Introduction
Osteosarcoma (OS), as the most common primary human
malignant tumor among children and adolescents, originating from
bone mesenchymal cells, has the characteristics of early distant
metastasis and a high local recurrence rate (1–3).
Currently, treatment of OS includes mainly neoadjuvant chemotherapy
combined with surgical treatment; however, the clinical prognosis
rate following current treatment strategies has not been largely
improved (4). The main reason for
the poor clinical prognosis is that the current treatments cannot
effectively inhibit distant metastasis and the recurrence of drug
resistance. In addition, the most fundamental reason is that the
pathogenesis of OS has not been well elucidated. With the rapid
development of molecular biology, new technologies provide
opportunities for a more in-depth study of the pathogenesis of
OS.
Long non-coding RNA (lncRNA) is a class of
endogenous RNA sequences >200 nucleotides in length, which
completely lacks or only has a small portion of protein encoding
capacity (5). The abnormal
sequences, abnormal spatial structure, abnormal protein binding and
expression of lncRNAs in the body are all closely associated with
the occurrence of human diseases. Numerous studies have shown that
lncRNAs have a wide range of functions in almost all physiologic
processes of an organism, and they do not only regulate cell
differentiation (6,7) and metabolism (8), but also participate in a variety of
pathological processes, such as cancer proliferation, invasion,
metastasis (9), apoptosis (10) and drug resistance (11). NR2F1-AS1, a recently discovered
lncRNA, has been found to enhance esophageal squamous cell
carcinoma (ESCC) progression by regulating the Hedgehog signaling
pathway (12). NR2F1-AS1 has also
been reported to regulate the microRNA (miR)-338-3p/cyclin D1 axis
to promote thyroid cancer progression (13). In papillary thyroid carcinoma,
NR2F1-AS1 has been confirmed to exacerbate cell proliferation by
regulating the miR-423-5p/SRY-box transcription factor (SOX)12
signaling pathway (14). In
addition, in endometrial cancer, NR2F1-AS1 has been reported to
sponge miR-363 and target SOX4 to play a role in cell proliferation
and migration (15). Notably, it
has been validated that NR2F1-AS1 facilitates the malignant
properties of OS via binding to miR-483-3p to enhance forkhead box
A1 (FOXA1) expression (16). These
findings demonstrate that NR2F1-AS1 acts as an oncogene in cancer.
However, the function of NR2F1-AS1 in OS is not well understood and
requires further investigation.
A number of studies have demonstrated that miRNAs
exhibit roles in numerous biological processes by regulating their
mRNA targets, including in cell development, tumorigenesis, cell
differentiation and aging (17–19).
In OS, the miRNA expression profile has been extensively studied,
and a variety of miRNAs has been identified to play a role in OS,
such as miR-422a, miR-145 and miR-194 (20). Furthermore, the aberrant expression
of miRNAs is closely associated with OS progression and prognosis.
miR-485-5p and miR-218-5p have been reported to inhibit the
malignant behavior of OS (21–23).
Therefore, the present study investigated these miRNAs to expand
our understanding of their functional effects in OS.
Baculoviral inhibitor of apoptosis repeat-containing
5 (BIRC5), also termed survivin, is a member of the family of
apoptotic inhibitors and plays an important regulatory role in
tumor progression in numerous types of cancers, including OS
(24–27). In addition, accumulating evidence
has confirmed that BIRC5 is negatively regulated by miRNAs in
different tumor cells to play functional roles. For example, in
breast cancer, BIRC5 has been demonstrated to be regulated by
miR-485-5p to suppress cancer progression and chemosensitivity
(24). Furthermore, in OS, miR-218
promotes apoptosis of U2OS osteosarcoma cells via targeting BIRC5
(22). However, in OS, whether
BIRC5 is controlled by miR-485-5p and miR-218-5p to serve roles in
tumor development and progression has not been fully elucidated, to
the best of our knowledge. It is well understood that lncRNAs can
combine with miRNAs to target mRNAs, which creates a network of
lncRNA-miRNA-mRNA and thus exerts biological functions in cancer.
Therefore, it is worth investigating whether NR2F1-AS1, miR-485-5p,
miR-218-5p and BIRC5 form an axis to affect the malignant phenotype
of OS.
The present study revealed that NR2F1-AS1 expression
was upregulated in OS. Further studies indicated that NR2F1-AS1
knockdown inhibited cell proliferation, migration and invasion, and
facilitated cell apoptosis by interacting with miR-485-5p and
miR-218-5p to downregulate BIRC5 expression. In summary, the
present study described the participation of the
NR2F1-AS1/miR-485-5p/miR-218-5p/BIRC5 axis in regulating the OS
malignant phenotype.
Materials and methods
Clinical samples
A total of 32 cases of OS tissue specimens and
corresponding adjacent normal tissue (age range, 8–50 years; mean
age 18.8±10.1 years) specimens were collected at the Zhengzhou
Orthopedic Hospital (Zhengzhou, Henan, China) between January 2016
and December 2018. All specimens were confirmed by pathological
biopsy, and none of the patients underwent chemotherapy and
radiation therapy before surgery. The tissue samples were quickly
frozen in liquid nitrogen in vitro. The patient samples were
collected with permission from the Medical Ethics Committee of
Zhengzhou Orthopedic Hospital, and written informed consent was
obtained from the patients before the operation.
Cell culture and cell
transfection
Human OS cell lines (HOS, MG63 and U2OS) and a
normal osteoblast cell line (hFOB1.19) were obtained from the
Chinese Academy of Sciences Cell Bank. The OS cells (HOS, MG63 and
U2OS) were cultured in RPMI-1640 medium (Sangon Biotech Co., Ltd.)
with 10% fetal bovine serum (FBS; Hyclone; Thermo Fisher
Scientific, Inc.) in a 5% CO2 atmosphere at 37°C. The
hFOB1.19 cells were cultured at 33.5°C with 5% CO2 in
RPMI-1640 medium with 10% FBS. The 293T cells were cultured at 37°C
with 5% CO2 in Gibco (no. 12800017, containing
NaHCO3 1.5 g/l) medium (Thermo Fisher Scientific, Inc.)
with 10% FBS. The small interfering RNA (siRNA) specifically
targeting NR2F1-AS1 (si-NR2F1-AS) and the negative control (si-NC)
were synthesized and purchased from Invitrogen; Thermo Fisher
Scientific, Inc. The sequences were as follows:
si-NR2F1-AS1#: 5′-GAAGAUAGUUUAUAAUUUAAA-3′;
si-NR2F1-AS2#: 5′-AGUUCAAGAAGAUAGUUUAUA-3′;
si-NR2F1-AS3#: 5′-GAUGUUCUCAAUAUUUCUAUU-3′; si-NC:
5′-CGAAACCUAGCGUGUACACAA-3′. miR-485-5p mimics and negative control
(NC), and miR-485-5p inhibitor (miR-485-5p in) and NC were also
obtained from Invitrogen; Thermo Fisher Scientific, Inc. The
sequences were as follows: miR-485-5p mimics:
5′-AGAGGCUGGCCGUGAUGAAUUC-3′; mimics NC:
5′-GUCCGCAGCACGCAUUAAGAUU-3′; miR-485-5p inhibitor:
5′-GAAUUCAUCACGGCCAGCCUCU-3′; inhibitor NC:
5′-UAAUUCGAGUCAUGAAUUUCA-3′. miR-218-5p mimics and negative control
(NC), and miR-218-5p inhibitor (miR-218-5p in) and NC were also
obtained from Invitrogen; Thermo Fisher Scientific, Inc. The
sequences were as follows: miR-218-5p mimics:
5′-UUGUGCUUGAUCUAACCAUGU-3′; mimics NC:
5′-UAAAUGACCGUCCCUCCGAGU-3′; miR-218-5p inhibitor:
5′-ACAUGGUUAGAUCAAGCACAA-3′; inhibitor NC:
5′-CAGUACUUUUGUGUAGUACAA-3′. A BIRC5 overexpression plasmid
pcDNA3.1-BIRC5 (pc-BIRC5) and empty pcDNA3.1 vector were purchased
from GeneChem, Inc. Using Lipofectamine 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.), siRNA and miRNAs at an appropriate
concentration mixed in 2 ml Opti-MEM plus 10% FBS were used to
transfect MG63 and U-2OS cells. The MG63 and U-2OS cells were
divided into different groups: i) lncRNA NR2F1-AS1 NC group
(transfected with 50 nM si-NR2F1-AS1 NC); ii)
si-NR2F1-AS11# group (transfected with 50 nM
si-NR2F1-AS11#); iii) si-NR2F1-AS12# group
(transfected with 50 nM si-NR2F1-AS12#); iv)
si-NR2F1-AS1 + miR-485-5p inh group (transfected with 50 nM
si-NR2F1-AS11# + miR-485-5p inhibitor); v) si-NR2F1-AS1
+ miR-485-5p inh NC group (transfected with 50 nM
si-NR2F1-AS11# + miR-485-5p inhibitor NC); vi)
si-NR2F1-AS1 + miR-218-5p inh group (transfected with 50 nM
si-NR2F1-AS11# + miR-218-5p inhibitor); vii)
si-NR2F1-AS1 + miR-218-5p inh NC group (transfected with 50 nM
si-NR2F1-AS11# + miR-218-5p inhibitor NC); viii)
si-NR2F1-AS1 + pc-BIRC5 group (transfected with 50 nM
si-NR2F1-AS11# + pcDNA3.1-BIRC5); and ix) si-NR2F1-AS1 +
pc-DNA3.1 group (transfected with 50 nM si-NR2F1-AS11# +
pcDNA3.1). Subsequently, 24 h after transfection, the cells were
prepared for the subsequent experiments.
Total RNA isolation and reverse
transcription-quantitative PCR (RT-qPCR)
Using TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc.), total RNA was isolated. Subsequently, RT
(RT-Reagent Kit, Takara Bio) of RNA to cDNA (Invitrogen; Thermo
Fisher Scientific, Inc.) and qPCR analysis were performed. The
thermocycling conditions of qPCR for NR2F1-AS1 and BIRC5 were as
follows: 5 min at 95°C, followed by 40 cycles of 95°C for 30 sec
and 65°C for 45 sec. For the measurement of miR-485-5p and
miR-218-5p expression, the thermocycling conditions were as
follows: Pre-denaturation at 95°C for 1 min, followed by 40 cycles
at 95°C for 15 sec, 60°C for 30 sec, and 72°C for 30 sec, which was
performed on an ABI 7500 qPCR instrument (Applied Biosystems;
Thermo Fisher Scientific, Inc.). Relative expression levels of
NR2F1-AS1, miR-218-5p and miR-485-5p were calculated using the
2−ΔΔCq method (28). The
NR2F1-AS1 and BIRC5 expression levels were made relative to GAPDH,
and the miR-485-5p and miR-218-5p expression levels were made
relative to U6. The sequences of all primers are presented in
Table I.
 | Table I.Primer sequences. |
Table I.
Primer sequences.
| Gene | Position | Sequence (5′ to
3′) |
|---|
| NR2F1-AS1 | Forward |
AGCGGTGCAAACCATGTG |
|
| Reverse |
CAAGTTGGCTGAACCAAAT |
| miR-218-5p | Forward |
AAGACACCCTGGACGAAGCC |
|
| Reverse |
ACAACCAGAGTCCACCGGCG |
| miR-485-5p | Forward |
ACACTCCAGCTGGGAAGACGGGAGGAAAGAA |
|
| Reverse |
CTCAACTGGTGTCGTGGA |
| GAPDH | Forward |
CAGTGCCAGCCTCGTCTAT |
|
| Reverse |
AGGGGCCATCCACAGTCTTC |
| U6 | Forward |
CTCGCTTCGGCAGCACATA |
|
| Reverse |
AACGATTCACGAATTTGCGT |
| BIRC5 | Forward |
AGGACCACCGCATCTCTACAT |
|
| Reverse |
AAGTCTGGCTCGTTCTCAGTG |
Cell Counting Kit-8 (CCK-8) assay
The transfected MG63 and U-2OS cells with good
growth status were seeded in a 96-well plate at a density of
1×104/well and incubated. CCK-8 reagent (10 µl) was then
added to each well at 0, 24, 48 and 72 h, and the cells were
further incubated for 2 h. The absorbance of the samples was then
determined at a wavelength of 450 nm.
Flow cytometric analysis of
apoptosis
Following transfection for 24 h, MG63 and U-2OS
cells were digested with 0.25% trypsin and harvested. Subsequently,
the cells were re-suspended with pre-cooled 1X PBS, centrifuged for
10 min (1,200 × g), and washed. The cells were stained with Annexin
V-FITC and propidium iodide (PI) for 10 min and incubated at room
temperature. Finally, the apoptosis rate was detected using a flow
cytometer and analyzed on FACSCalibur (BD Biosciences).
Transwell assay
Following transfection for 24 h, MG63 and U-2OS
cells were digested and re-suspended with FBS-free RPMI-1640
medium. For the invasion assay, the upper chamber of a Transwell
plate was pre-coated with Matrigel (BD Biosciences) overnight. The
next day, a 100-µl cell suspension (5×104 cells) was
placed into the upper chamber. For the migration assay, there is no
need to pre-coated with Matrigel. The same number of cells was
placed into the upper chamber of the Transwell plate. For both
assays, the lower chamber was filled with RPMI-1640 medium
containing 20% of FBS. Then, the Transwell plate was cultured in an
incubator containing 5% CO2. Following 24 h, the cells
were wished, fixed with 70% ethanol for 15 min, stained with
crystal violet (0.1%) or 30 min at 25°C and counted with an Olympus
IX51 inverted microscope (Olympus Corp.; magnification, ×200).
Bioinformatics analysis
To investigate how NR2F1-AS1 regulates miR-485-5p
and miR-218-5p, and how miR-485-5p and miR-218-5p regulate BIRC5,
bioinformatics websites were used for analysis. The websites used
were as follows: Diana Tools (http://carolina.imis.athena-innovation.gr/diana_tools/web/index.php?r=lncbasev2/index),
miRwalk (http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2/),
Target Scan (http://www.targetscan.org/vert_71/) and miRbase
(http://www.mirbase.org/).
Luciferase reporter assay
The NR2F1-AS1 fragments containing the predicted
miR-485-5p and miR-218-5p binding sites were separately amplified
by PCR (PCR thermocycling condition: 3 min at 94°C, followed by 35
cycles of 94°C for 30 sec, 58°C for 30 sec and 72°C for 45 sec) and
cloned into a pmirGLO dual luciferase miRNA target expression
vector (Promega Corp.) to create a wild-type NR2F1-AS1 reporter
vector (NR2F1-AS1-wild-type; NR2F1-AS1-Wt). To mutate the putative
miR-485-5p and miR-218-5p binding sites in NR2F1-AS1, the sequences
of the putative binding site were changed and replaced to form a
NR2F1-AS1-mutated-type (NR2F1-AS1-Mut). Subsequently, the
recombinant plasmids and miRNAs (miR-485-5p mimics, miR-218-5p
mimics or miR-NC) were co-transfected into 293T cells using
Lipofectamine™ 2000. After 48 h, the luciferase activities were
measured using the Dual Luciferase Reporter Gene Assay System
(Promega Corp.). To investigate whether BIRC5 is a functional
target of miR-485-5p and miR-218-5p, the aforementioned methods
were followed and recombinant plasmids
pmirGLO-BIRC5-wt/pmirGLO-BIRC5-mut were constructed. These
recombinant plasmids were then co-transfected with miRNAs into 293T
cells and luciferase activities were determined.
Western blotting
MG63 and U-2OS cells were transfected for 48 h and
were then harvested and lysed with RIPA lysis buffer (Thermo Fisher
Scientific, Inc.). The protein concentration was measured by BCA
(Thermo Fisher Scientific, Inc.) and then a 40 µg protein sample of
each group was separated by 12% SDS-PAGE and transferred to a
nitrocellulose membrane. Following blocking with 5% skim milk, the
membrane was incubated with GAPDH and BIRC5 primary antibodies:
GAPDH (cat. no. ab128915; 1:10,000 dilution in TBST; Abcam), BIRC5
(cat. no. ab76424; 1:5,000 dilution; Abcam) overnight at 4°C,
followed by incubation with horseradish peroxidase-labeled goat
anti-Rabbit secondary antibodies (cat. no. ab205718; 1:5,000
dilution; Abcam) for 2 h at room temperature. Finally, the immune
reactive bands were visualized with a chemiluminescent kit. GAPDH
was used as a reference gene.
Statistical analysis
SPSS 24.0 (IBM Corp.) and GraphPad Prism 5 (GraphPad
Software, Inc.) were used for data analysis. All data are expressed
as mean ± standard deviation. Student's t-test was used for the
comparison of two groups. Univariate ANOVA followed by Fisher's LSD
post hoc tests was used for comparisons between multiple groups.
Correlation between NR2F1-AS1 and miRNA (miR-218-5p and miR-485-5p)
expression was determined using Spearman's correlation analysis.
The correlation between NR2F1-AS1 expression and clinical
parameters of patients with OS was employed by a χ2
test. P<0.05 was considered to indicate a statistically
significant difference.
Results
NR2F1-AS1 expression is upregulated
and exerts an oncogenic function in OS
NR2F1-AS1 has been confirmed to facilitate the
malignancy of multiple tumors. Therefore, the present study
investigated the NR2F1-AS1 expression in OS. Consistent with
previous studies, it was demonstrated that NR2F1-AS expression was
significantly increased in OS (Fig.
1A). Furthermore, among the 32 patients, a high level of
NR2F1-AS1 was closely associated with more advanced Enneking stage
(P=0.029) and distant metastasis (P=0.004; Table II). In addition, NR2F1-AS1 was
significantly overexpressed in OS cell lines, especially in MG63
and U-2OS cells when compared with the normal osteoblast cell line
hFOB1.19 (Fig. 1B). Thus, MG63 and
U-2OS cells were selected for use in subsequent functional studies.
Subsequently, the functions of NR2F1-AS1 in OS were evaluated and
three siRNAs (si-NR2F1-AS11#, si-NR2F1-AS12#,
si-NR2F1-AS13#) were assessed to detect their knockdown
efficiency. The results demonstrated that si-NR2F1-AS11#
exhibited the highest knockdown efficiency, followed by
si-NR2F1-AS12# and finally si-NR2F1-AS13#,
which also revealed that the transfections were successful
(Fig. 2A). Therefore,
si-NR2F1-AS11# and si-NR2F1-AS12# were
selected for further experiments. Cell proliferation, invasion,
migration and apoptosis were observed following
NR2F1-AS1-knockdown. By performing a CCK-8 assay, it was determined
that NR2F1-AS1-knockdown markedly reduced the proliferative
capacity of the OS MG63 and U-2OS cells cells (Fig. 2B and C). Furthermore, the data of
flow cytometry demonstrated that the apoptosis of OS cells was
significantly increased following silencing of NR2F1-AS1 expression
(Fig. 2D). Furthermore, it was
identified that the migration and invasion of OS cells were
significantly attenuated following NR2F1-AS1-knockdown (Fig. 3A and B). In summary, these findings
indicated that NR2F1-AS1 expression is upregulated in OS and that
NR2F1-AS1 exerts an oncogenic function in OS.
 | Table II.Association between NR2F1-AS1
expression level and clinical parameters of the patients with
OS. |
Table II.
Association between NR2F1-AS1
expression level and clinical parameters of the patients with
OS.
|
| NR2F1-AS1
expression |
|
|---|
| Parameters | Low (n=15) | High (n=17) | P-value |
|---|
| Age (years) |
|
| 0.589 |
|
<18 | 11 | 12 |
|
|
≥18 | 4 | 5 |
|
| Sex |
|
| 0.354 |
|
Male | 6 | 9 |
|
|
Female | 9 | 8 |
|
| Tumor site |
|
| 0.517 |
|
Femur/Tibia | 8 | 10 |
|
|
Other | 7 | 7 |
|
| Enneking stage |
|
| 0.029a |
|
I–II | 12 | 7 |
|
|
III | 3 | 10 |
|
| Distant
metastasis |
|
| 0.004 a |
|
Present | 13 | 6 |
|
|
Absent | 2 | 11 |
|
miR-485-5p and miR-218-5p are direct
targets of NR2F1-AS1 in OS cells
A number of studies have demonstrated that lncRNAs
combined with miRNAs exert biological functions in cancer (29), cardiovascular disease (30), acute megakaryoblastic leukemia
(31) and diabetes mellitus
(32). Therefore, the present study
attempted to identify the potential binding sites of NR2F1-AS1.
With biological software analysis, miR-485-5p and miR-218-5p were
identified and it was determined that their seed regions could
recognize NR2F1-AS1 sequences (Fig.
4A). The subsequent luciferase assays performed demonstrated
the interactions of miR-485-5p with NR2F1-AS1, and miR-218-5p with
NR2F1-AS1 (Fig. 4B). Subsequently,
it was observed that miR-485-5p and miR-218-5p were expressed at
significantly low levels in OS tissues, which was in contrast to
NR2F1-AS1 expression. Furthermore, negative correlations were
identified between miR-485-5p and NR2F1-AS1, and miR-218-5p and
NR2F1-AS1 (Fig. 4C and D). Finally,
it was validated that NR2F1-AS1 inhibition could result in a
significant increase in miR-218-5p and miR-485-5p (Fig. 4E). In summary, these findings
suggest that NR2F1-AS1 may interact with miR-485-5p and miR-218-5p
to serve a role in the biological functions in OS.
NR2F1-AS1 affects OS cell biological
behaviors by regulating miR-485-5p and miR-218-5p
First, we transfected miR-485-5p mimics and the
negative control (NC), miR-485-5p inhibitor and the negative
control (NC) to MG63 and U-2OS cells to detect whether the
transfections were successful. From the results of qPCR, we
revealed the transfections were successful (Fig. 5A). Subsequently, to investigate
whether NR2F1-AS1 affects the biological behaviors of OS cells via
miR-485-5p, the inhibitor of miR-485-5p (miR-485-5p inh) and the
negative control (NC inh) were co-transfected with
si-NR2F1-AS11# (si-NR2F1-AS1) into MG63 and U-2OS cells.
A series of experiments were then performed. As presented in
Figs. 5B and C and 6A and B, NR2F1-AS1-knockdown markedly
reduced cell proliferation, migration and invasion, and stimulated
apoptosis. However, these effects were markedly attenuated by
concurrent miR-485-5p-knockdown. In addition, we transfected
miR-218-5p mimics and the negative control (NC), miR-218-5p
inhibitor and the negative control (NC) to MG63 and U-2OS cells to
detect whether the transfections were successful. The results of
qPCR showed that the transfections were successful (Fig. 7A). To validate that NR2F1-AS1
affects OS cell biological behaviors by regulating miR-218-5p, the
inhibitor of miR-218-5p (miR-218-5p inh) and the negative control
(NC inh) were con-transfected with si-NR2F1-AS1 into MG63 and U-2OS
cells. As presented in Figs. 7B and
C and 8A and B, it was
demonstrated that suppression of NR2F1-AS1 could hinder the
proliferation, migration and invasion, and promote the apoptosis of
MG63 and U-2OS cells. Furthermore, the effects were substantially
attenuated by the inhibition of miR-218-5p. These observations
indicated that NR2F1-AS1 affects OS cell biological behaviors by
targeting miR-485-5p and miR-218-5p.
BIRC5 is a direct target of miR-485-5p
and miR-218-5p in OS cells
A number of studies have confirmed that miRNAs serve
roles in numerous biological processes by regulating their mRNA
targets. Therefore, the present study searched for potential
targets of miR-485-5p and miR-218-5p. Through biological software
analysis, BIRC5, a member of the apoptosis suppressor (IAP) gene
family, was identified. It was revealed that the 3′-untranslated
region of BIRC5 mRNA directly interacts with miR-485-5p and
miR-218-5p (Fig. 9A). Subsequently,
luciferase assays verified the interactions of miR-485-5p with
BIRC5, and miR-218-5p with BIRC5 (Fig.
9B). It was then validated that inhibition of miR-485-5p and
miR-218-5p resulted in increased BIRC5 expression (Fig. 9C and D). In addition, BIRC5
expression was observed to be expressed at a high level in OS
tissues as shown in Fig. 9E.
Pearson's analysis demonstrated that BIRC5 expression was
negatively correlated with miR-485-5p and miR-218-5p expression,
but positively correlated with NR2F1-AS1 expression in OS tissues
(Fig. 9F-H). As a whole, these
observations demonstrated that BIRC5 is a direct target of
miR-485-5p and miR-218-5p in OS cells.
 | Figure 9.BIRC5 is the direct target of
miR-485-5p and miR-218-5p in OS cells. (A) Sequences matching
between BIRC5 and miR-485-5p, and BIRC5 and miR-218-5p. (B)
Luciferase assay was used to verify the binding between BIRC5 and
miR-485-5p, and BIRC5 and miR-218-5p. **P<0.01 compared with
miR-485-5p-NC group (verifying the binding between BIRC5 and
miR-485-5p, the left panel); **P<0.01 compared with
miR-218-5p-NC group (verifying the binding between BIRC5 and
miR-218-5p, the right panel). (C) RT-qPCR assay was performed to
detect BIRC5 expression in MG63 and U-2OS cells respectively
transfected with miR-485-5p inhibitor (inh), and miR-218-5p
inhibitor (inh). **P<0.01, vs. NC group. (D) Western blot assay
was performed to detect BIRC5 expression in MG63 and U-2OS cells,
respectively, transfected with miR-485-5p inhibitor and miR-218-5p
inhibitor. (E) Expression of BIRC5 in 32 OS tissues. **P<0.01.
(F) Correlation between BIRC5 and miR-485-5p expression in 32 OS
tissues. (G) Correlation between BIRC5 and miR-218-5p expression in
32 OS tissues. (H) Correlation between BIRC5 and NR2F1-AS1
expression in 32 OS tissues. OS, osteosarcoma; NR2F1-AS1, long
non-coding RNA NR2F1 antisense RNA 1; BIRC5, baculoviral inhibitor
of apoptosis repeat-containing 5. |
NR2F1-AS1 affects OS cell biological
behaviors by regulating BIRC5
As aforementioned, NR2F1-AS1 affects OS cell
biological behaviors by regulating miR-485-5p and miR-218-5p, and
miR-485-5p and miR-218-5p could target BIRC5. Therefore, it was
next investigated with a number of experiments whether NR2F1-AS1
affects OS cell biological behaviors by regulating BIRC5 in OS
cells. We first transfected pcDNA3.1-BIRC5 and pcDNA3.1 into cells
and the results of qPCR showed that the transfections were
successful (Fig. 10A). And then
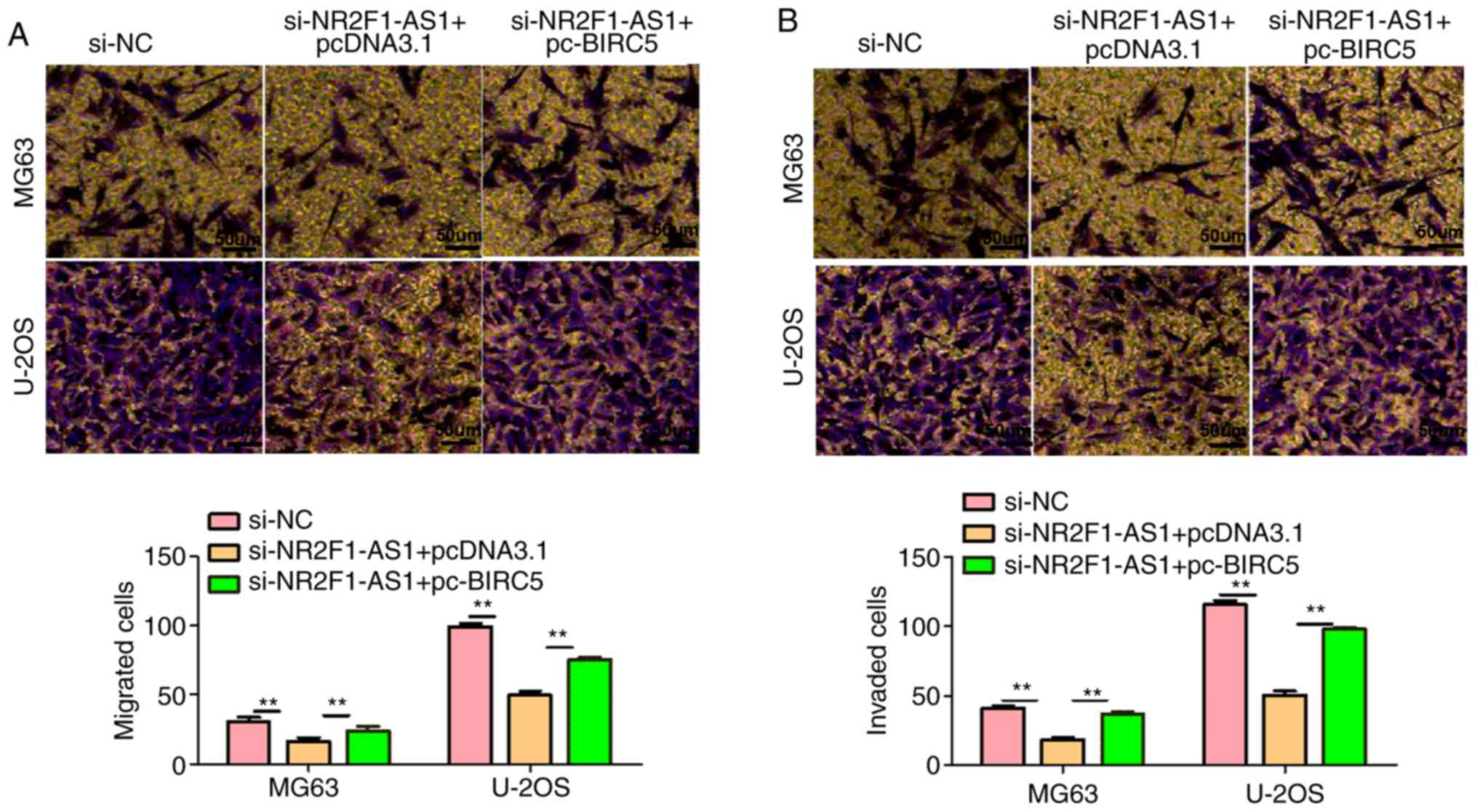
through a series of experiments, we demonstrated that
NR2F1-AS1-knockdown markedly decreased cell proliferation,
migration and invasion, and promoted cell apoptosis (Figs. 10B and C and 11A and B). However, this influence was
substantially weakened following overexpression of BIRC5, which
suggests that NR2F1-AS1 affects OS cell biological behaviors by
regulating BIRC5. Combined with previous studies, it was concluded
that NR2F1-AS1 facilitates the OS cell malignant phenotype by
downregulating miR-485-5p and miR-218-5p, and upregulating BIRC5
expression.
Discussion
Previously, abnormal expression of long non-coding
RNAs (lncRNAs) and microRNAs (miRNAs) has been reported to
contribute to tumor progression. For example, in esophageal
squamous cell carcinoma (ESCC), SNHG6 was identified to enhance
cell malignancy (33). In
osteosarcoma (OS), ANRIL has been reported to be closely associated
with the malignant behavior of OS cells (9). In addition, APTR has been reported to
inhibit miR-132-3p and upregulate YAP1 to accelerate OS progression
(34). In hepatocellular carcinoma,
DLGAP1-AS1 has been confirmed to facilitate cell proliferation via
the miR-486-5p/H3F3B axis (35).
NR2F1-AS1 has been reported to play a role in papillary thyroid
carcinoma (14), ESCC (12), endometrial cancer (15), hepatocellular carcinoma (36) and OS (16). In OS, NR2F1-AS1 has been identified
to sponge miR-483-3p to upregulate FOXA1 and enhance the malignant
properties of OS cells (16).
However, the mechanism of NR2F1-AS1 in OS remains unknown.
Initially, the present study determined that
NR2F1-AS1 expression was upregulated in OS tissues. Furthermore, a
high level of NR2F1-AS1 was closely correlated with a more advanced
Enneking stage and distant metastasis. Subsequently, NR2F1-AS1
expression was found to be present at a high level in OS cells. To
determine the roles of NR2F1-AS1, three siRNAs were first
transfected into OS cells in order to identify the most effective
siRNA for further functional experiments. The results demonstrated
that knockdown of NR2F1-AS1 significantly repressed the
proliferation, migration and invasion, and promoted the apoptosis
of OS cells, which indicated that NR2F1-AS1 exerts an oncogenic
function in OS, consistent with other studies.
A number of studies have reported that lncRNAs exert
biological functions in cancer by interacting with miRNAs to
regulate their target genes (10,13,30).
Therefore, the present study attempted to identify the potential
binding sites of NR2F1-AS1 using biological software analysis, and
it was identified that the seed regions of miR-485-5p and
miR-218-5p could recognize NR2F1-AS1 sequences. The subsequent
luciferase assays confirmed the interactions of miR-485-5p with
NR2F1-AS1, and miR-218-5p with NR2F1-AS1. miR-485-5p has been
recognized as an oncogene in non-small cell lung cancer (37), glioma (38), breast cancer (39), colorectal cancer (40), OS (41) and esophageal cancer (42). miR-218-5p has been reported to
inhibit the malignancy of cervical cancer (43), oral squamous cell carcinoma
(44) and non-small cell lung
cancer (45). To detect the
functional roles of miR-485-5p and miR-218-5p in OS, the expression
levels of miR-485-5p and miR-218-5p were first determined in OS
tissues. The data revealed that miR-485-5p and miR-218-5p
expression levels were low in OS tissues, which were in contrast to
NR2F1-AS1 expression, and both miR-485-5p and miR-218-5p were
negatively correlated with NR2F1-AS1 expression. Finally, it was
validated that NR2F1-AS1 inhibition led to an increase in both
miR-218-5p and miR-485-5p. In summary, these findings demonstrated
that NR2F1-AS1 may interact with miR-485-5p and miR-218-5p to exert
biological roles in OS.
It has been reported that in papillary thyroid
cancer (PTC), miR-485-5p is sponged by LINC00460 and thereby
upregulates Raf1 expression to facilitate PTC progression (46). In addition, in cholangiocarcinoma,
miR-485-5p has been demonstrated to negatively regulate FLVCR1-AS1
(47). Furthermore, in non-small
cell lung cancer, DGCR5 has been identified to sponge miR-218-5p to
promote cancer progression (48).
In prostate cancer, PCA3 sponges miR-218-5p and modulated HMGB1
(49). These findings demonstrate
that lncRNAs can affect the behavior of tumor cells via miR-485-5p
or miR-218-5p. The present study revealed that inhibition of
miR-485-5p or miR-218-5p could attenuate the effects of
NR2F1-AS1-knockdown on cell proliferation, migration and invasion,
and apoptosis, which indicated that NR2F1-AS1 affects the
malignancy of OS cells via miR-485-5p and miR-218-5p. Notably,
baculoviral inhibitor of apoptosis repeat-containing 5 (BIRC5) was
identified as a target gene of both miR-485-5p and miR-218-5p.
BIRC5, a member of the apoptosis suppressor (IAP) gene family, is
closely associated with tumor apoptosis. The present study
demonstrated that BIRC5 not only has a higher expression level in
OS tissues, but also that BIRC5 expression is negatively associated
with miR-485-5p and miR-218-5p, and positively correlated with
NR2F1-AS1. Furthermore, it was observed that NR2F1-AS1 affects the
proliferation, migration, invasion and apoptosis of OS cells via
BIRC5. Therefore, these data demonstrated that NR2F1-AS1
facilitates the OS cell malignant phenotype via downregulating
miR-485-5p and miR-218-5p, and then upregulating BIRC5 expression.
Notably, NR2F1-AS1 was reported to enhance the malignant properties
of OS by increasing FOXA1 expression via sponging of
microRNA-483-3p (16). In our
manuscript, we revealed that NR2F1-AS1 acted as an oncogene in
osteosarcoma that facilitated osteosarcoma cell proliferation and
migration through targeting miR-485-5p and miR-218-5p and then
targeting BIRC5. The difference from that study was that we
revealed NR2F1-AS1 exert roles by acting on two miRNAs
simultaneously, which confirmed the diversity of target genes. And
more importantly, we demonstrated that a new axis
NR2F1-AS1/miR-485-5p/miR-218-5p/BIRC5 is involved in the
tumorigenesis of OS, which revealed the multi-targeting and network
regulation of lncRNA-miRNA-mRNA. However, there are some
limitations in the present study. For example, only 32 OS tissues
were utilized and the number of samples was small. In addition,
in vivo results to confirm the effects of the investigated
NR2F1-AS1 axis on tumor growth were not obtained at present.
Therefore, the focus of future studies will be to collect more
tissue specimens and simultaneity to illuminate the effect of the
NR2F1-AS1/miR-485-5p/miR-218-5p/BIRC5 axis on tumor growth in
vivo.
In conclusion, the present study provides further
understanding of the oncogenic functions of NR2F1-AS1 in OS. To the
best of our knowledge, the present study was the first to
demonstrate that the NR2F1-AS1/miR-485-5p/miR-218-5p/BIRC5 axis is
involved in the tumorigenesis of OS. These findings revealed a
novel regulatory network in OS and may contribute to the
identification of new therapeutic targets for OS.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
GJ performed the experiments and drafted the
manuscript. YW performed the data analysis and figure design. YY
concepted the study design. ZL carried out the sample selection. XW
managed the project administration. All authors read and approved
the manuscript and agree to be accountable for all aspects of the
research in ensuring that the accuracy or integrity of any part of
the work are appropriately investigated and resolved.
Ethics approval and consent to
participate
The patient samples were collected and the protocol
was granted permission of the Medical Ethics Committee of Zhengzhou
Orthopedic Hospital, and informed consents of all of the patients
were obtained before the operation.
Patient consent for publication
Not applicable
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ritter J and Bielack SS: Osteosarcoma. Ann
Oncol. 21 (Suppl 7):vii320–vii325. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Arndt CA, Rose PS, Folpe AL and Laack NN:
Common musculoskeletal tumors of childhood and adolescence. Mayo
Clin Proc. 87:475–487. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gianferante DM, Mirabello L and Savage SA:
Germline and somatic genetics of osteosarcoma-connecting aetiology,
biology and therapy. Nat Rev Endocrinol. 13:480–491. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ferrari S and Serra M: An update on
chemotherapy for osteosarcoma. Expert Opin Pharmacother.
16:2727–2736. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lee C and Kikyo N: Strategies to identify
long noncoding RNAs involved in gene regulation. Cell Biosci.
2:372012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chen X, Li J, Liang D, Zhang L and Wang Q:
LncRNA AWPPH participates in the development of non-traumatic
osteonecrosis of femoral head by upregulating Runx2. Exp Ther Med.
19:153–159. 2020.PubMed/NCBI
|
|
7
|
Wang Y, Mei C, Su X, Wang H, Yang W and
Zan L: MEF2A regulates the MEG3-DIO3 miRNA mega cluster-targeted
PP2A signaling in bovine skeletal myoblast differentiation. Int J
Mol Sci. 20:27482019. View Article : Google Scholar
|
|
8
|
Nolte W, Weikard R, Brunner RM, Albrecht
E, Hammon HM, Reverter A and Kühn C: Biological network approach
for the identification of regulatory long non-coding RNAs
associated with metabolic efficiency in cattle. Front Genet.
10:11302019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Guan H, Mei Y, Mi Y, Li C, Sun X, Zhao X,
Liu J, Cao W, Li Y and Wang Y: Downregulation of lncRNA ANRIL
suppresses growth and metastasis in human osteosarcoma cells.
OncoTargets Ther. 11:4893–4899. 2018. View Article : Google Scholar
|
|
10
|
Liu D, Zhang H, Cong J, Cui M, Ma M, Zhang
F, Sun H and Chen C: H3K27 acetylation-induced lncRNA EIF3J-AS1
improved proliferation and impeded apoptosis of colorectal cancer
through miR-3163/YAP1 axis. J Cell Biochem. 121:1923–1933. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu X, Huang Z, Qian W, Zhang Q and Sun J:
Silence of lncRNA UCA1 rescues drug resistance of cisplatin to
non-small-cell lung cancer cells. J Cell Biochem. 120:9243–9249.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Han N, Zuo L, Chen H, Zhang C, He P and
Yan H: Long non-coding RNA homeobox A11 antisense RNA (HOXA11-AS)
promotes retinoblastoma progression via sponging miR-506-3p.
OncoTargets Ther. 12:3509–3517. 2019. View Article : Google Scholar
|
|
13
|
Guo F, Fu Q, Wang Y and Sui G: Long
non-coding RNA NR2F1-AS1 promoted proliferation and migration yet
suppressed apoptosis of thyroid cancer cells through regulating
miRNA-338-3p/CCND1 axis. J Cell Mol Med. 23:5907–5919. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yang C, Liu Z, Chang X, Xu W, Gong J, Chai
F and Cui D: NR2F1-AS1 regulated miR-423-5p/SOX12 to promote
proliferation and invasion of papillary thyroid carcinoma. J Cell
Biochem. 121:2009–2018. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang L, Zhao S and Mingxin YU: LncRNA
NR2F1-AS1 is involved in the progression of endometrial cancer by
sponging miR-363 to target SOX4. Pharmazie. 74:295–300.
2019.PubMed/NCBI
|
|
16
|
Li S, Zheng K, Pei Y, Wang W and Zhang X:
Long noncoding RNA NR2F1-AS1 enhances the malignant properties of
osteosarcoma by increasing forkhead box A1 expression via sponging
of microRNA-483-3p. Aging (Albany NY). 11:11609–11623. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gulino R, Forte S, Parenti R, Memeo L and
Gulisano M: MicroRNA and pediatric tumors: Future perspectives.
Acta Histochem. 117:339–354. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Meng YC, Lin T, Jiang H, Zhang Z, Shu L,
Yin J, Ma X, Wang C, Gao R and Zhou XH: MiR-122 Exerts inhibitory
effects on osteoblast proliferation/differentiation in osteoporosis
by activating the PCP4-mediated JNK pathway. Mol Ther Nucleic
Acids. 20:345–358. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tan YX, Hong Y, Jiang S, Lu MN, Li S, Chen
B, Zhang L, Hu T, Mao R, Mei R and Xiyang YB: MicroRNA-449a
regulates the progression of brain aging by targeting SCN2B in
SAMP8 mice. Int J Mol Med. 45:1091–1102. 2020.PubMed/NCBI
|
|
20
|
Fan H, Lu S, Wang S and Zhang S:
Identification of critical genes associated with human osteosarcoma
metastasis based on integrated gene expression profiling. Mol Med
Rep. 20:915–930. 2019.PubMed/NCBI
|
|
21
|
Xuan C, Jin M, Gao Y, Xu S, Wang L, Wang
Y, Han R and An Q: MiR-218 suppresses the proliferation of
osteosarcoma through downregulation of E2F2. Oncol Lett.
17:571–577. 2019.PubMed/NCBI
|
|
22
|
Wang DZ, Jing SF, Hao SB, Huang XY, Miao
QT and Gao JF: MiR-218 promotes apoptosis of U2OS osteosarcoma
cells through targeting BIRC5. Eur Rev Med Pharmacol Sci.
22:6650–6657. 2018.PubMed/NCBI
|
|
23
|
Jin J, Cai L, Liu ZM and Zhou XS:
MiRNA-218 inhibits osteosarcoma cell migration and invasion by
down-regulating of TIAM1, MMP2 and MMP9. Asian Pac J Cancer Prev.
14:3681–3684. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang M, Cai WR, Meng R, Chi JR, Li YR,
Chen AX, Yu Y and Cao XC: MiR-485-5p suppresses breast cancer
progression and chemosensitivity by targeting survivin. Biochem
Biophys Res Commun. 501:48–54. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yun S, Kim WK, Kwon Y, Jang M, Bauer S and
Kim H: Survivin is a novel transcription regulator of KIT and is
downregulated by miRNA-494 in gastrointestinal stromal tumors. Int
J Cancer. 142:2080–2093. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang XP, Yao J, Guan J, Zhou ZQ, Zhang ZY
and Yang J: MicroRNA-542-3p functions as a tumor suppressor via
directly targeting survivin in hepatocellular carcinoma. Biomed
Pharmacother. 99:817–824. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yu X, Zhang Y, Cavazos D, Ma X, Zhao Z, Du
L and Pertsemlidis A: MiR-195 targets cyclin D3 and survivin to
modulate the tumorigenesis of non-small cell lung cancer. Cell
Death Dis. 9:1932018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bhan A, Soleimani M and Mandal SS: Long
noncoding RNA and cancer: A new paradigm. Cancer Res. 77:3965–3981.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liu DC, Song LL, Liang Q, Hao L, Zhang ZG
and Han CH: Long noncoding RNA LEF1-AS1 silencing suppresses the
initiation and development of prostate cancer by acting as a
molecular sponge of miR-330-5p via LEF1 repression. J Cell Physiol.
234:12727–12744. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Emmrich S, Streltsov A, Schmidt F,
Thangapandi VR, Reinhardt D and Klusmann JH: LincRNAs MONC and
MIR100HG act as oncogenes in acute megakaryoblastic leukemia. Mol
Cancer. 13:1712014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Suwal A, Hao JL, Liu XF, Zhou DD, Pant OP,
Gao Y, Hui P, Dai XX and Lu CW: NONRATT021972 long-noncoding RNA: A
promising lncRNA in diabetes-related diseases. Int J Med Sci.
16:902–908. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang Y, Li R, Ding X, Zhang K and Qin W:
Upregulation of long non-coding RNA SNHG6 promote esophageal
squamous cell carcinoma cell malignancy and its diagnostic value.
Am J Transl Res. 11:1084–1091. 2019.PubMed/NCBI
|
|
34
|
Guan H, Shang G, Cui Y, Liu J, Sun X, Cao
W, Wang Y and Li Y: Long noncoding RNA APTR contributes to
osteosarcoma progression through repression of miR-132-3p and
upregulation of yes-associated protein 1. J Cell Physiol.
234:8998–9007. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Peng X, Wei F and Hu X: Long noncoding RNA
DLGAP1-AS1 promotes cell proliferation in hepatocellular carcinoma
via sequestering miR-486-5p. J Cell Biochem. 121:1953–1962. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Huang H, Chen J, Ding CM, Jin X, Jia ZM
and Peng J: LncRNA NR2F1-AS1 regulates hepatocellular carcinoma
oxaliplatin resistance by targeting ABCC1 via miR-363. J Cell Mol
Med. 22:3238–3245. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Gao F, Wu H, Wang R, Guo Y, Zhang Z, Wang
T, Zhang G, Liu C and Liu J: MicroRNA-485-5p suppresses the
proliferation, migration and invasion of small cell lung cancer
cells by targeting flotillin-2. Bioengineered. 10:1–12. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wang R, Zuo X, Wang K, Han Q, Zuo J, Ni H,
Liu W, Bao H, Tu Y and Xie P: MicroRNA-485-5p attenuates cell
proliferation in glioma by directly targeting paired box 3. Am J
Cancer Res. 8:2507–2517. 2018.PubMed/NCBI
|
|
39
|
Zhang JX, Chen ZH, Chen DL, Tian XP, Wang
CY, Zhou ZW, Gao Y, Xu Y, Chen C, Zheng ZS, et al:
LINC01410-miR-532-NCF2-NF-κB feedback loop promotes gastric cancer
angiogenesis and metastasis. Oncogene. 37:2660–2675. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Hu XX, Xu XN, He BS, Sun HL, Xu T, Liu XX,
Chen XX, Zeng KX, Wang SK and Pan YQ: MicroRNA-485-5p functions as
a tumor suppressor in colorectal cancer cells by targeting CD147. J
Cancer. 9:2603–2611. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wang FR, Xu SH, Wang BM and Wang F:
MiR-485-5p inhibits metastasis and proliferation of osteosarcoma by
targeting CX3CL1. Eur Rev Med Pharmacol Sci. 22:7197–7204.
2018.PubMed/NCBI
|
|
42
|
Han DL, Wang LL, Zhang GF, Yang WF, Chai
J, Lin HM, Fu Z and Yu JM: MiRNA-485-5p, inhibits esophageal cancer
cells proliferation and invasion by down-regulating O-linked
N-acetylglucosamine transferase. Eur Rev Med Pharmacol Sci.
23:2809–2816. 2019.PubMed/NCBI
|
|
43
|
Xu Y, He Q, Lu Y, Tao F, Zhao L and Ou R:
MicroRNA-218-5p inhibits cell growth and metastasis in cervical
cancer via LYN/NF-κB signaling pathway. Cancer Cell Int.
18:1982018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Li X, He J, Shao M, Cui B, Peng F, Li J,
Ran Y, Jin D, Kong J, Chang J, et al: Downregulation of miR-218-5p
promotes invasion of oral squamous cell carcinoma cells via
activation of CD44-ROCK signaling. Biomed Pharmacother.
106:646–654. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zhu K, Ding H, Wang W, Liao Z, Fu Z, Hong
Y, Zhou Y, Zhang CY and Chen X: Tumor-suppressive miR-218-5p
inhibits cancer cell proliferation and migration via EGFR in
non-small cell lung cancer. Oncotarget. 7:28075–28085. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Li G and Kong Q: LncRNA LINC00460 promotes
the papillary thyroid cancer progression by regulating the
LINC00460/miR-485-5p/Raf1 axis. Biol Res. 52:612019. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Bao W, Cao F, Ni S, Yang J, Li H, Su Z and
Zhao B: lncRNA FLVCR1-AS1 regulates cell proliferation, migration
and invasion by sponging miR-485-5p in human cholangiocarcinoma.
Oncol Lett. 18:2240–2247. 2019.PubMed/NCBI
|
|
48
|
Wang J, Shu HZ, Xu CY and Guo SG: LncRNA
DGCR5 promotes non-small cell lung cancer progression via sponging
miR-218-5p. Eur Rev Med Pharmacol Sci. 23:9947–9954.
2019.PubMed/NCBI
|
|
49
|
Zhang G, He X, Ren C, Lin J and Wang Q:
Long noncoding RNA PCA3 regulates prostate cancer through sponging
miR-218-5p and modulating high mobility group box 1. J Cell
Physiol. 234:13097–13109. 2019. View Article : Google Scholar : PubMed/NCBI
|