hausdorf D, Pelekanou V, Rehman J, Velcheti V, Herbst R,
LoRusso P and Rimm DL: Quantitative assessment of the heterogeneity
of PD-L1 expression in non-small-cell lung cancer. JAMA Oncol.
2:46–54. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Daud AI, Wolchok JD, Robert C, Hwu WJ,
Weber JS, Ribas A, Hodi FS, Joshua AM, Kefford R, Hersey P, et al:
Programmed Death-Ligand 1 expression and response to the
anti-programmed Death 1 antibody pembrolizumab in melanoma. J Clin
Oncol. 34:4102–4109. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Weber JS, Kudchadkar RR, Yu B, Gallenstein
D, Horak CE, Inzunza HD, Zhao X, Martinez AJ, Wang W, Gibney G, et
al: Safety, efficacy, and biomarkers of nivolumab with vaccine in
ipilimumab-refractory or -naive melanoma. J Clin Oncol.
31:4311–4318. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Topalian SL, Taube JM, Anders RA and
Pardoll DM: Mechanism-driven biomarkers to guide immune checkpoint
blockade in cancer therapy. Nat Rev Cancer. 16:275–287. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zou W: Immunosuppressive networks in the
tumour environment and their therapeutic relevance. Nat Rev Cancer.
5:263–274. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Greenwald RJ, Freeman GJ and Sharpe AH:
The B7 family revisited. Annu Rev Immunol. 23:515–548. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Keir ME, Butte MJ, Freeman GJ and Sharpe
AH: PD-1 and its ligands in tolerance and immunity. Annu Rev
Immunol. 26:677–704. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Freeman GJ: Structures of PD-1 with its
ligands: Sideways and dancing cheek to cheek. Proc Natl Acad Sci
USA. 105:10275–10276. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chemnitz JM, Parry RV, Nichols KE, June CH
and Riley JL: SHP-1 and SHP-2 associate with immunoreceptor
tyrosine-based switch motif of programmed death 1 upon primary
human T cell stimulation, but only receptor ligation prevents T
cell activation. J Immunol. 173:945–954. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Reck M, Rodriguez-Abreu D, Robinson AG,
Hui R, Csoszi T, Fulop A, Gottfried M, Peled N, Tafreshi A, Cuffe
S, et al: Pembrolizumab versus chemotherapy for PD-L1-positive
non-small-cell lung cancer. N Engl J Med. 375:1823–1833. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Weber JS, D'Angelo SP, Minor D, Hodi FS,
Gutzmer R, Neyns B, Hoeller C, Khushalani NI, Miller WH Jr, Lao CD,
et al: Nivolumab versus chemotherapy in patients with advanced
melanoma who progressed after anti-CTLA-4 treatment (CheckMate
037): A randomised, controlled, open-label, phase 3 trial. Lancet
Oncol. 16:375–384. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Rizvi NA, Mazieres J, Planchard D,
Stinchcombe TE, Dy GK, Antonia SJ, Horn L, Lena H, Minenza E,
Mennecier B, et al: Activity and safety of nivolumab, an anti-PD-1
immune checkpoint inhibitor, for patients with advanced, refractory
squamous non-small-cell lung cancer (CheckMate 063): A phase 2,
single-arm trial. Lancet Oncol. 16:257–265. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ferris RL, Blumenschein G Jr, Fayette J,
Guigay J, Colevas AD, Licitra L, Harrington K, Kasper S, Vokes EE,
Even C, et al: Nivolumab for recurrent squamous-cell carcinoma of
the head and neck. N Engl J Med. 375:1856–1867. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Carbone DP, Reck M, Paz-Ares L, Creelan B,
Horn L, Steins M, Felip E, van den Heuvel MM, Ciuleanu TE, Badin F,
et al: First-line nivolumab in stage IV or recurrent non-small-cell
lung cancer. N Engl J Med. 376:2415–2426. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hellmann MD, Ciuleanu TE, Pluzanski A, Lee
JS, Otterson GA, Audigier-Valette C, Minenza E, Linardou H, Burgers
S, Salman P, et al: Nivolumab plus Ipilimumab in lung cancer with a
high tumor mutational burden. N Engl J Med. 378:2093–2104. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hellmann MD, Paz-Ares L, Bernabe Caro R,
Zurawski B, Kim SW, Carcereny Costa E, Park K, Alexandru A,
Lupinacci L, de la Mora Jimenez E, et al: Nivolumab plus Ipilimumab
in advanced non-small-cell lung cancer. N Engl J Med.
381:2020–2031. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Herbst RS, Baas P, Kim DW, Felip E,
Perez-Gracia JL, Han JY, Molina J, Kim JH, Arvis CD, Ahn MJ, et al:
Pembrolizumab versus docetaxel for previously treated,
PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010):
A randomised controlled trial. Lancet. 387:1540–1550. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Eggermont AMM, Blank CU, Mandala M, Long
GV, Atkinson V, Dalle S, Haydon A, Lichinitser M, Khattak A,
Carlino MS, et al: Adjuvant pembrolizumab versus placebo in
resected stage III melanoma. N Engl J Med. 378:1789–1801. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Mok TSK, Wu YL, Kudaba I, Kowalski DM, Cho
BC, Turna HZ, Castro G Jr, Srimuninnimit V, Laktionov KK,
Bondarenko I, et al: Pembrolizumab versus chemotherapy for
previously untreated, PD-L1-expressing, locally advanced or
metastatic non-small-cell lung cancer (KEYNOTE-042): A randomised,
open-label, controlled, phase 3 trial. Lancet. 393:1819–1830. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Rosenberg JE, Hoffman-Censits J, Powles T,
van der Heijden MS, Balar AV, Necchi A, Dawson N, O'Donnell PH,
Balmanoukian A, Loriot Y, et al: Atezolizumab in patients with
locally advanced and metastatic urothelial carcinoma who have
progressed following treatment with platinum-based chemotherapy: A
single-arm, multicentre, phase 2 trial. Lancet. 387:1909–1920.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Balar AV, Galsky MD, Rosenberg JE, Powles
T, Petrylak DP, Bellmunt J, Loriot Y, Necchi A, Hoffman-Censits J,
Perez- Gracia JL, et al: Atezolizumab as first-line treatment in
cisplatin-ineligible patients with locally advanced and metastatic
urothelial carcinoma: A single-arm, multicentre, phase 2 trial.
Lancet. 389:67–76. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Petrylak DP, Powles T, Bellmunt J, Braiteh
F, Loriot Y, Morales-Barrera R, Burris HA, Kim JW, Ding B, Kaiser
C, et al: Atezolizumab (MPDL3280A) monotherapy for patients with
metastatic urothelial cancer: Long-term outcomes from a phase 1
study. JAMA Oncol. 4:537–544. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Socinski MA, Jotte RM, Cappuzzo F, Orlandi
F, Stroyakovskiy D, Nogami N, Rodriguez-Abreu D, Moro-Sibilot D,
Thomas CA, Barlesi F, et al: Atezolizumab for first-line treatment
of metastatic Nonsquamous NSCLC. N Engl J Med. 378:2288–2301. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Powles T, O'Donnell PH, Massard C, Arkenau
HT, Friedlander TW, Hoimes CJ, Lee JL, Ong M, Sridhar SS, Vogelzang
NJ, et al: Efficacy and safety of durvalumab in locally advanced or
metastatic urothelial carcinoma: Updated results from a phase 1/2
open-label study. JAMA Oncol. 3:e1724112017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Taube JM, Klein A, Brahmer JR, Xu H, Pan
X, Kim JH, Chen L, Pardoll DM, Topalian SL and Anders RA:
Association of PD-1, PD-1 ligands, and other features of the tumor
immune microenvironment with response to anti-PD-1 therapy. Clin
Cancer Res. 20:5064–5074. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Chen G, Huang AC, Zhang W, Zhang G, Wu M,
Xu W, Yu Z, Yang J, Wang B, Sun H, et al: Exosomal PD-L1
contributes to immunosuppression and is associated with anti-PD-1
response. Nature. 560:382–386. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Okuma Y, Wakui H, Utsumi H, Sagawa Y,
Hosomi Y, Kuwano K and Homma S: Soluble programmed cell death
ligand 1 as a novel biomarker for nivolumab therapy for
non-small-cell lung cancer. Clin Lung Cancer. 19:410–417.e1. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Ritprajak P and Azuma M: Intrinsic and
extrinsic control of expression of the immunoregulatory molecule
PD-L1 in epithelial cells and squamous cell carcinoma. Oral Oncol.
51:221–228. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Brahmer JR, Tykodi SS, Chow LQ, Hwu WJ,
Topalian SL, Hwu P, Drake CG, Camacho LH, Kauh J, Odunsi K, et al:
Safety and activity of anti-PD-L1 antibody in patients with
advanced cancer. N Engl J Med. 366:2455–2465. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
You W, Li L, Sun D, Liu X, Xia Z, Xue S,
Chen B, Qin H, Ai J and Jiang H: Farnesoid X receptor constructs an
immunosuppressive microenvironment and sensitizes
FXRhigh PD-L1low NSCLC to Anti-PD-1
immunotherapy. Cancer Immunol Res. 7:990–1000. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Ratcliffe MJ, Sharpe A, Barker C, Scorer P
and Walker J: Abstract LB-094: A comparative study of PD-L1
diagnostic assays and the classification of patients as PD-L1
positive and PD-L1 negative. Cancer Res. 76 (14
Suppl):LB-094-LB-094. 2016.
|
|
51
|
Fujimoto D, Yamashita D, Fukuoka J,
Kitamura Y, Hosoya K, Kawachi H, Sato Y, Nagata K, Nakagawa A,
Tachikawa R, et al: Comparison of PD-L1 assays in non-small cell
lung cancer: 22C3 pharmDx and SP263. Anticancer Res. 38:6891–6895.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Lim JS, Sundar R, Chenard-Poirier M, Lopez
J and Yap TA: Emerging biomarkers for PD-1 pathway cancer therapy.
Biomark Med. 11:53–67. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Noman MZ and Chouaib S: Targeting hypoxia
at the forefront of anticancer immune responses. Oncoimmunology.
3:e9544632014. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Gowrishankar K, Gunatilake D, Gallagher
SJ, Tiffen J, Rizos H and Hersey P: Inducible but not constitutive
expression of PD-L1 in human melanoma cells is dependent on
activation of NF-κB. PLoS One. 10:e01234102015. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Liu J, Hamrouni A, Wolowiec D, Coiteux V,
Kuliczkowski K, Hetuin D, Saudemont A and Quesnel B: Plasma cells
from multiple myeloma patients express B7-H1 (PD-L1) and increase
expression after stimulation with IFN-{gamma} and TLR ligands via a
MyD88-, TRAF6-, and MEK-dependent pathway. Blood. 110:296–304.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Lee SJ, Jang BC, Lee SW, Yang YI, Suh SI,
Park YM, Oh S, Shin JG, Yao S, Chen L, et al: Interferon regulatory
factor-1 is prerequisite to the constitutive expression and
IFN-gamma-induced upregulation of B7-H1 (CD274). FEBS Lett.
580:755–762. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Kondo A, Yamashita T, Tamura H, Zhao W,
Tsuji T, Shimizu M, Shinya E, Takahashi H, Tamada K, Chen L, et al:
Interferon-gamma and tumor necrosis factor-alpha induce an
immunoinhibitory molecule, B7-H1, via nuclear factor-kappaB
activation in blasts in myelodysplastic syndromes. Blood.
116:1124–1131. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Jin YH, Hou W, Kang HS, Koh CS and Kim BS:
The role of interleukin-6 in the expression of PD-1 and PDL-1 on
central nervous system cells following infection with Theiler's
murine encephalomyelitis virus. J Virol. 87:11538–11551. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Xiong HY, Ma TT, Wu BT, Lin Y and Tu ZG:
IL-12 regulates B7-H1 expression in ovarian cancer-associated
macrophages by effects on NF-κB signalling. Asian Pac J Cancer
Prev. 15:5767–5772. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Akbay EA, Koyama S, Carretero J, Altabef
A, Tchaicha JH, Christensen CL, Mikse OR, Cherniack AD, Beauchamp
EM, Pugh TJ, et al: Activation of the PD-1 pathway contributes to
immune escape in EGFR-driven lung tumors. Cancer Discov.
3:1355–1363. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Hecht M, Buttner-Herold M,
Erlenbach-Wunsch K, Haderlein M, Croner R, Grutzmann R, Hartmann A,
Fietkau R and Distel LV: PD-L1 is upregulated by radiochemotherapy
in rectal adenocarcinoma patients and associated with a favourable
prognosis. Eur J Cancer. 65:52–60. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Katsuya Y, Horinouchi H, Asao T, Kitahara
S, Goto Y, Kanda S, Fujiwara Y, Nokihara H, Yamamoto N, Watanabe S,
et al: Expression of programmed death 1 (PD-1) and its ligand
(PD-L1) in thymic epithelial tumors: Impact on treatment efficacy
and alteration in expression after chemotherapy. Lung Cancer.
99:4–10. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Takamori S, Takada K, Tagawa T, Toyokawa
G, Hirai F, Yamashita N, Okamoto T, Oki E, Yoshizumi T, Oda Y, et
al: Differences in PD-L1 expression on tumor and immune cells
between lung metastases and corresponding primary tumors. Surg
Oncol. 27:637–641. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Kandoth C, McLellan MD, Vandin F, Ye K,
Niu B, Lu C, Xie M, Zhang Q, McMichael JF, Wyczalkowski MA, et al:
Mutational landscape and significance across 12 major cancer types.
Nature. 502:333–339. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Alexandrov LB, Nik-Zainal S, Wedge DC,
Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A,
Borresen-Dale AL, et al: Signatures of mutational processes in
human cancer. Nature. 500:415–421. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Herbst RS, Soria JC, Kowanetz M, Fine GD,
Hamid O, Gordon MS, Sosman JA, McDermott DF, Powderly JD, Gettinger
SN, et al: Predictive correlates of response to the anti-PD-L1
antibody MPDL3280A in cancer patients. Nature. 515:563–567. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Rizvi NA, Hellmann MD, Snyder A, Kvistborg
P, Makarov V, Havel JJ, Lee W, Yuan J, Wong P, Ho TS, et al: Cancer
immunology. Mutational landscape determines sensitivity to PD-1
blockade in non-small cell lung cancer. Science. 348:124–128. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Forde PM, Chaft JE, Smith KN, Anagnostou
V, Cottrell TR, Hellmann MD, Zahurak M, Yang SC, Jones DR,
Broderick S, et al: Neoadjuvant PD-1 blockade in resectable lung
cancer. N Engl J Med. 378:1976–1986. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Wang Z, Duan J, Cai S, Han M, Dong H, Zhao
J, Zhu B, Wang S, Zhuo M, Sun J, et al: Assessment of blood tumor
mutational burden as a potential biomarker for immunotherapy in
patients with non-small cell lung cancer with use of a
next-generation sequencing cancer gene panel. JAMA Oncol.
5:696–702. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Lipson EJ, Sharfman WH, Drake CG, Wollner
I, Taube JM, Anders RA, Xu H, Yao S, Pons A, Chen L, et al: Durable
cancer regression off-treatment and effective reinduction therapy
with an anti-PD-1 antibody. Clin Cancer Res. 19:462–468. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Li GM: Mechanisms and functions of DNA
mismatch repair. Cell Res. 18:85–98. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Le DT, Uram JN, Wang H, Bartlett BR,
Kemberling H, Eyring AD, Skora AD, Luber BS, Azad NS, Laheru D, et
al: PD-1 blockade in tumors with mismatch-repair deficiency. N Engl
J Med. 372:2509–2520. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Le DT, Durham JN, Smith KN, Wang H,
Bartlett BR, Aulakh LK, Lu S, Kemberling H, Wilt C, Luber BS, et
al: Mismatch repair deficiency predicts response of solid tumors to
PD-1 blockade. Science. 357:409–413. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Schumacher TN and Schreiber RD:
Neoantigens in cancer immunotherapy. Science. 348:69–74. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
75
|
McGranahan N, Furness AJ, Rosenthal R,
Ramskov S, Lyngaa R, Saini SK, Jamal-Hanjani M, Wilson GA, Birkbak
NJ, Hiley CT, et al: Clonal neoantigens elicit T cell
immunoreactivity and sensitivity to immune checkpoint blockade.
Science. 351:1463–1469. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Van Allen EM, Golay HG, Liu Y, Koyama S,
Wong K, Taylor-Weiner A, Giannakis M, Harden M, Rojas-Rudilla V,
Chevalier A, et al: Long-term benefit of PD-L1 blockade in lung
cancer associated with JAK3 activation. Cancer Immunol Res.
3:855–863. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Azuma K, Ota K, Kawahara A, Hattori S,
Iwama E, Harada T, Matsumoto K, Takayama K, Takamori S, Kage M, et
al: Association of PD-L1 overexpression with activating EGFR
mutations in surgically resected nonsmall-cell lung cancer. Ann
Oncol. 25:1935–1940. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Parsa AT, Waldron JS, Panner A, Crane CA,
Parney IF, Barry JJ, Cachola KE, Murray JC, Tihan T, Jensen MC, et
al: Loss of tumor suppressor PTEN function increases B7-H1
expression and immunoresistance in glioma. Nat Med. 13:84–88. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Marzec M, Zhang Q, Goradia A, Raghunath
PN, Liu X, Paessler M, Wang HY, Wysocka M, Cheng M, Ruggeri BA, et
al: Oncogenic kinase NPM/ALK induces through STAT3 expression of
immunosuppressive protein CD274 (PD-L1, B7-H1). Proc Natl Acad Sci
USA. 105:20852–20857. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Yadav M, Jhunjhunwala S, Phung QT,
Lupardus P, Tanguay J, Bumbaca S, Franci C, Cheung TK, Fritsche J,
Weinschenk T, et al: Predicting immunogenic tumour mutations by
combining mass spectrometry and exome sequencing. Nature.
515:572–576. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Zaretsky JM, Garcia-Diaz A, Shin DS,
Escuin-Ordinas H, Hugo W, Hu-Lieskovan S, Torrejon DY,
Abril-Rodriguez G, Sandoval S, Barthly L, et al: Mutations
associated with acquired resistance to PD-1 blockade in melanoma. N
Engl J Med. 375:819–829. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Skoulidis F, Goldberg ME, Greenawalt DM,
Hellmann MD, Awad MM, Gainor JF, Schrock AB, Hartmaier RJ, Trabucco
SE, Gay L, et al: STK11/LKB1 mutations and PD-1 inhibitor
resistance in KRAS-mutant lung adenocarcinoma. Cancer Discov.
8:822–835. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Chen F, Zhuang X, Lin L, Yu P, Wang Y, Shi
Y, Hu G and Sun Y: New horizons in tumor microenvironment biology:
Challenges and opportunities. BMC Med. 13:452015. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Mantovani A, Allavena P, Sica A and
Balkwill F: Cancer-related inflammation. Nature. 454:436–444. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Yang L, Pang Y and Moses HL: TGF-beta and
immune cells: An important regulatory axis in the tumor
microenvironment and progression. Trends Immunol. 31:220–227. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Pauken KE and Wherry EJ: Overcoming T cell
exhaustion in infection and cancer. Trends Immunol. 36:265–276.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Li HY, McSharry M, Bullock B, Nguyen TT,
Kwak J, Poczobutt JM, Sippel TR, Heasley LE, Weiser-Evans MC,
Clambey ET, et al: The tumor microenvironment regulates sensitivity
of murine lung tumors to PD-1/PD-L1 antibody blockade. Cancer
Immunol Res. 5:767–777. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Smyth MJ, Ngiow SF, Ribas A and Teng MW:
Combination cancer immunotherapies tailored to the tumour
microenvironment. Nat Rev Clin Oncol. 13:143–158. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Tumeh PC, Harview CL, Yearley JH, Shintaku
IP, Taylor EJ, Robert L, Chmielowski B, Spasic M, Henry G, Ciobanu
V, et al: PD-1 blockade induces responses by inhibiting adaptive
immune resistance. Nature. 515:568–571. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Chen PL, Roh W, Reuben A, Cooper ZA,
Spencer CN, Prieto PA, Miller JP, Bassett RL, Gopalakrishnan V,
Wani K, et al: Analysis of immune signatures in longitudinal tumor
samples yields insight into biomarkers of response and mechanisms
of resistance to immune checkpoint blockade. Cancer Discov.
6:827–837. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Daud AI, Loo K, Pauli ML,
Sanchez-Rodriguez R, Sandoval PM, Taravati K, Tsai K, Nosrati A,
Nardo L, Alvarado MD, et al: Tumor immune profiling predicts
response to anti-PD-1 therapy in human melanoma. J Clin Invest.
126:3447–3452. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Kim R, Keam B, Kim S, Kim M, Kim SH, Kim
JW, Kim YJ, Kim TM, Jeon YK, Kim DW, et al: Differences in tumor
microenvironments between primary lung tumors and brain metastases
in lung cancer patients: Therapeutic implications for immune
checkpoint inhibitors. BMC Cancer. 19:192019. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Chen DS and Mellman I: Elements of cancer
immunity and the cancer-immune set point. Nature. 541:321–330.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Kim CG, Jang M, Kim Y, Leem G, Kim KH, Lee
H, Kim TS, Choi SJ, Kim HD, Han JW, et al: VEGF-A drives
TOX-dependent T cell exhaustion in anti-PD-1-resistant
microsatellite stable colorectal cancers. Sci Immunol.
4:eaay05552019. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Ngiow SF, Young A, Jacquelot N, Yamazaki
T, Enot D, Zitvogel L and Smyth MJ: A threshold level of intratumor
CD8+ T-cell PD1 expression dictates therapeutic response to
Anti-PD1. Cancer Res. 75:3800–3811. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Davis RJ, Moore EC, Clavijo PE, Friedman
J, Cash H, Chen Z, Silvin C, Van Waes C and Allen C: Anti-PD-L1
efficacy Can Be enhanced by inhibition of myeloid-derived
suppressor cells with a selective inhibitor of PI3Kδ/γ. Cancer Res.
77:2607–2619. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Mao Y, Eissler N, Blanc KL, Johnsen JI,
Kogner P and Kiessling R: Targeting suppressive myeloid cells
potentiates checkpoint inhibitors to control spontaneous
neuroblastoma. Clin Cancer Res. 22:3849–3859. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Stone TW and Darlington LG: Endogenous
kynurenines as targets for drug discovery and development. Nat Rev
Drug Discov. 1:609–620. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
99
|
Llosa NJ, Cruise M, Tam A, Wicks EC,
Hechenbleikner EM, Taube JM, Blosser RL, Fan H, Wang H, Luber BS,
et al: The vigorous immune microenvironment of microsatellite
instable colon cancer is balanced by multiple counter-inhibitory
checkpoints. Cancer Discov. 5:43–51. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Higgs BW, Robbins PB, Blake-Haskins JA,
Zhu W, Morehouse C, Brohawn PZ, Rebelatto MC, Yao Y, Jin X, Shi L
and Ranade K: 15LBA High tumoral IFNγ mRNA, PD-L1 protein, and
combined IFNγ mRNA/PD-L1 protein expression associates with
response to durvalumab (anti-PD-L1) monotherapy in NSCLC patients.
Eur J Cancer. 51 (Suppl):S7172015. View Article : Google Scholar
|
|
101
|
Ascierto ML, McMiller TL, Berger AE,
Danilova L, Anders RA, Netto GJ, Xu H, Pritchard TS, Fan J, Cheadle
C, et al: The intratumoral balance between metabolic and
immunologic gene expression is associated with Anti-PD-1 response
in patients with renal cell carcinoma. Cancer Immunol Res.
4:726–733. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Francisco LM, Salinas VH, Brown KE,
Vanguri VK, Freeman GJ, Kuchroo VK and Sharpe AH: PD-L1 regulates
the development, maintenance, and function of induced regulatory T
cells. J Exp Med. 206:3015–3029. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Ding ZC, Lu X, Yu M, Lemos H, Huang L,
Chandler P, Liu K, Walters M, Krasinski A, Mack M, et al:
Immunosuppressive myeloid cells induced by chemotherapy attenuate
antitumor CD4+ T-cell responses through the PD-1-PD-L1 axis. Cancer
Res. 74:3441–3453. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Platten M, Wick W and Van den Eynde BJ:
Tryptophan catabolism in cancer: Beyond IDO and tryptophan
depletion. Cancer Res. 72:5435–5440. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Ayers M, Lunceford J, Nebozhyn M, Murphy
E, Loboda A, Kaufman DR, Albright A, Cheng JD, Kang SP, Shankaran
V, et al: IFN-γ-related mRNA profile predicts clinical response to
PD-1 blockade. J Clin Invest. 127:2930–2940. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Zhang X, Zeng Y, Qu Q, Zhu J, Liu Z, Ning
W, Zeng H, Zhang N, Du W, Chen C, et al: PD-L1 induced by IFN-γ
from tumor-associated macrophages via the JAK/STAT3 and PI3K/AKT
signaling pathways promoted progression of lung cancer. Int J Clin
Oncol. 22:1026–1033. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Quandt D, Jasinski-Bergner S, Muller U,
Schulze B and Seliger B: Synergistic effects of IL-4 and TNFα on
the induction of B7-H1 in renal cell carcinoma cells inhibiting
allogeneic T cell proliferation. J Transl Med. 12:1512014.
View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Ni XY, Sui HX, Liu Y, Ke SZ, Wang YN and
Gao FG: TGF-β of lung cancer microenvironment upregulates B7H1 and
GITRL expression in dendritic cells and is associated with
regulatory T cell generation. Oncol Rep. 28:615–621. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Rodriguez-Garcia M, Porichis F, de Jong
OG, Levi K, Diefenbach TJ, Lifson JD, Freeman GJ, Walker BD,
Kaufmann DE and Kavanagh DG: Expression of PD-L1 and PD-L2 on human
macrophages is up-regulated by HIV-1 and differentially modulated
by IL-10. J Leukoc Biol. 89:507–515. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Kondou R, Iizuka A, Nonomura C, Miyata H,
Ashizawa T, Nagashima T, Ohshima K, Urakami K, Kusuhara M,
Yamaguchi K, et al: Classification of tumor microenvironment immune
types based on immune response-associated gene expression. Int J
Oncol. 54:219–228. 2019.PubMed/NCBI
|
|
111
|
Spranger S, Spaapen RM, Zha Y, Williams J,
Meng Y, Ha TT and Gajewski TF: Up-regulation of PD-L1, IDO, and
T(regs) in the melanoma tumor microenvironment is driven by CD8(+)
T cells. Sci Transl Med. 5:200ra1162013. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Zhao J, Wen X, Tian L, Li T, Xu C, Wen X,
Melancon MP, Gupta S, Shen B, Peng W, et al: Irreversible
electroporation reverses resistance to immune checkpoint blockade
in pancreatic cancer. Nat Commun. 10:8992019. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Liu P, Zhao L, Pol J, Levesque S,
Petrazzuolo A, Pfirschke C, Engblom C, Rickelt S, Yamazaki T,
Iribarren K, et al: Crizotinib-induced immunogenic cell death in
non-small cell lung cancer. Nat Commun. 10:14862019. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Nikolich-Zugich J, Slifka MK and Messaoudi
I: The many important facets of T-cell repertoire diversity. Nat
Rev Immunol. 4:123–132. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Ruggiero E, Nicolay JP, Fronza R, Arens A,
Paruzynski A, Nowrouzi A, Urenden G, Lulay C, Schneider S, Goerdt
S, et al: High-resolution analysis of the human T-cell receptor
repertoire. Nat Commun. 6:80812015. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Zheng Y, Wang T, Tu X, Huang Y, Zhang H,
Tan D, Jiang W, Cai S, Zhao P, Song R, et al: Gut microbiome
affects the response to anti-PD-1 immunotherapy in patients with
hepatocellular carcinoma. J Immunother Cancer. 7:1932019.
View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Sunakawa Y, Inoue E, Matoba R, Kawakami H,
Sato Y, Nakajima TE, Muro K, Ichikawa W and Fujii M: DELIVER
(JACCRO GC-08) trial: Discover novel host-related immune-biomarkers
for nivolumab in advanced gastric cancer. Future Oncol.
15:2441–2447. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Matson V, Fessler J, Bao R, Chongsuwat T,
Zha Y, Alegre ML, Luke JJ and Gajewski TF: The commensal microbiome
is associated with anti-PD-1 efficacy in metastatic melanoma
patients. Science. 359:104–108. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Jin Y, Dong H, Xia L, Yang Y, Zhu Y, Shen
Y, Zheng H, Yao C, Wang Y and Lu S: The diversity of gut microbiome
is associated with favorable responses to anti-programmed death 1
immunotherapy in Chinese patients with NSCLC. J Thorac Oncol.
14:1378–1389. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Ivanov II and Littman DR: Modulation of
immune homeostasis by commensal bacteria. Curr Opin Microbiol.
14:106–114. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Sivan A, Corrales L, Hubert N, Williams
JB, Aquino-Michaels K, Earley ZM, Benyamin FW, Lei YM, Jabri B,
Alegre ML, et al: Commensal Bifidobacterium promotes antitumor
immunity and facilitates anti-PD-L1 efficacy. Science.
350:1084–1089. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Vetizou M, Pitt JM, Daillere R, Lepage P,
Waldschmitt N, Flament C, Rusakiewicz S, Routy B, Roberti MP, Duong
CP, et al: Anticancer immunotherapy by CTLA-4 blockade relies on
the gut microbiota. Science. 350:1079–1084. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Dong M, Meng Z, Kuerban K, Qi F, Liu J,
Wei Y, Wang Q, Jiang S, Feng M and Ye L: Diosgenin promotes
antitumor immunity and PD-1 antibody efficacy against melanoma by
regulating intestinal microbiota. Cell Death Dis. 9:10392018.
View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Weide B, Martens A, Hassel JC, Berking C,
Postow MA, Bisschop K, Simeone E, Mangana J, Schilling B, Di
Giacomo AM, et al: Baseline biomarkers for outcome of melanoma
patients treated with pembrolizumab. Clin Cancer Res. 22:5487–5496.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Zer A, Sung MR, Walia P, Khoja L, Maganti
M, Labbe C, Shepherd FA, Bradbury PA, Feld R, Liu G, et al:
Correlation of neutrophil to lymphocyte ratio and absolute
neutrophil count with outcomes with PD-1 axis inhibitors in
patients with advanced non-small-cell lung cancer. Clin Lung
Cancer. 19:426–434.e1. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Diem S, Kasenda B, Spain L, Martin-Liberal
J, Marconcini R, Gore M and Larkin J: Serum lactate dehydrogenase
as an early marker for outcome in patients treated with anti-PD-1
therapy in metastatic melanoma. Br J Cancer. 114:256–261. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Gibney GT, Kudchadkar RR, DeConti RC,
Thebeau MS, Czupryn MP, Tetteh L, Eysmans C, Richards A, Schell MJ,
Fisher KJ, et al: Safety, correlative markers, and clinical results
of adjuvant nivolumab in combination with vaccine in resected
high-risk metastatic melanoma. Clin Cancer Res. 21:712–720. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Niemeijer AN, Leung D, Huisman MC, Bahce
I, Hoekstra OS, van Dongen G, Boellaard R, Du S, Hayes W, Smith R,
et al: Whole body PD-1 and PD-L1 positron emission tomography in
patients with non-small-cell lung cancer. Nat Commun. 9:46642018.
View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Bensch F, van der Veen EL, Lub-de Hooge
MN, Jorritsma- Smit A, Boellaard R, Kok IC, Oosting SF, Schroder
CP, Hiltermann TJN, van der Wekken AJ, et al:
89Zr-atezolizumab imaging as a non-invasive approach to
assess clinical response to PD-L1 blockade in cancer. Nat Med.
24:1852–1858. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Sun R, Limkin EJ, Vakalopoulou M, Dercle
L, Champiat S, Han SR, Verlingue L, Brandao D, Lancia A, Ammari S,
et al: A radiomics approach to assess tumour-infiltrating CD8 cells
and response to anti-PD-1 or anti-PD-L1 immunotherapy: An imaging
biomarker, retrospective multicohort study. Lancet Oncol.
19:1180–1191. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Joyce JA and Fearon DT: T cell exclusion,
immune privilege, and the tumor microenvironment. Science.
348:74–80. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Kluger HM, Zito CR, Barr ML, Baine MK,
Chiang VL, Sznol M, Rimm DL, Chen L and Jilaveanu LB:
Characterization of PD-L1 expression and associated T-cell
infiltrates in metastatic melanoma samples from variable anatomic
sites. Clin Cancer Res. 21:3052–3060. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Rooney MS, Shukla SA, Wu CJ, Getz G and
Hacohen N: Molecular and genetic properties of tumors associated
with local immune cytolytic activity. Cell. 160:48–61. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Feng Z, Puri S, Moudgil T, Wood W, Hoyt
CC, Wang C, Urba WJ, Curti BD, Bifulco CB and Fox BA: Multispectral
imaging of formalin-fixed tissue predicts ability to generate
tumor-infiltrating lymphocytes from melanoma. J Immunother Cancer.
3:472015. View Article : Google Scholar : PubMed/NCBI
|