Introduction
Breast cancer (BC) is one of the most commonly
diagnosed malignancy in the world. The mortality rate for BC ranks
first among all female malignant tumors (1). Globally, the number of newly diagnosed
BC cases reached approximately 2.1 million in 2018, accounting for
almost 25% of cancer cases among women (2). BC exhibits a complex pathogenesis and
is a clinically heterogeneous disease with a wide range of clinical
behaviors and treatment responses (1). Although many dysregulated molecular
pathways have been discovered in BC, the development of effective
therapeutic methods has been limited (3). It is urgent to discover novel
molecules to suppress BC proliferation, induce apoptosis and
inhibit invasion, and provide potential therapeutic strategies to
improve the survival and quality of life of BC patients (4).
MicroRNAs (miRNAs/miRs) are a class of endogenous
non-coding RNAs of approximately 19–24 nucleotides in length, which
are generally located in unstable regions of the human genome and
are usually dysregulated in malignant tumors to regulate gene
functions (5). miRNAs regulate
target genes through binding to the 3′untranslated regions (3′UTRs)
of the target mRNAs, subsequently inhibiting gene expression
(6). Through regulation of the
targeted proteins, miRNAs play an important role in many tumor
cellular processes such as proliferation, cell cycle, apoptosis,
invasion and metastasis, and participate in almost all signaling
pathways. miR-188-5p has been reported to be an inhibitor of tumor
growth and metastasis in prostate cancer (7) and hepatocellular carcinoma (8). However, to the best of our knowledge,
the functions of miR-188-5p in BC remain elusive.
In the present study, we detected the expression of
miR-188-5p in tumor tissues of BC patient tissues and several BC
cell lines. Furthermore, we investigated its regulatory role in BC
proliferation, apoptosis and invasion. We also predicted and
confirmed the targeted protein of miR-188-5p, transcription factor
zinc finger protein 91 (ZFP91), elucidating the regulatory
mechanisms of miR-188-5p in BC.
Materials and methods
Patients and tissues
One hundred paired BC tissue specimens including
malignant and normal tissues used in this study were obtained from
Shengjing Hospital of China Medical University (Shenyang, Liaoning,
China) during the period from January 2017 to December 2018 with
the informed consent of patients. The age range of the patients was
from 31 to 75 years, with a mean age of 48.28 years. All
experiments were approved by the Ethics Committee of Shengjing
Hospital of China Medical University (no. 2016PS18J). The samples
were snap-frozen in liquid nitrogen and stored at −80°C. TNM
staging system was performed for tumor grading of BC and for
evaluating and staging of patients, respectively, which was carried
out according to the 7th edition of the American Joint Committee on
Cancer (AJCC) TNM classification system (9).
Cell cultivation
The BC cell lines MDA-MB-231 (ATCC, CRM-HTB-26),
BT-549 (ATCC, HTB-122), and MCF-7 (ATCC, CRL-3435) were cultured in
RPMI-1640 medium (Sigma-Aldrich; Merck KGaA) containing 10%
heat-inactivated fetal bovine serum (FBS) (MP Biomedicals), 1%
penicillin-streptomycin (Invitrogen; Thermo Fisher Scientific,
Inc.; no. 15070-063). The nonmalignant mammary epithelial cell
line, MCF 10A (ATCC, CRL-10317), was cultivated in DMEM/F12 Ham's
Mixture supplemented with 5% Equine Serum (Hyclone; GE Healthcare),
EGF (20 ng/ml), insulin (10 µg/ml), hydrocortisone (0.5 mg/ml), and
cholera toxin (100 ng/ml) (all from Sigma-Aldrich; Merck KGaA). All
cells were incubated at 37°C in a humidified 5% CO2
atmosphere.
RNA isolation and quantitative
(q)PCR
Total RNAs from tissues or cells were isolated using
RNX™-Plus Reagent (Cinnagen) and cDNA was synthesized using the
PrimeScript™ RT Reagent Kit (Takara) according to the
manufacturer's instructions. qPCR was performed using a SYBR Premix
ExTaq™ kit (Takara), with the following primer sets on the ABI 7300
qPCR system (Applied Biosystems). β-actin was used to normalize the
relative expression of the target genes. miRNAs were detected
through a miScript II RT kit (Qiagen) in a fluorescence thermal
cycler (Bio-Rad Laboratories, Inc.). The primers for miR-188-5p and
the reference gene U6 were purchased from Novland Biopharm. The
thermocycling condition were 95°C for 10 min, followed by 40 cycles
of 95°C for 15 sec and 60°C for 1min, followed by a hold at 4°C.
The relative expression ratio of miR-188-5p was quantified using
the 2−ΔΔCq method (10).
The relative expression of miR-188-5p was normalized to U6. The
primer sequences are listed in Table
I.
 | Table I.Primer information. |
Table I.
Primer information.
| Gene name | Bi-directional
primer sequence |
|---|
| miR-188-5p | F:
5′-CCCTCTCTCACATCCCTTGCAT-3′ |
|
| R:
5′-ATCCTGCAAACCCTGCATGTG-3′ |
| ZFP91 | F:
5′-TGAGACCTACAAACCCCACTT-3′ |
|
| R:
5′-CCTTTTGGGTAAACGTGGACTTT-3′ |
| Homo-β-actin | F:
5′-TTCCTCCGCAAGGATGACACGC-3′ |
|
| R:
5′-CCTTTTGGGTAAACGTGGACTTT-3′ |
| U6 snRNA | F:
5′-CGGGTTTGTTTTGCATTTCT-3′ |
|
| F:
5′-AGTCCCAGCATGAACAGCTT-3′ |
Plasmid preparation
The coding region of human ZFP91 was amplified from
human breast cancer cell MDA-MB-231 cDNA library by PCR. Then we
cloned the prepared ZFP91 fragment into pCMV-Tag2B (Stratagene) to
obtain pCMV-Tag2B-ZFP91. The primer sequences are listed in
Table I.
Cell transfection
The miR-188-5p mimics and miR-NC were purchased from
Thermo Fisher Scientific, Inc. Firstly, Lipofectamine 2000
transfection reagent (Thermo Fisher Scientific, Inc.) was used to
transfect miR-188-5p mimics, miR-NC into MDA-MB-231 cells in
accordance with the manufacturer's instructions.
After the whole detection, the pCMV-Tag2B vector
(PC) was transfected into MDA-MB-231 cells with miR-NC (NC+PC) or
miR-188-5p mimics (PC+miR-188-5p); pCMV-Tag2B-ZFP91 was
transfected into MDA-MB-231 cells with miR-188-5p mimics
(miR-188-5p+ZFP91). As the cells were grown to 90%, miR mimics or
vector was transfected into cells according to the manufacturer's
instructions. Then cells were cultivated for up to 72 h. Finally,
the total RNA and protein were extracted and properly stored for
further research.
Cell proliferation assay
Cell Counting Kit-8 (CCK-8 Kit, Dojindo) was
performed to detect cell proliferation based on the reduction of
WST-8 to WST-8 formazan. Briefly, the MDA-MB-231 cells were seeded
in a 96-well plate at a density of 5×103 cells/well. On
the day of the experiment, the cells were transfected with empty
vector and miR-188-5p. CCK-8 reagent was added into the culture
medium at the indicated time and incubated for 60 min; the
absorbance at 450 nm was measured by a microplate reader.
Colony formation assay
MDA-MB-231 cells from the different treated groups
were seeded in a 60-mm dish containing 200 cells, followed by a
14-day cultivation at 37°C with 5% CO2. The supernatant
was discarded and cells were washed twice with PBS. The colonies
were fixed in 4% paraformaldehyde for 15 min and then stained with
Giemsa staining solution (Solarbio Science & Technology, Co.,
Ltd., Beijing, China) for 20 min. Colonies were counted and images
were captured under an inverted microscope (Nikon, Tokyo, Japan).
This assay was repeated 3 times.
Cell apoptosis assay
MDA-MB-231 cells were stained by Annexin V-Alexa
Fluor-488/propidium iodide (PI) staining to identify the apoptotic
MDA-MB-231 cells. After transfection with miR-188-5p for 48 h,
MDA-MB-231 cells were stained with Annexin V-Alexa Fluor-488 for 15
min on ice, followed by the addition of PI solution for the
secondary staining process. All experimental procedures were
strictly protected from lights. The data were calculated by FlowJo
software v.8.7 (Tree Star) after FACs Calibur (BD Biosciences)
analysis.
Cell migration and invasion
assays
After the counting, MDA-MB-231 cells in the
different groups were inoculated equally at a density of
5×105 cells/ml in the upper compartments of
polycarbonate membrane filters. Cell migration and invasion assays
were performed; uncoated for the migration assay and coated with
Matrigel (1:8, BD Biosciences) for the invasion assay. After 24 h,
the migrated and invaded cells in the membrane were fixed with
methanol, and then stained with 0.1% crystal violet for 20 min at
room temperature. Cells were observed under a light microscope with
magnification, ×100.
Western blotting
Protein samples extracted from tissues or cultivated
cells were lysed in RIPA buffer containing protease and phosphatase
inhibitor cocktail and incubated at 4°C, followed by the quantified
measurement of protein using BCA kit (FUJIFILM Wako Pure Chemical
Corp.). After protein samples (40 µg/each sample) were loaded and
separated on 10% SDS-PAGE gels for electrophoresis, the proteins
were then transferred onto a polyvinylidene difluoride (PVDF)
membranes (Millipore, USA). The membranes were blocked in 5% (w/v)
skim milk for 1 h at room temperature and incubated at 4°C
overnight with primary antibodies anti-ZFP91 (dilution 1:1,000,
Abcam, ab30970) and anti-vimentin (dilution 1:1,000, Cell Signaling
Technology, Inc., #3932), anti-E-cadherin (dilution 1:1,000, Cell
Signaling Technology, Inc., #3195), N-cadherin (dilution 1:1,000,
Cell Signaling Technology, Inc., #13116), matrix metalloproteinase
(MMP)-2 (dilution 1:1,000, Cell Signaling Technology, Inc.,
#40994), MMP-9 (dilution 1:1,000, Cell Signaling Technology, Inc.,
#13667), NF-κB p65 (dilution 1:1,000, Cell Signaling Technology,
Inc., #8242), RelB (dilution 1:1,000, Cell Signaling Technology,
Inc, #4954) and GAPDH (dilution 1:5,000, Cell Signaling Technology,
Inc, #5174) as internal control. On the following day, all
membranes were incubated with anti-rabbit IgG HRP-labeled secondary
antibodies (dilution 1:2,000, Cell Signaling Technology, Inc.,
#7074). Finally, the signals were detected and analyzed with the
application of Luminata Forte Western HRP Substrate (Millipore) in
the Bio-Rad ChemiDox XRS+ imaging system (Bio-Rad
Laboratories).
Luciferase reporter assay
To further investigate the specific correlation
between miR-188-5p and ZFP91, Targetscan (www.targetscan.org/mamm_31/) and miRanda (www.microrna.org/microrna/home.do) were
performed. The ZFP91 was selected to be the predicted targeting of
miR-188-5p. The fragments of the 3′UTR of ZFP91 containing
miR-188-5p binding sites and its mutants were amplified by PCR, and
then the PCR products were inserted into pmirGLO dual-luciferase
miRNA target expression vector (Promega Corp.). The reporter and
control vector were transfected into 293T cells using Lipofectamine
3000 (Thermo Fisher Scientific, Inc.). After cultivation for 48 h,
the relative luciferase activity was examined by the
Dual-Luciferase Reporter Assay Kit (Thermo Fisher Scientific, Inc.)
according to the manufacturer's instructions.
Preparation of tumor xenograft animal
model and treatment with miR-188-5p mimics
Thirty-six nude mice, female, weighing 20±5 g, were
purchased from Huafukang Biotech (Beijing, China). The experiments
were performed in the animal facility at the Department of
Laboratory Animal Science of China Medical University and approved
by the Animal Ethics Committee of Shengjing Hospital (approval no.
2019218). Nude mice were randomly divided into a control group
(n=12), miR-188-5p group (n=12), and NC group (n=12). A density of
5×106 cells in logarithmic phase were transfected with
1X PBS (control group), NC or miR-188-5p. Then the different groups
of cells were resuspended in 1X PBS and injected into the nude
mice, respectively. Then tumor size was measured every 3 days using
a slide caliper and the tumor volume (V) was calculated using the
formula: V=length × width2/2. After 21 days, the mice
were euthanasia by cervical dislocation and the tumors were
excised, imaged, weighed and stored properly for further
investigations.
Statistical analysis
GraphPad Prism 8 (GraphPad Software, Inc.) was used
to perform statistical analysis. The results are represented as
mean ± SD of at least 3 independent experiments. The comparisons
between groups were evaluated by Student' t-test. One-way ANOVA
followed by Tukey test was used to evaluate the differences for
multiple comparisons. The statistical significance of correlations
between miR-188-5p and ZFP91 expression in BC tissue were analyzed
by Pearson's correlation coefficient. P<0.05 was considered to
indicate a statistically significant difference.
Results
miR-188-5p is significantly decreased
in BC tissue and cell lines
Firstly, we analyzed the expression level of
miR-188-5p in 100 cases of BC tissues and adjacent counterparts by
RT-qPCR. The results showed that the level of miR-188-5p in BC
tissues was significantly lower than that in the normal adjacent
counterparts (Fig. 1A, P<0.05).
We also found that miR-188-5p was correlated with BC TNM stage
(Fig. 1B, P<0.05). The
expression level of miR-188-5p in advanced BC tumors was lower than
that in early stage tumors, suggesting that miR-188-5p is inversely
correlated with the malignancy of BC.
We also compared the expression level of miR-188-5p
in the non-malignant mammary epithelial cell line MCF-10A and BC
cell lines MDA-MB-231, BT-549 and MCF-7. Our data showed that the
levels of miR-188-5p in the MDA-MB-231, BT-549 and MCF-7 cells were
lower than that in the MCF-10A cells (Fig. 1C, P<0.05). Meanwhile, the lowest
miR-188-5p expression was detected in MDA-MB-231, therefore,
MDA-MB-231 cells were selected for further experiments.
miR-188-5p inhibits proliferation,
induces cell apoptosis and suppresses migration and invasion of BC
cells
As the expression of miR-188-5p in both BC cell
lines and tumor tissues of BC patients were clearly downregulated,
we sought to investigate the effects of miR-188-5p on BC
development by using both in vitro BC cell line cultivation
and in vivo mouse tumor xenografts. As shown in Fig. 2A, transfection of MDA-MB-231 cells
with miR-188-5p mimics significantly elevated the expression level
of miR-188-5p when compared to the control and miR-NC groups
(P<0.05). Importantly, the increased level of miR-188-5p in
MDA-MB-231 cells significantly inhibited the cell proliferation
when compared to the control and miR-NC groups (Fig. 2B and C, P<0.05). It was also
observed that the apoptotic MDA-MB-231 cell numbers were
significantly increased by the upregulation of miR-188-5p when
compared to the control and miR-NC groups (Fig. 2D, P<0.05). Importantly,
miR-188-5p mimics significantly inhibited the invasion and
migration abilities of the MDA-MB-23 cells under Transwell assay
detection when compared to the control and miR-NC groups (Fig. 2E, P<0.05). Moreover, miR-188-5p
mimics significantly enhanced the expression of vimentin and
N-cadherin and reduced the level of E-cadherin when compared to the
control and miR-NC groups (Fig. 2F,
P<0.05). The matrix metalloproteinases, MMP2 and MMP9 (MMP2/9),
possess the ability to hydrolyze components of the basement
membrane and stimulate tumor growth, metastasis and
epithelial-mesenchymal transition (EMT) (11). miR-188-5p mimics were demonstrated
to significantly inhibit the expression of MMP2 and MMP9 (Fig. 2F, P<0.05). These data provide
robust evidence that miR-188-5p inhibits the tumor proliferation,
induces apoptosis, reduces tumor invasion and migration and
inhibits EMT of BC, which may be through the regulation of MMP2/9
expression.
ZFP91 is the downstream target of
miR-188-5p
To further investigate the specific correlation
between miR-188-5p and ZFP91, Targetscan (www.targetscan.org/mamm_31/) and miRanda (www.microrna.org/microrna/home.do) were
performed. The results predicted that miR-188-5p possesses the
binding sites of ZFP91 (Fig. 3A).
Hence, we sought to discover the regulatory mechanisms of
miR-188-5p on BC development through targeting on ZFP91. As
hypothesized, the upregulation of miR-188-5p in MDA-MB-231 cells
decreased ZFP91 mRNA and protein levels when compared to the miR-NC
and control groups (Fig. 3B,
P<0.05). Moreover, the luciferase assay confirmed that
miR-188-5p specifically binds to the 3′UTR of ZFP91 (Fig. 3C, P<0.05). It was also discovered
that the injection of MDA-MB-231 transfected with miR-188-5p mimics
in tumor xenograft mice inhibited ZFP91 expression (Fig. 6B, P<0.05). These results
suggested that ZFP91 is the downstream target gene of miR-188-5p in
BC.
miR-188-5p regulates BC cell
progression through targeting ZFP91
To further investigate the biological functions of
miR-188-5p in BC development, we established a ZFP91-overexpressing
MDA-MB-231 cell line. The expression of ZFP91 was confirmed by
RT-qPCR (Fig. S1, P<0.05). With
this system, inhibition of ZFP91 by miR-188-5p mimics was reversed
(Fig. 4A, P<0.05). Then it was
found that the co-transfection of MDA-MB-231 cells,
miR-188-5p+ZFP91 group, significantly enhanced the cell
proliferation compared to that in PC+miR-188-5p group (Fig. 4B and C, P<0.05), significantly
suppressed cell apoptosis (Fig. 4D,
P<0.05) and significantly promoted invasion, migration (Fig. 4E, P<0.05) and EMT (Fig. 4F, P<0.05), in contrast with the
mono-transfection of miR-188-5p mimics in MDA-MB-231 cells.
Moreover, the regulatory role of miR-188-5p on MMP2 and MMP9 was
also reversed by overexpression of ZFP91.
 | Figure 4.MDA-MB-231 cell proliferation and
apoptosis are regulated by miR-188-5p/ZFP91. pCMV-Tag2B vector (PC)
was transfected into BC MDA-MB-231 cells with miR-NC (NC+PC) or
miR-188-5p mimics (PC+miR-188-5p); pCMV-Tag2B-ZFP91 was transfected
into MDA-MB-231 cells with miR-188-5p mimics (miR-188-5p+ZFP91).
(A) The mRNA levels of miR-188-5p in the co-transfected
overexpressing ZFP91/MDA-MB-231 cells were quantified by RT-qPCR.
*P<0.05. Cell proliferation was measured by (B) the CCK-8 Kit
and (C) colony formation assay. *P<0.05, compared to control
group; #P<0.05, compared to miR-NC group. (D) Cell
apoptosis was detected by Annexin V/PI staining and FACs.
*P<0.05. (E) The invasion and migration capability of MDA-MB-231
cells were detected by Transwell assay. *P<0.05. (F) Expression
of E-cadherin, N-cadherin, Vimentin, MMP2, MMP9 and NF-κB p65(Rel)
were detected by western blotting. Relative genes expression was
normalized by GAPDH expression. The data are represented as the
mean ± SD (n=3). *P<0.05. One-way ANOVA followed by Tukey's test
was used to evaluate the difference for multiple comparisons. BC,
breast cancer; ZFP91, zinc finger protein 91; MMP, matrix
metalloproteinase; NC, negative control. |
miR-188-5p and ZFP91 are correlated in
tumor tissues of BC patients
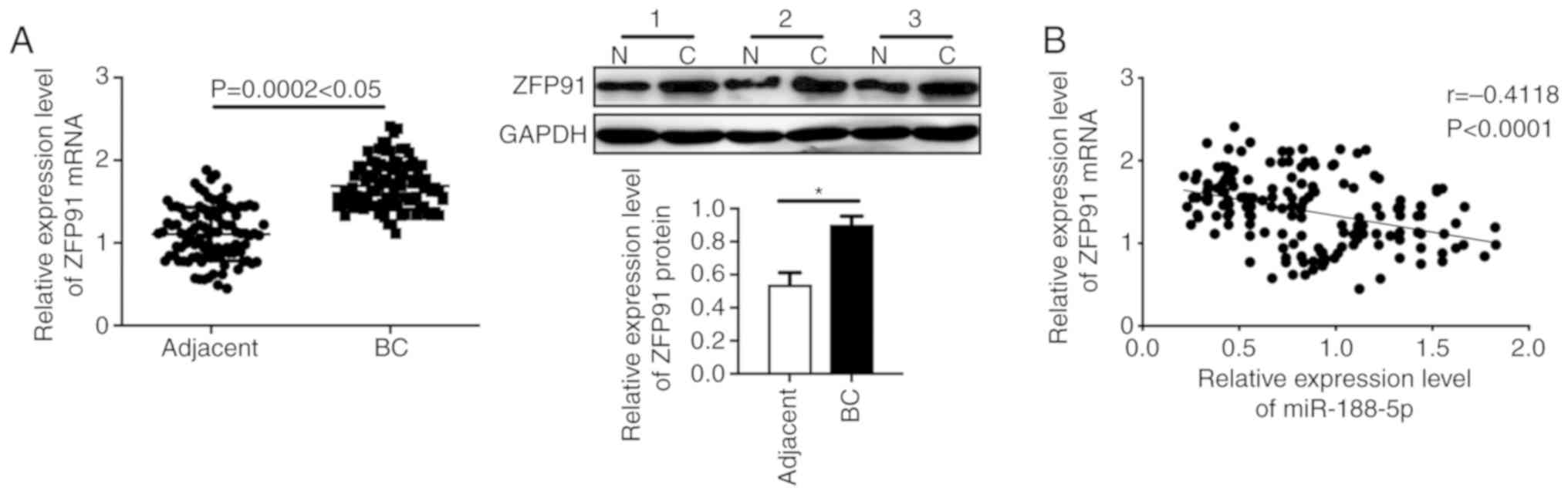
Furthermore, we examined the levels of ZFP91 in the
tumor tissues and adjacent normal tissues of BC patients. The
aberrantly high level of ZFP91 was observed in the tumor tissues of
the BC patients. (Fig. 5A,
P<0.05). Spearman's correlation analysis showed a significantly
inverse correlation between miR-188-5p and ZFP91 in the BC patient
tissues (Fig. 5B, P<0.05). Taken
together, these results further confirmed that the proliferation
and apoptosis of BC is regulated by miR-188-5p/ZFP91.
miR-188-5p inhibits the proliferation
of MDA-MB-231 cells and reduces the expression of ZPF91 in a BC
xenograft mouse model
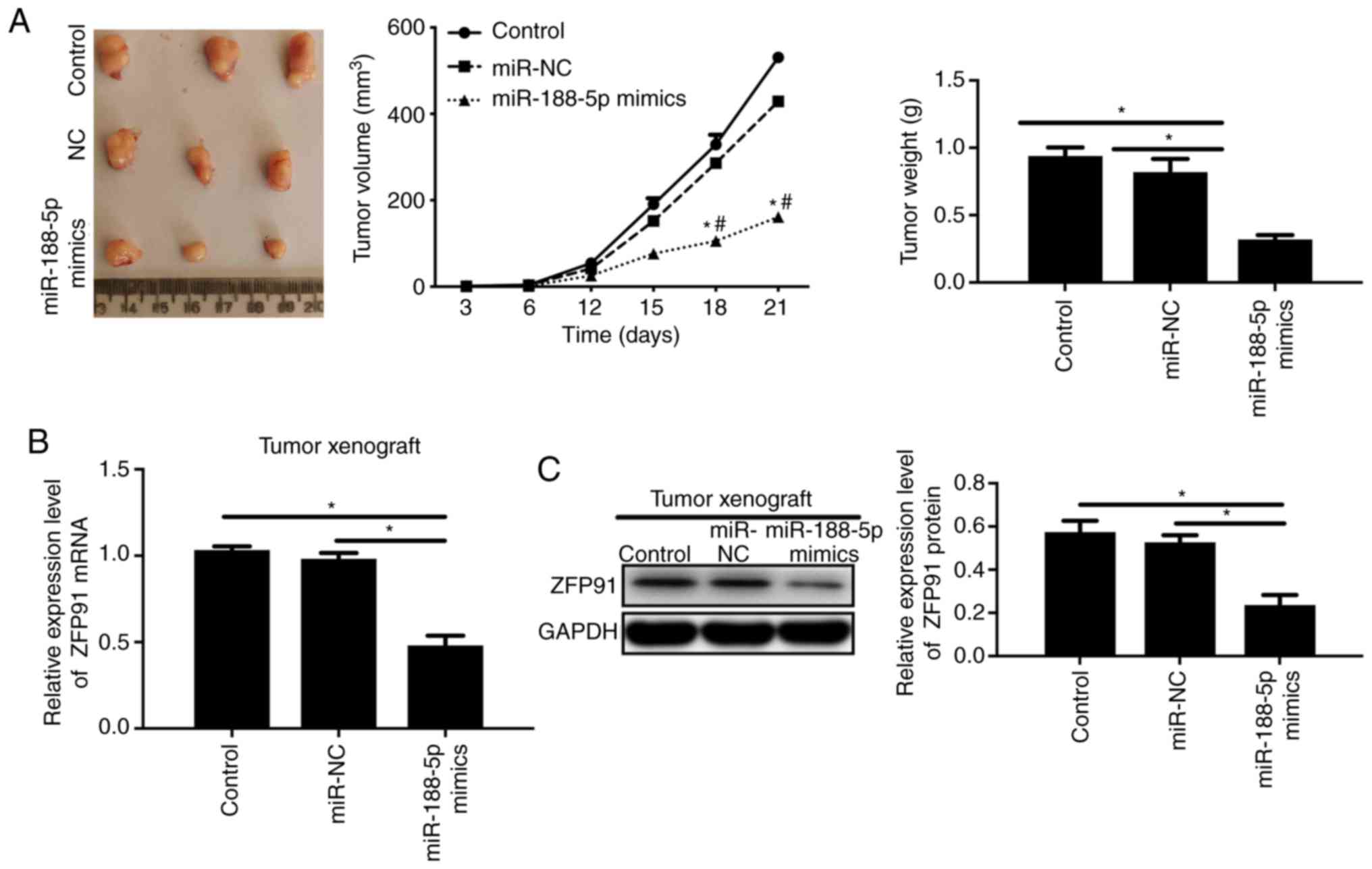
Moreover, to evaluate the regulatory role of
miR-188-5p in a BC xenograft mouse model, we injected the
MDA-MB-231 cells transfected with miR-188-5p mimics or miR-NC into
nude mice. The results showed that miR-188-5p mimics inhibited the
tumor volume and weight compared to the miR-NC group (Fig. 6A, P<0.05). Protein expression and
the mRNA level of ZPF91 were also suppressed by miR-188-5p mimics
in the tumor tissues of the xenograft mouse model when compared
with the miR-NC and control groups (Fig. 6B, P<0.05).
miR-188-5p/ZFP91 axis regulates
NF-kB/P65 and RelB expression
Numerous studies have reported that zinc finger
protein 91 (ZFP91) promotes proliferation and tumorigenesis of
different cancer types via regulation of the NF-κB p65 pathway
(12–14). Therefore, to further investigate the
regulatory mechanism of the miR-188-5p/ZFP91 axis, we detected the
expression of NF-κBp65 and RelB in BC cells. The results showed
that miR-188-5p mimics significantly reduced the expression of
NF-κBp65 and RelB together (Fig.
S2A, P<0.05). Moreover, the co-transfection of miR-188-5p
mimics and ZFP91 also upregulated the expression levels of NF-κBp65
and RelB compared to the mono-transfection of miR-188-5p mimics
(Fig. S2B, P<0.05). In summary,
these results illustrated that the miR-188-5p/ZFP91 axis regulates
the progression of BC via the non-canonical NF-κB signaling
pathway.
Discussion
Breast cancer (BC) is one of the most common types
of tumors diagnosed in women worldwide (1). BC is the second leading cause of
cancer-related mortality worldwide (1) (15).
In 2015, more than 3,00,000 women were diagnosed with BC in China,
and almost 17 percent of all newly diagnosed cancer cases were in
women (1). However, the molecular
mechanisms of BC still await elucidation, and effective molecular
targets for the diagnosis and treatment of BC are urgently required
(16). Recently, research has
reported that miRNAs are small non-coding RNA molecules which
regulate target protein expression to play critical roles as
tumor-promotors or suppressors (17). Several studies have demonstrated
that miR-188-5p promotes cell proliferation, migration and
metastasis in gastric cancer (18,19)
and hepatocellular carcinoma (8).
Moreover, Iwakawa et al detected higher expression of
miR-188-5p in stage III breast cancer and TNBC (20). Wang et al reported that
circulating miR-188-5p was upregulated in BC patients and
associated with TNM of BC; interestingly, miR-188-5p was
downregulated in BC MDA-MB-231 and MCF-7 cells. Moreover, using
gain-of- and loss-of-function analyses of miR-188-5p in breast
cancer cells the authors demonstrated that miR-188-5p inhibited the
proliferation and invasion of BC MDA-MB-231 cells via targeting
IL6ST (21). However, we
demonstrated that the expression of miR-188-5p was drastically
downregulated in BC tissue specimens, which was also decreased in
BC cell lines, MDA-MB-231, BT549 and MCF-7, compared to normal
breast epithelial cell line MCF-10A. Moreover, the downregulation
of miR-188-5p was significantly associated with advanced TNM stage.
However, to investigate the relationship of miR-188-5p and BC
patient prognosis, we found that Kaplan-Meier analysis of
miR-188-5p was limited due to the small sample size in TCGA. These
results illustrated that the downregulation of miR-188-5p may be
related with BC progression in the clinic, suggesting that
miR-188-5p may be a valuable BC diagnostic indicator. Moreover, we
confirmed that miR-188-5p mimics considerably inhibited the
proliferation, induced the apoptosis and inhibited the invasion of
BC cells, suggesting that miR-188-5p plays an inhibitory role in BC
cells.
Transcription factor zinc finger protein 91 (ZFP91)
was firstly identified in the mouse in 1995 (22), which was found to be overexpressed
in colon (12), liver, prostate,
stomach (23) and breast cancer
(24). ZFP91 has a molecular mass
of 63.4 kDa with 570 amino acids, containing five zinc-finger
motives, a leucine zipper, a coiled-coil structure and nuclear
localization sequences. ZFP91 was confirmed to be a transcription
factor located in the cellular nucleus (25). Ma et al reported that ZFP91
functions as an oncogene in cancer development by activating HIF-1α
transcription (12). The
overexpression of ZFP91 was also found to result in the promotion
of NK-κB signaling pathway activation through increasing NK-κB
inducing kinase (NIK) (14), whose
activity and overexpression are related to cancer progression in
melanoma, pancreatic, breast- and lung cancer (26). The inhibition of ZFP91 was
demonstrated to promote apoptosis in BC, stomach cancer cells
(23), colon cancer and endometrial
cancer (25). In addition, the
overexpression of ZFP91 was found to increase the cancer cell
growth rate and metastatic capability (23). ZFP91 was also reported to interact
with cyclin-dependent kinase inhibitor 2A (CDKN2A), which is an
alternative reading frame (ARF) tumor suppressor, inhibiting the
induction of p53-dependent cell death (27). To illuminate the molecular
mechanisms of miR-188, we predicted that IL6ST, FOXN2, ZFP91 may be
the targets of miR-188-5p using Targetscan and miRanda.
Furthermore, Peng et al reported that ZFP91 is the target
protein of miR-188-5p in gastric cancer (28). In addition, overexpression of
miR-188-5p was confirmed to inhibit the progression of breast
cancer. Thus, we chose the reported oncogene ZFP91 for further
investigation. In the present study, we confirmed that the
3′untranslated region (3′UTR) of ZFP91 was bound by miR-188-5p
through Dual luciferase assay. Moreover, transfection of miR-188-5p
mimics in MDA-MB-231 cells reduced the ZFP91 mRNA and protein
levels together. miRNAs usually bind to the 3′UTRs of target mRNAs
and do not reduce the level of mRNAs; however, miRNAs also were
reported to decay the target mRNAs and decease mRNA level (20,29).
Restoration of ZFP91 largely reversed the decreased proliferation
and induced apoptosis which were both regulated by miR-188-5p
overexpression. Moreover, in the tumor xenograft mouse model, we
observed that the expression of ZFP91 was downregulated by an
increased level of miR-188-5p. Furthermore, the expression of
miR-188-5p and ZFP91 were negatively correlated in BC patient
tissues. Therefore, our studies confirmed that miR-188-5p can
inhibit the progression of human BC via targeting ZFP91.
ZFP91 has been reported to promote proliferation in
colon cancer (12), prostate cancer
(13) and gastric cancer (28). Ma et al reported that ZFP91
activates NF-kappaB/p65 to promote proliferation and tumorigenesis
of colon cancer (12). Paschke
et al identified that ZFP91 is a noncanonical NF-κB
signaling pathway regulator with oncogenic properties in prostate
cancer (13). In the present study,
we also confirmed that a decrease in ZFP91 could significantly
inhibit NF-κB/p65 and RelB expression in BC cells. Therefore
miR-188-5p overexpression reduced ZFP91 via the noncanonical NF-κB
signaling pathway to inhibit the progression of BC.
In conclusion, our data showed that miR-188-5p is
downregulated in BC cell lines and tissues, and the downregulated
expression of miR-188-5p is associated with the poor prognosis of
patients with BC. We further investigated that overexpression of
miR-188-5p could inhibit proliferation and induce the apoptosis of
MDA-MB-231 cells. Furthermore, ZFP91 was predicted and confirmed as
a target gene of miRNA-188-5p, and the effects of miR-188-5p on BC
cells were dependent on the inhibition of ZFP91. Additionally, a
decrease in ZFP91 significantly inhibited the NF-κB/p65 and RelB
expression in BC cells. Moreover, the expression levels of
miR-188-5p and ZFP91 were highly correlated with BC progression.
Therefore, we suggest that miR-188-5p can inhibit breast cancer
progression via the ZFP91/NF-κB/p65 axis and may be a potential
diagnostic indicator for BC.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This study was supported by the National Natural
Science Foundation of China (grant nos. 81871465 and 81470086) and
345 Talent Project.
Availability of data and materials
The datasets used and analyzed during the current
study are available from the corresponding author upon reasonable
request.
Authors' contributions
ZY and ZL conceived and designed the study. ZY, ZC,
GY performed the experiments. ZY wrote the paper. ZY, ZL, ZC and GY
reviewed the results and data and edited the manuscript. All
authors read and approved the manuscript and agree to be
accountable for all aspects of the research in ensuring that the
accuracy or integrity of any part of the work are appropriately
investigated and resolved.
Ethics approval and consent to
participate
Tissues used in this study were obtained from
Shengjing Hospital of China Medical University (Shenyang, Liaoning,
China) with the informed consent of patients, and all experiments
were approved by the Ethics Committee of Shengjing Hospital of
China Medical University (no. 2016PS18J).
Patient consent for publication
Not applicable
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Davidson NE, Armstrong SA, Coussens LM,
Cruz-Correa MR, DeBerardinis RJ, Doroshow JH, Foti M, Hwu P,
Kensler TW, Morrow M, et al: AACR cancer progress report 2016. Clin
Cancer Res. 22 (Suppl 19):S1–S137. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ueno NT, Fernandez JRE, Cristofanilli M,
Overmoyer B, Rea D, Berdichevski F, El-Shinawi M, Bellon J,
Le-Petross HT, Lucci A, et al: International consensus on the
clinical management of inflammatory breast cancer from the morgan
welch inflammatory breast cancer research program 10th anniversary
conference. J Cancer. 9:1437–1447. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lim B, Woodward WA, Wang X, Reuben JM and
Ueno NT: Inflammatory breast cancer biology: The tumour
microenvironment is key. Nat Rev Cancer. 18:485–499. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Patnaik A, Rosen LS, Tolaney SM, Tolcher
AW, Goldman JW, Gandhi L, Papadopoulos KP, Beeram M, Rasco DW,
Hilton JF, et al: Efficacy and safety of abemaciclib, an inhibitor
of CDK4 and CDK6, for patients with breast cancer, non-small cell
lung cancer, and other solid tumors. Cancer Discov. 6:740–753.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Filipowicz W, Bhattacharyya SN and
Sonenberg N: Mechanisms of post-transcriptional regulation by
microRNAs: Are the answers in sight? Nat Rev Genet. 9:102–114.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang H, Qi S, Zhang T, Wang A, Liu R, Guo
J, Wang Y and Xu Y: miR-188-5p inhibits tumour growth and
metastasis in prostate cancer by repressing LAPTM4B expression.
Oncotarget. 6:6092–6104. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Fang F, Chang RM, Yu L, Lei X, Xiao S,
Yang H and Yang LY: MicroRNA-188-5p suppresses tumor cell
proliferation and metastasis by directly targeting FGF5 in
hepatocellular carcinoma. J Hepatol. 63:874–885. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Edge SB, Byrd DR, Compton CC, Fritz AG,
Greene FL and Trotti A: AJCC cancer staging manual. 7. Springer;
New York, NY: 2010, PubMed/NCBI
|
|
10
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jacob A, Jing J, Lee J, Schedin P, Gilbert
SM, Peden AA, Junutula JR and Prekeris R: Rab40b regulates MMP2 and
MMP9 trafficking during invadopodia formation and breast cancer
cell invasion. J Cell Sci. 126:4647–4658. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ma J, Mi C, Wang KS, Lee JJ and Jin X:
Zinc finger protein 91 (ZFP91) activates HIF-1α via NF-κB/p65 to
promote proliferation and tumorigenesis of colon cancer.
Oncotarget. 7:365512016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Paschke L, Jopek K, Szyszka M, Tyczewska
M, Ziolkowska A, Rucinski M and Malendowicz LK: ZFP91: A
noncanonical NF-κB signaling pathway regulator with oncogenic
properties is overexpressed in prostate cancer. Biomed Res Int.
2016:69635822016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Jin X, Jin HR, Jung HS, Lee SJ, Lee JH and
Lee JJ: An atypical E3 ligase zinc finger protein 91 stabilizes and
activates NF-kappaB-inducing kinase via Lys63-linked
ubiquitination. J Biol Chem. 285:30539–30547. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
McGuire A, Brown JAL, Malone C, McLaughlin
R and Kerin MJ: Effects of age on the detection and management of
breast cancer. Cancers (Basel). 7:908–929. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
McLaughlin SA, Staley AC, Vicini F,
Thiruchelvam P, Hutchison NA, Mendez J, MacNeill F, Rockson SG,
DeSnyder SM, Klimberg S, et al: Considerations for clinicians in
the diagnosis, prevention, and treatment of breast cancer-related
lymphedema: Recommendations from a multidisciplinary expert ASBrS
panel: Part 1: Definitions, assessments, education, and future
directions. Ann Surg Oncol. 24:2818–2826. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Romano G, Veneziano D, Acunzo M and Croce
CM: Small non-coding RNA and cancer. Carcinogenesis. 38:485–491.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang M, Qiu R, Gong Z, Zhao X, Wang T,
Zhou L, Lu W, Shen B, Zhu W and Xu W: miR-188-5p emerges as an
oncomiRNA to promote gastric cancer cell proliferation and
migration via upregulation of SALL4. J Cell Biochem.
120:15027–15037. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li Y, Yan X, Shi J, He Y, Xu J, Lin L,
Chen W and Lin X and Lin X: Aberrantly expressed miR-188-5p
promotes gastric cancer metastasis by activating Wnt/β-catenin
signaling. BMC Cancer. 19:5052019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Iwakawa HO and Tomari Y: The functions of
microRNAs: mRNA decay and translational repression. Trends Cell
Biol. 25:651–665. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang M, Zhang H, Yang F, Qiu R, Zhao X,
Gong Z, Yu W, Zhou B, Shen B and Zhu W: miR-188-5p suppresses
cellular proliferation and migration via IL6ST: A potential
noninvasive diagnostic biomarker for breast cancer. J Cell Physiol.
235:4890–4901. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Saotome Y, Winter CG and Hirsh D: A widely
expressed novel C2H2 zinc-finger protein with multiple consensus
phosphorylation sites is conserved in mouse and man. Gene.
152:233–238. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lee JJ, Lee JH, Lee K, Hong YS and Jin XJ:
Therapeutic agent for cancer, inflammation, and auto-immune disease
containing inhibitor of zinc finger protein 91. US Patent
20080248024A1. Filed February 28, 2008; issued October 9, 2008.
|
|
24
|
Paschke L, Jopek K, Szyszka M, Tyczewska
M, Malendowicz LK and Rucinski M: ZFP91 zinc finger protein
expression pattern in normal tissues and cancers. Oncol Lett.
17:3599–3606. 2019.PubMed/NCBI
|
|
25
|
Unoki M, Okutsu J and Nakamura Y:
Identification of a novel human gene, ZFP91, involved in acute
myelogenous leukemia. Int J Oncol. 22:1217–1223. 2003.PubMed/NCBI
|
|
26
|
Xiao G and Fu J: NF-κB and cancer: A
paradigm of Yin-Yang. Am J Cancer Res. 1:192–221. 2011.PubMed/NCBI
|
|
27
|
Tompkins V, Hagen J, Zediak VP and Quelle
DE: Identification of novel ARF binding proteins by two-hybrid
screening. Cell Cycle. 5:641–646. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Peng Y, Shen X, Jiang H, Chen Z, Wu J, Zhu
Y, Zhou Y and Li J: miR-188-5p suppresses gastric cancer cell
proliferation and invasion via targeting ZFP91. Oncol Res.
27:65–71. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Behm-Ansmant I, Rehwinkel J and Izaurralde
E: MicroRNAs silence gene expression by repressing protein
expression and/or by promoting mRNA decay. Cold Spring Harb Symp
Quant Biol. 71:523–530. 2006. View Article : Google Scholar : PubMed/NCBI
|