Introduction
As one of the most frequently diagnosed urological
malignancies of the urinary system, bladder cancer (BC) is also the
10th most commonly occurring type of cancer worldwide (1) and ranks 11th in terms of mortality
(2). Despite advances in the
treatment for BC, including adjuvant chemoradiotherapies and
surgical techniques, the recurrence and progression rates of BC
remain high, and the quality of life and 5-year survival rates of
BC exhibit no signs of significant improvement (3–7).
Consequently, it is still urgent to investigate the underlying
molecular mechanisms, new prognostic and predictive biomarkers of
malignant progression and metastasis of bladder cancer.
Long non-coding RNAs (lncRNA), usually more than 200
bases in length, have become a new field of research (8–10).
Compared with microRNAs (miRNAs) and protein-coding messenger RNAs
(mRNAs), lncRNAs, owing to the greater tissue specificity, can be
used for novel biomarkers of cancer diagnosis and prognosis
(11). Recently, emerging studies
have demonstrated that lncRNAs act as key regulators through their
involvement in gene modulation at the transcriptional,
post-transcriptional and chromosomal levels (9,12,13).
Furthermore, numerous studies have also revealed
that lncRNAs play significant roles as oncogenes or suppressors in
the carcinogenesis and evolution of various types of human
malignancies, including colorectal (14,15),
hepatocellular (16,17), gastric (18), breast (11), esophageal (19) and small-cell lung (20) cancers. Notably, several lncRNAs,
such as DGCR5 (21), FAM83H-AS1
(22), GAS6-AS2 (23), HCG18 (24) and SNHG16 (25) have served as critical regulators in
the biological behavior processes of BC over the past decades. For
instance, overexpression of lncRNA-ATB markedly facilitated cell
viability, migration, and invasion in bladder cancer cells and it
may provide a potential prognostic biomarker and a therapeutic
target for bladder cancer (26).
Chen et al demonstrated that lncRNA-LNMAT1 accelerated
lymphatic metastasis of bladder cancer via CCL2-dependent
macrophage recruitment, and that it could serve as a potential
therapeutic target for clinical intervention in LN-metastatic
bladder cancer (12). Furthermore,
lncRNAs could also act as anti-oncogenes. Tuo et al argued
that lncRNA TP73-AS1 may serve as a tumor suppressor that
participates in bladder cancer progression by inhibiting the
epithelial-mesenchymal transition (EMT) process (27).
Appreciable progress has been made to comprehend
lncRNA biology for the past few years, but the expression pattern,
clinical value for BC diagnosis, therapy, and prognosis, as well as
the possible molecular mechanisms of lncRNAs in BC remain largely
unknown and warrant further study. Thus, the present study
performed an investigation into lncRNAs and mRNAs in five pairs of
BC and adjacent non-tumor tissues by RNA-sequencing (RNA-seq) in
order to improve the current understanding of BC pathogenesis or
development, and identified 103 markedly differentially expressed
(DE) lncRNAs, and 2,756 significantly dysregulated mRNAs. Based on
a bioinformatics analysis, the present study focused on the four
lncRNAs (upregulated ENST00000433108; downregulated
ENST00000598996, ENST00000524265 and ENST00000398461) and two mRNAs
(namely CDK1 and CCNB1, both upregulated) in BC that are
potentially associated with known pathogenic genes. These selected
lncRNAs and mRNAs were validated via reverse
transcription-quantitative PCR (RT-qPCR), and their dysregulation
was conspicuously associated with certain clinicopathological
characteristics and a worse prognosis in patients with BC.
Furthermore, the present study also analyzed the sensitivity and
specificity of these lncRNAs in the diagnosis of BC by constructing
a ROC curve. According to the bioinformatics analysis, the screened
lncRNAs have a competing endogenous (ce) RNA network regulation
relationship with a variety of tumor-associated genes obtained from
sequencing data of mRNAs; involving diverse tumor-associated
signaling pathways, and then suggesting that these lncRNAs may be
closely associated with BC progression through the aforementioned
pathways. Notably, ectopic expression of ENST00000598996 or
ENST00000524265 significantly suppressed cell migration, invasion
and proliferation of BC cells.
The present study profiled the expression of lncRNAs
and mRNAs in BC tissues and matched normal tissues, and examined
the fundamental functional phenotypes of strictly selected lncRNAs
(ENST00000598996 and ENST00000524265) in BC cells, with the aim of
improving the current understanding of tumorigenesis and
progression, but also to facilitate the prognosis prediction and
clinical treatment management of BC.
Materials and methods
Clinical specimens
A total of 64 BC and matched non-tumorous tissues
were collected from patients with BC who had undergone surgical
resection without preoperative radiotherapy or chemotherapy at the
Department of Urology of the Second Affiliated Hospital of Tianjin
Medical University (China) between November 2015 and June 2019. The
final diagnosis of each patient was determined histopathologically.
The detailed clinical information of these patients is listed in
Table SI. Overall survival (OS)
was updated on 1 October, 2019 and was defined as the time from
recruitment to death for any reason. Progression-free survival
(PFS) was defined as the time from inclusion to recurrence or
metastasis progression. In the present study, written informed
consent was obtained from every patient or their guardians based on
the guidelines approved by the Medical Ethics Committee of the
Second Hospital of Tianjin Medical University.
Cell culture and transfection
Human normal bladder epithelium immortalized cells
SV-HUC-1 and four bladder cancer cell lines (EJ-1, HTB-9, T24 and
253J-BV) were purchased from the Chinese Academy of Sciences Cell
Bank (Shanghai, China); and cell lines were authenticated using STR
DNA profiling. The EJ-1, HTB-9, T24, and 253J-BV cells were
cultured in the RPMI-1640 medium (Biological Industries)
supplemented with 10% fetal bovine serum (FBS) and 100 U/ml of
penicillin, 100 mg/ml of streptomycin in a humidified incubator
containing 5% CO2 at 37°C. The SV-HUC-1 cell line was
cultured in the Ham's F-12K Nutrient Mixture (Gibco; Thermo Fisher
Scientific, Inc.) with the same aforementioned culture
conditions.
The ENST00000598996 and ENST00000524265
overexpression plasmid and the corresponding empty vectors (NC)
were synthesized by Hanbio Biotechnology Co., Ltd. The
Lipofectamine® 2000 Reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) was used for transfection of the aforementioned
plasmids. After 24 h of plating, transfection was performed in
serum-free medium using Lipofectamine® 2000. After 6 h,
the transfected cells were placed in complete medium (RPMI-1640)
containing 10% FBS and maintained at 37°C in 5% CO2.
RNA extraction and quality control
(QC)
Total RNA was isolated from the bladder cancer and
adjacent normal bladder tissues using TRIzol® reagent
(Ambion) or an HP Total RNA kit (Omega Bio-Tek, Inc.) according to
the manufacturer's protocol. RNA concentration and purity for
tissues of sequencing analysis and subsequent verification was
measured using a NanoDrop ND-1000 (Thermo Fisher Scientific, Inc.).
In the present study, all RNA samples passed the quality standard
based on a qualified ratio of optical density (OD) 260 to OD280
(1.8–2.1). Briefly based on denaturing agarose gel electrophoresis,
RNA integrity and quality for tissues of sequencing analysis were
validated by the presence of sharp clear bands of 28S and 18S
ribosomal RNA, with a 28S:18S ratio of 2:1, along with the absence
of genomic DNA and degraded RNA.
High-throughput sequencing
First, ribosomal (r)RNAs were removed from the total
RNA using Ribo-Zero rRNA Removal kits (Illumina, Inc.) according to
the manufacturer's instructions. RNA libraries were constructed
using a TruSeq Stranded Total RNA Library Prep kit (Illumina, Inc.)
from the rRNA-depleted RNA based on the manufacturer's protocols.
In the libraries, RNA quality and quantity were controlled using
the BioAnalyzer 2100 system (Agilent Technologies; Thermo Fisher
Scientific, Inc.). Subsequently, libraries were denatured to
single-stranded DNA molecules, captured on Illumina flow cells,
amplified in situ as clusters and finally sequenced for 150
cycles utilizing an Illumina HiSeq Sequencer according to the
manufacturer's protocols.
Sequencing analysis of lncRNAs and
mRNAs
The original reads were obtained from the Illumina
HiSeq 4000 sequencer and 3′ adaptor-trimming and removal of
low-quality reads were performed using Cutadapt software (v1.9.3)
(https://cutadapt.readthedocs.io/en/stable/). The
resulting high-quality clean reads were utilized to analyze lncRNAs
and mRNAs. The high-quality reads were aligned to the human
reference genome (UCSC HG19) using HISAT2 software (https://ccb.jhu.edu/software/hisat2).
Identification of DE lncRNAs and mRNAs
and RT-qPCR validation
Next, guided by the Ensembl GTF gene annotation file
(http://ftp.ensembl.org/pub/release-75/gtf/homo_sapiens/),
Cuffdiff software (v2.2.1, part of cufflinks) (http://cole-trapnell-lab.github.io/cufflinks/cuffdiff/index.html)
was used to obtain the FPKM as the expression profiles of lncRNAs
and mRNAs, and the fold change (FC) and P-values were calculated
based on the FPKM; differentially expressed lncRNAs and mRNAs (FC
≥2.0 and P-values ≤0.05) were classified as significant. Scatter
plots were drawn to reveal the different lncRNA or mRNA expression
patterns in the BC and matched adjacent non-cancerous specimens by
the CRAN packages ggplot2 in R (https://www.r-project.org/).
RT-qPCR was performed to validate the expression of
the selected DE lncRNAs and mRNAs from RNA sequencing. The qPCR
thermocycling conditions were as follows: Initial denaturation at
95°C for 10 min, followed by 40 cycles at 95°C for 10 sec and 60°C
for 50 sec. Following the manufacturer's instructions, total RNA
was extracted using TRIzol® reagent (Ambion) or an HP
Total RNA kit (Omega Bio-Tek, Inc.) and then reverse-transcribed by
Transcriptor First Strand cDNA Synthesis kit and amplified in
triplicate on an Applied Biosystems 7500 Fast Real-Time PCR System
(Applied Biosystems; Thermo Fisher Scientific, Inc.) using the
FastStart Universal SYBR Green Master with ROX (Roche Diagnostics).
GAPDH was used as an internal reference for normalization. The
relative expression ratio of these lncRNAs and mRNAs was calculated
through the 2−ΔΔCq method (28). All primers were synthesized by
Sangon Biotech Co. Ltd. and the sequences are presented in Table I.
 | Table I.Primers used to validate lncRNA and
mRNA expression. |
Table I.
Primers used to validate lncRNA and
mRNA expression.
| Gene name | Primer type | Primer
sequence | Product length
(bp) |
|---|
|
ENST00000433108 | Forward |
5′-AGGGGCAAAAGAAACATCCAA-3′ | 237 |
|
| Reverse |
5′-CCCCTGATCTAGCCACTCATC-3′ |
|
|
ENST00000398461 | Forward |
5′-GCCCTAGGGGAGTGACTACA-3′ | 155 |
|
| Reverse |
5′-CCCCTGTGCCCAATTTCAAC-3′ |
|
|
ENST00000524265 | Forward |
5′-ACAAGAAGCAGAATGGGCCT-3′ | 192 |
|
| Reverse |
5′-AATCACACGGGCTTTGCATT-3′ |
|
|
ENST00000598996 | Forward |
5′-CCGGTCACTTCACGATGACA-3′ | 193 |
|
| Reverse |
5′-CAGGACCTCCAGGGAAACAC-3′ |
|
| CCNB1 | Forward |
5′-AACTTTCGCCTGAGCCTATTTT-3′ | 228 |
|
| Reverse |
5′-TTGGTCTGACTGCTTGCTCTT-3′ |
|
| CDK1 | Forward |
5′-AAACTACAGGTCAAGTGGTAGCC-3′ | 148 |
|
| Reverse |
5′-TCCTGCATAAGCACATCCTGA-3′ |
|
| GAPDH | Forward |
5′-CGGAGTCAACGGATTTGGTC-3′ | 180 |
|
| Reverse |
5′-TTCCCGTTCTCAGCCTTGAC-3′ |
|
Bioinformatic analyses
Gene Ontology (GO) and Kyoto Encyclopedia of Genes
and Genomes (KEGG) analyses were performed in order to determine
the underlying functions for the differentially expressed
lncRNA-associated intersection genes (downstream mRNAs predicted by
lncRNAs vs. sequencing DE mRNAs) using the Database for Annotation,
Visualization, and Integrated Discovery (DAVID) Bioinformatics
resources (http://david.abcc.ncifcrf.gov/) (29). GO analysis can be classified into
biological processes (BP), cellular components (CC), and molecular
functions (MF). P<0.05 was considered to indicate a
statistically significant difference.
Construction of the ceRNA and
protein-protein interaction (PPI) networks
For determining the interactions between mRNAs and
lncRNAs, the lncRNA-miRNA-mRNA network was constructed. The miRanda
(http://www.microrna.org/microrna/home.do) and DIANA
(http://carolina.imis.athena-innovation.gr/) (30) tools were utilized to predict the
first top 5 miRNAs of the screened six DE lncRNAs. Subsequently,
the miRTarBase (http://mirtarbase.mbc.nctu.edu.tw/) (31) and TargetScan (http://www.targetscan.org/) (32) databases were performed to predict
the miRNA-targeted mRNAs, and an intersection with sequencing DE
mRNAs was constructed.
Then, according to these intersection genes, the PPI
network was performed using STRING (version 11.0) (https://string-db.org/) online analysis. The
construction and visualization of the lncRNA-miRNA-mRNA and PPI
networks were accomplished by Cytoscape v3.6.1 (http://www.cytoscape.org/).
Cell proliferation assay
Cell proliferation was investigated using a Cell
Counting Kit-8 (CCK-8) and colony-formation experiments. For the
CCK-8 assay, 2×103 cells/well were seeded into 96-well
plates. At the indicated time-points (0, 24, 48, 72 and 96 h), each
well had 10 µl CCK-8 solution (Beyotime Institute of Biotechnology)
added, and the plates were incubated at 37°C for another 3.5 h in
the dark. The absorbance was measured at a wavelength of 450 nm
using a microplate reader. For the clone formation assay, cells
(500/well) were seeded into 6-well plates and cultured for 10 days.
Next, the cell colonies with a diameter >1 mm were stained with
0.1% crystal violet at room temperature for 15 min and counted.
Cell migration and invasion
assays
Wound healing and Transwell assays (without Matrigel
coating) were performed to evaluate cell migration. The Transwell
assay [with Matrigel coating (BD Biosciences)] was performed to
assess cell invasion. For the wound healing assay, transfected
cells were cultured to 90–95% confluency in 6-well plates and
scratched using 200-µl pipette tips. Migration images were captured
at 0 and 36 h after scraping with an inverted microscope
(magnification ×200; Olympus Corporation) and analyzed objectively
via Image J v1.46 software (National Institutes of Health). For the
Transwell assay, 5×104 transfected cells in FBS-free
RPMI-1640 medium were seeded into the top chamber, while the lower
chamber was supplemented with RPMI-1640 medium containing 20% FBS.
After culturing for 36 h, the non-migrated or non-invasive cells
were removed with a cotton tip, and the remaining cells on the
bottom surface were fixed with 4% paraformaldehyde at room
temperature for 15 min and stained with 0.1% crystal violet for 15
min at room temperature. The stained cells were imaged in five
randomly selected fields under an optical microscope (magnification
×400; Olympus Corporation) and counted using ImageJ v1.46 software
(National Institutes of Health).
Statistical analysis
Statistical analyses were performed using GraphPad
Prism 7.0 software (GraphPad Software, Inc.) and SPSS 20.0 software
(IBM Corp.). All data are presented as the mean ± standard
deviation of at least three independent experiments. P<0.05 was
considered to indicate a statistically significant difference.
A paired t-test was used to analyze the differences
between BC tissues and paired normal tissues. Two-way analysis of
variance (ANOVA) followed by Dunnett's post hoc test was
implemented for comparisons of multiple groups, as noted in the
Figs. 5C and 9A-E. The associations between
ENST00000598996 or ENST00000524265 levels and clinicopathological
parameters of patients with BC were further analyzed via a Fisher's
exact test. A ROC curve was drawn to estimate diagnostic
performance. Survival analysis was conducted by Kaplan-Meier method
and log-rank test for significance in GraphPad Prism 7.0. Survival
data were further evaluated using the univariate and multivariate
Cox proportional hazards model.
 | Figure 5.Verification of candidate four
lncRNAs and two mRNAs by RT-qPCR (A) Relative expression levels in
64 pairs of BC tissues and non-cancerous tissues. (B) Comparison of
fold change (2−ΔΔCq) of these lncRNAs and mRNAs between
the sequencing expression profiles (n=5 paired tissues) and RT-qPCR
results (n=64 paired tissues). (C) Determination of the expression
of these selected lncRNAs and mRNAs in four malignant BC cell lines
compared with normal urothelial cell line SV-HUC-1. The results are
presented as the mean ± SD, **P<0.01, ***P<0.001 and
****P<0.0001. lncRNA, long non-coding RNA; mRNA, messenger RNA;
BC, bladder cancer; RT-qPCR, reverse transcription-quantitative
PCR; lnc-3108, ENST00000433108; lnc-4265, ENST00000524265;
lnc-8461, ENST00000398461; lnc-8996, ENST00000598996; NS,
non-significant; Nor, normal. |
 | Figure 9.Overexpression of lnc8996 and lnc4265
weakens the malignant biological behaviors of BC cells. (A) BC cell
lines (EJ-1 and HTB-9) were transfected with plasmids and the
expression levels of lnc8996 and lnc4265 were significantly
increased as revealed by RT-qPCR. (B) CCK-8 (n=5) and (C)
colony-formation (n=3) assays revealed that ectopic lnc8996 and
lnc4265 expression inhibited EJ-1 and HTB-9 cell proliferation.
Bars represent the number of colonies. Overexpression of lnc8996
and lnc4265 weakens the malignant biological behaviors of BC cells.
The effect of lnc8996 and lnc4265 on cell migration and invasion
capabilities was evaluated by (D) scratch wound healing, (E)
Transwell migration and invasion assays in EJ-1 and HTB-9 cells,
respectively. Bars represent (D) the migration rate, and (E) the
number of migrated and invasive cells. Scale bar, 50 µm. Data are
presented as the means ± SD of three independent experiments;
*P<0.05, **P<0.01, ***P<0.001 and ****P<0.0001.
lnc-8996, ENST00000598996; lnc-4265, ENST00000524265; BC, bladder
cancer; RT-qPCR, reverse transcription-quantitative PCR; CCK-8,
Cell Counting Kit-8; OE, overexpression; NC, negative control. |
Results
Overview of lncRNA and mRNA profiles
in BC tissues
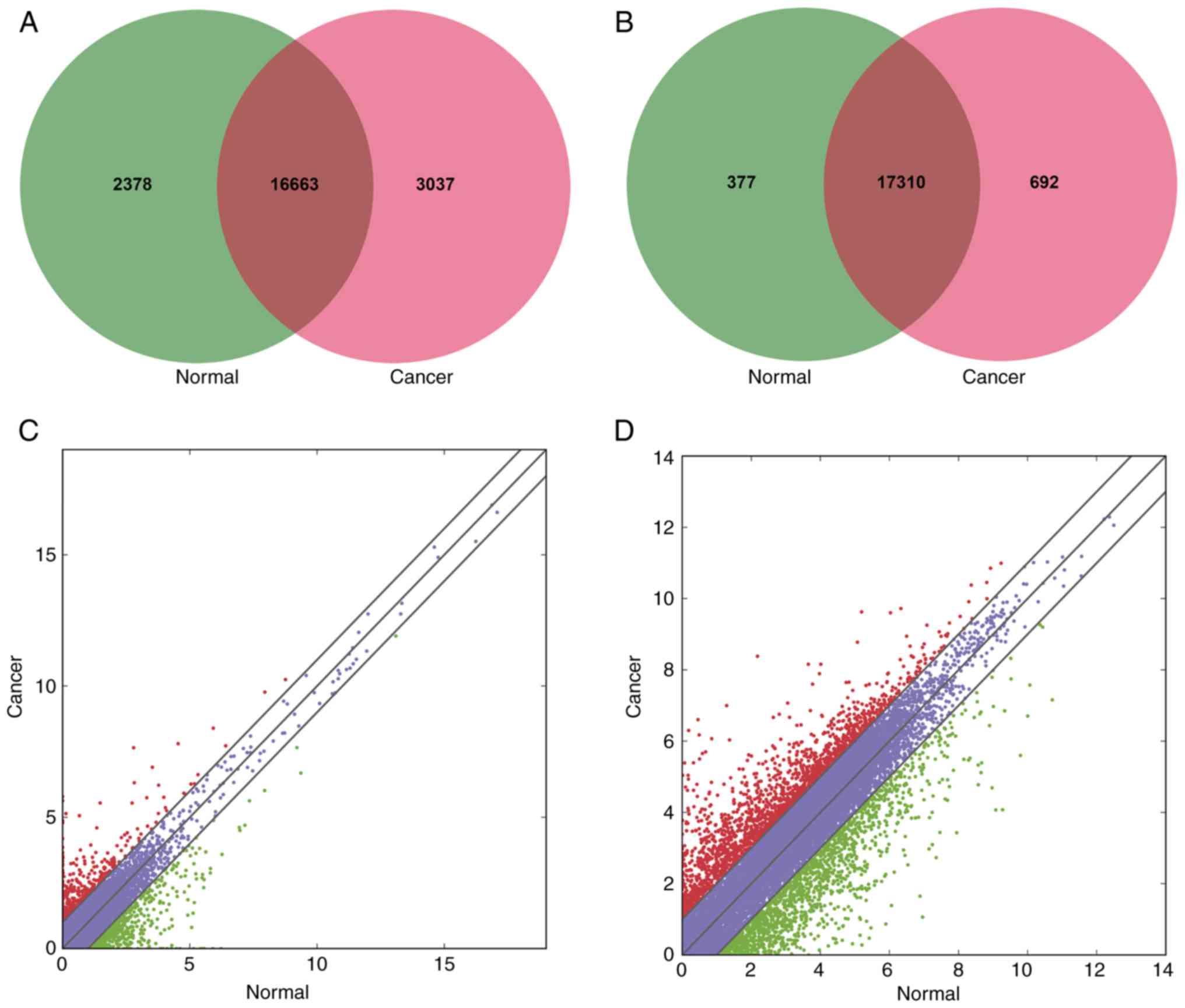
The present study first analyzed the profiling of
lncRNAs and mRNAs in five paired human BC and adjacent
non-cancerous tissues via RNA-Seq. A total of 37,684 lncRNAs and
20,266 mRNAs were detected in both groups. For lncRNAs, 2,378 were
only detected in the paired control tissues, 3,037 were only
detected in BC tissues and 16,663 were detected in both groups
(Fig. 1A). Of the total 37,684
lncRNAs, 35 upregulated and 68 downregulated lncRNAs were
significantly differentially expressed in BC specimens when
compared with controls (P<0.05, fold change ≥2.0) (Table II; Figs. 1C and 2A). Based on the sequencing data, there
were more downregulated lncRNAs than upregulated. For mRNAs, 377
were only discovered in the matched control tissues, 692 were only
detected in BC tissues and 17,310 were discovered in both groups
(Fig. 1B). Among the whole 20,266
mRNAs, 1,467 upregulated and 1,289 downregulated mRNAs were
significantly aberrantly expressed in BC tissues (P<0.05, fold
change ≥2.0) (Figs. 1D and 2A).
 | Table II.Upregulated and downregulated lncRNAs
in whole are listed. |
Table II.
Upregulated and downregulated lncRNAs
in whole are listed.
| Transcript ID | Regulation |
Log2Fc | P-value | Chr | Strand | Biotype |
|---|
|
ENST00000410910 | Up | inf | 0.01745 |
chr2:17304107-17304407 | – | misc_RNA |
|
ENST00000434300 | Up | inf | 0.02275 |
chr1:192486768-192536347 | – | antisense |
|
ENST00000438158 | Up | inf | 0.04595 |
chr1:222215046-222238105 | – | lincRNA |
|
ENST00000522365 | Up | inf | 0.0214 |
chr8:84315992-84321135 | + | lincRNA |
|
ENST00000538428 | Up | inf | 0.02985 |
chr1:98453555-98515419 | – | lincRNA |
|
ENST00000548280 | Up | inf | 0.03395 |
chr14:28787346-28887818 | + | lincRNA |
|
ENST00000566101 | Up | inf | 0.00985 |
chr18:37646249-37679197 | + | lincRNA |
|
ENST00000566876 | Up | inf | 0.00765 |
chr16:50424826-50429044 | – | lincRNA |
|
ENST00000602912 | Up | inf | 0.0165 |
chr11:67227338-67227784 | + | lincRNA |
| TCONS_00017351 | Up | inf | 0.0294 |
chrX:86979060-86983512 | – | long_nonc. |
| TCONS_00017630 | Up | inf | 0.01895 |
chrY:7302523-7388562 | + | long_nonc. |
| TCONS_00020860 | Up | inf | 0.0048 |
chr12:88006397-88043289 | – | long_nonc. |
| TCONS_00020861 | Up | inf | 0.00695 |
chr12:88045658-88064325 | – | long_nonc. |
| TCONS_00022565 | Up | inf | 0.0313 |
chr14:80931125-80938379 | + | long_nonc. |
|
TCONS_l2_00030270 | Up | inf | 0.0043 |
chrX:97098491-97101678 | + | long_nonc. |
|
TCONS_l2_00005321 | Up | 147.61 | 0.013 |
chr11:65184125-65213011 | + | long_nonc. |
| TCONS_00001651 | Up | 5.08 | 0.00025 |
chr1:121484056-121485434 | – | long_nonc. |
| uc001ycm.1 | Up | 3.70 | 0.0328 |
chr14:94571181-94583033 | + | long_nonc. |
|
ENST00000429829 | Up | 3.57 | 0.01185 |
chrX:73012039-73072588 | – | lincRNA |
|
ENST00000562848 | Up | 3.39 | 0.04505 |
chr12:54375292-54449814 | + | lincRNA |
|
TCONS_l2_00027748 | Up | 3.17 | 0.04745 |
chr8:49277961-49337122 | + | long_nonc. |
|
ENST00000525217 | Up | 3.02 | 0.0245 |
chr11:287304-288987 | + | antisense |
|
ENST00000587187 | Up | 3.01 | 0.0078 |
chr19:4903091-4962165 | + | processed_tr. |
|
ENST00000448587 | Up | 2.75 | 0.0105 |
chr19:5558177-5581226 | – | lincRNA |
|
ENST00000605886 | Up | 2.47 | 0.0232 |
chr1:156611181-156631216 | – | antisense |
|
ENST00000570148 | Up | 2.00 | 0.01645 |
chr16:81344335-81347028 | – | antisense |
| TCONS_00006377 | Up | 1.73 | 0.00415 |
chr3:195366860-195378976 | + | long_nonc. |
|
TCONS_l2_00025984 | Up | 1.67 | 0.02905 |
chr7:65300346-65310515 | + | long_nonc. |
|
ENST00000439156 | Up | 1.65 | 0.01925 |
chr1:61005920-61106163 | – | lincRNA |
|
ENST00000400385 | Up | 1.65 | 0.0472 |
chr21:45225638-45232448 | – | antisense |
|
ENST00000606434 | Up | 1.62 | 0.0259 |
chr5:6671218-6674499 | + | lincRNA |
|
TCONS_l2_00025849 | Up | 1.59 | 0.0122 |
chr7:45831387-45863181 | + | long_nonc. |
| NR_104643 | Up | 1.51 | 0.01445 |
chr5:42985500-42993435 | – | long_nonc. |
|
ENST00000566699 | Up | 1.48 | 0.04175 |
chr7:139877060-139879440 | + | antisense |
|
ENST00000433108 | Up | 1.33 | 0.01015 |
chr1:209602164-209606183 | + | lincRNA |
| TCONS_00006771 | Up | 1.24 | 0.0104 |
chr3:197360473-197374168 | – | long_nonc. |
| GU228573 | Down | −52.13 | 0.02865 |
chr7:55086713-55324313 | + | long_nonc. |
| uc021×rd.1 | Down | −5.44 | 0.0098 |
chr4:119809995-119982402 | + | long_nonc. |
|
ENST00000474768 | Down | −5.24 | 0.01835 |
chr3:64501332-64997143 | + | antisense |
|
ENST00000433310 | Down | −5.11 | 0.00005 |
chr21:29811666-30047170 | – | lincRNA |
|
ENST00000460833 | Down | −5.10 | 0.00625 |
chr3:64501332-64997143 | + | antisense |
|
ENST00000481312 | Down | −5.09 | 0.0287 |
chr3:64501332-64997143 | + | antisense |
|
ENST00000555383 | Down | −4.80 | 0.0094 |
chr14:96671015-96735304 | + | sense_intr. |
|
ENST00000515734 | Down | −4.77 | 0.03565 |
chr5:58264864-59843484 | + | lincRNA |
|
ENST00000593604 | Down | −4.54 | 0.02775 |
chr16:86508134-86542705 | – | lincRNA |
|
ENST00000598996 | Down | −4.53 | 0.0471 |
chr16:86508134-86542705 | – | lincRNA |
|
ENST00000426585 | Down | −4.47 | 0.03795 |
chr22:17082776-17185367 | + | lincRNA |
|
ENST00000608684 | Down | −4.46 | 0.0486 |
chr1:78444858-78604133 | – | antisense |
|
ENST00000602478 | Down | −4.37 | 0.00165 |
chr22:43011249-43011913 | + | lincRNA |
|
ENST00000398460 | Down | −4.18 | 0.0053 |
chr14:101245746-101327368 | + | lincRNA |
| uc022buq.1 | Down | −4.02 | 0.0404 |
chrX:31115793-33357558 | – | long_nonc. |
|
ENST00000562393 | Down | −4.00 | 0.0376 |
chr16:84853589-84954374 | + | long_nonc. |
| AL049990 | Down | −3.99 | 0.0392 |
chr4:121952575-121954272 | – | long_nonc. |
|
ENST00000522358 | Down | −3.89 | 0.00175 |
chr5:148786251-148812399 | + | lincRNA |
| uc021pwm.1 | Down | −3.88 | 0.00725 |
chr10:97071527-97321171 | – | long_nonc. |
|
ENST00000421206 | Down | −3.87 | 0.0343 |
chr10:120967100-121215131 | + | sense_intr. |
| antipeg11_dna | Down | −3.87 | 0.00005 |
chr14:101346991-101351184 | + | long_nonc. |
|
ENST00000398878 | Down | −3.52 | 0.004 |
chr16:29262828-30215631 | + | lincRNA |
| uc003bgk.1 | Down | −3.50 | 0.0189 |
chr22:45898117-45998697 | + | long_nonc. |
|
ENST00000604430 | Down | −3.40 | 0.02785 |
chr16:29262828-30215631 | + | lincRNA |
|
ENST00000445466 | Down | −3.38 | 0.02205 |
chr3:151961616-152214602 | – | antisense |
|
ENST00000505254 | Down | −3.20 | 0.01775 |
chr5:148786251-148812399 | + | lincRNA |
|
ENST00000415513 | Down | −3.10 | 0.0329 |
chr7:104581509-105039755 | + | lincRNA |
| NR_024344 | Down | −3.10 | 0.0002 |
chr11:133766329-133771635 | – | long_nonc. |
| uc001rpy.3 | Down | −3.03 | 0.03735 |
chr12:48099867-48231681 | – | long_nonc. |
| uc002vgv.1 | Down | −2.99 | 0.0171 |
chr2:218664511-218867718 | – | long_nonc. |
|
ENST00000573866 | Down | −2.95 | 0.00015 |
chr17:19015312-19015949 | – | lincRNA |
|
ENST00000452120 | Down | −2.92 | 0.00005 |
chr14:101245746-101327368 | + | lincRNA |
| U54776 | Down | −2.88 | 0.00045 |
chr6:136172833-136546762 | + | long_nonc. |
|
ENST00000524265 | Down | −2.87 | 0.01335 |
chr5:148786251-148812399 | + | lincRNA |
|
ENST00000398461 | Down | −2.87 | 0.00045 |
chr14:101245746-101327368 | + | lincRNA |
| TCONS_00024657 | Down | −2.79 | 0.0003 |
chr16:53069601-53086785 | – | long_nonc. |
|
ENST00000443576 | Down | −2.73 | 0.04575 |
chr13:33677271-33924767 | – | lincRNA |
|
ENST00000452320 | Down | −2.66 | 0.03175 |
chr7:77646392-79100524 | + | processed_tr. |
|
ENST00000533633 | Down | −2.60 | 0.00005 |
chr11:13983913-14295237 | + | processed_tr. |
| NR_034108 | Down | −2.52 | 0.03375 |
chr6:111620233-111927481 | + | long_nonc. |
|
ENST00000587762 | Down | −2.52 | 0.00005 |
chr19:13945329-13947173 | – | lincRNA |
|
ENST00000604052 | Down | −2.52 | 0.0484 |
chr2:28607275-28640179 | + | sense_intr. |
|
ENST00000571722 | Down | −2.49 | 0.0154 |
chr17:18966761-18967449 | – | lincRNA |
|
ENST00000563424 | Down | −2.44 | 0.0017 |
chr16:50679719-50683160 | + | lincRNA |
| uc002aqh.1 | Down | −2.33 | 0.0031 |
chr15:67152640-67156987 | + | long_nonc. |
| uc002aqi.3 | Down | −2.14 | 0.0026 |
chr15:67156988-67159361 | + | long_nonc. |
|
ENST00000608023 | Down | −2.02 | 0.0001 |
chr1:207974862-208042495 | – | processed_tr. |
|
ENST00000568809 | Down | −2.00 | 0.0244 |
chrX:99928326-99928978 | + | antisense |
| uc001kct.3 | Down | −1.90 | 0.0163 |
chr10:85926983-85931832 | – | long_nonc. |
| NR_121662 | Down | −1.90 | 0.00015 |
chr3:114033346-114866118 | – | long_nonc. |
| AF052103 | Down | −1.80 | 0.02055 |
chr16:85399033-85400346 | + | long_nonc. |
| uc010yvs.2 | Down | −1.79 | 0.0181 |
chr2:100889752-100939195 | – | long_nonc. |
| TCONS_00026354 | Down | −1.76 | 0.03035 |
chr18:66817066-66832387 | + | long_nonc. |
| ciRS-7 | Down | −1.74 | 0.00035 |
chrX:139863223-139866829 | + | long_nonc. |
|
TCONS_l2_00017201 | Down | −1.72 | 0.04045 |
chr21:33937195-33940382 | – | long_nonc. |
|
ENST00000606034 | Down | −1.70 | 0.0252 |
chr1:931345-933431 | – | lincRNA |
|
ENST00000448490 | Down | −1.62 | 0.00395 |
chr10:91589266-91600618 | + | lincRNA |
|
ENST00000432045 | Down | −1.53 | 0.01185 |
chr7:130538478-130618587 | – | lincRNA |
|
ENST00000500358 | Down | −1.53 | 0.0482 |
chr4:100010007-100222513 | + | antisense |
|
ENST00000565297 | Down | −1.51 | 0.0404 |
chr8:79749763-79752757 | + | lincRNA |
|
ENST00000453660 | Down | −1.24 | 0.01495 |
chr9:136125787-136150617 | – | processed_tr. |
|
ENST00000584058 | Down | −1.19 | 0.0355 |
chr3:15708742-15901278 | + | misc_RNA |
|
ENST00000437646 | Down | -inf | 0.00005 |
chr17:19322347-19327670 | + | lincRNA |
|
ENST00000446601 | Down | -inf | 0.0128 |
chr3:26753797-26754855 | + | lincRNA |
|
ENST00000592429 | Down | -inf | 0.04485 |
chr19:5158505-5340814 | + | antisense |
| TCONS_00006712 | Down | -inf | 0.0396 |
chr3:179243601-179245572 | – | long_nonc. |
| TCONS_00016239 | Down | -inf | 0.00525 |
chr9:10095-10799 | – | long_nonc. |
| TCONS_00022925 | Down | -inf | 0.0455 |
chr14:24096369-24096959 | + | long_nonc. |
Subsequently, the present study predicted their
respective sponge miRNAs and target mRNAs based on the abnormally
expressed lncRNA data. Next, the present study compared these mRNA
predictions with the mRNA sequencing results (Fig. 2B). Hence, 236 targeted DE-mRNAs were
identified, including 120 upregulated and 116 downregulated
(Table SII). Then, the
aforementioned resulting mRNAs (236) were utilized for the GO, KEGG
pathway and ceRNA network analyses.
Functional analysis of miRNA target
genes
It is well known that lncRNAs are crucial
transcriptional regulators of gene expression involved in the
nosogenesis of multiple types of cancer. In order to investigate
the underlying mechanisms of dyseregulated lncRNAs in the
carcinogenesis or development of BC, GO enrichment analysis of the
aforementioned 236 target DE-mRNAs was conducted to estimate the
functional performance of the 103 DE lncRNAs.
GO enrichment analysis were classified as three
aspects: BP, MF and CC. The most enriched and meaningful BP were
associated with ‘gene regulation of apoptotic process’
(GO:0042981), ‘regulation of cell cycle’ (GO:0051726), ‘positive
regulation of gene expression’ (GO:0010628), ‘positive regulation
of cell proliferation’ (GO:0008284) and ‘regulation of
angiogenesis’ (GO:0045765) (Fig.
3A). The most enriched GO terms under MF were ‘DNA binding’
(GO:0003677), ‘protein kinase activity’ (GO:0004672),
‘ubiquitin-like protein ligase binding’ (GO:0044389), ‘MAP kinase
phosphatase activity’ (GO:0033549) and ‘transcription regulatory
region DNA binding’ (GO:0044212) (Fig.
3B). In the CC analysis, ‘nuclear chromosome part’
(GO:0044454), ‘chromosome, telomeric region’ (GO:0000781),
‘sarcoplasm’ (GO:0016528), ‘integral component of plasma membrane’
(GO:0005887) and ‘focal adhesion’ (GO:0005925) were the
significantly enriched terms (Fig.
3C).
Next, in order to identify the lncRNA
indirectly-regulated signaling pathways, the present study
performed a KEGG analysis on these 236 DE-mRNAs. The significantly
enriched KEGG pathways are presented in Fig. 4A and B. Among them, ‘pathways in
cancer’, ‘cell cycle’, ‘MAPK signaling pathway’, ‘HIF-1 signaling
pathway’, ‘PI3K-Akt signaling pathway’, ‘transcriptional
misregulation in cancer’, ‘cAMP signaling pathway’, ‘p53 signaling
pathway and NF-κB signaling pathway’, were reported as various
cancer-associated pathways. The results indicated that these
pathways may significantly contribute to the pathogenesis and
development of BC.
RT-qPCR verification of lncRNA and
mRNA expression
Subsequently, to confirm the reliability and
validity of the aforementioned analyses, the present study randomly
selected four lncRNAs (upregulated ENST00000433108; downregulated
ENST00000598996, ENST00000524265 and ENST00000398461), and two
upregulated mRNAs (CDK1 and CCNB1) and determined their practical
variations in the 64 paired non-cancerous and cancer samples from
the 64 newly diagnosed BC patients via RT-qPCR (Fig. 5A and Table SIII). The RT-qPCR validation data
for aforementioned screened lncRNAs and mRNAs and the sequencing
results were relatively consistent (Fig. 5B). For ENST00000598996 and
ENST00000524265, the present study also performed RT-qPCR
verification in four BC cell lines with different degrees of
malignancy and a normal cell line (Fig.
5C).
Prediction of miRNA binding sites,
ceRNA and PPI network construction
In the present study, 103 BC-associated lncRNAs were
identified that were differentially expressed between BC tissues
vs. paired adjacent non-tumor tissues. Then, the 5 vital miRNAs
were predicted though miRanda and DIANA tools (30) (Table
III). Target genes that were predicted to bind to these miRNAs
were further identified using miRTarBase (31) and TargetScan (32). Eventually, based on the
aforementioned analysis, the present study identified an
intersection between the mRNAs predicted by lncRNAs and sequencing
DE mRNAs. The results presented the 236 intersection mRNAs
(Table SII), and certain of them
have been affirmed as cancer-associated genes, for instance CDK1,
CCNB1, BUB1, EGF, E2F1, E2F2, FEN1, CD36, PARP1 and EFNA1 (33,34).
 | Table III.Five cancer-related key miRNAs
identified for each of the selected four DE lncRNAs. |
Table III.
Five cancer-related key miRNAs
identified for each of the selected four DE lncRNAs.
| lncRNAs | MRE1 | MRE2 | MRE3 | MRE4 | MRE5 |
|---|
| lnc-3108 | hsa-miR-1827 | hsa-miR-1253 | hsa-miR-558 | hsa-miR-1276 | hsa-miR-661 |
| lnc-8996 | hsa-miR-331-3p | hsa-miR-346 | hsa-miR-1299 | hsa-miR-384 | hsa-miR-1231 |
| lnc-4265 | hsa-miR-578 | hsa-miR-602 |
hsa-miR-92a-2-5p | hsa-miR-23b-5p |
hsa-miR-5088-3p |
| lnc-8461 | hsa-miR-198 | hsa-miR-326 |
hsa-miR-146b-3p | hsa-miR-421 | hsa-miR-1294 |
To date, the functions of most lncRNAs have not been
determined. Therefore, the present study constructed a ceRNA and
PPI network according to predicted lncRNA-miRNA, miRNA-mRNA and
protein-protein degree of interaction, in order to investigate the
critical lncRNAs in BC tissues. The present study selected all
dysregulated lncRNAs and their 236 targeted DE-mRNAs to construct
the ceRNA global network (Fig.
S1). The miRNAs hsa-miR-93-5p and hsa-miR-17-5p were equally
associated with the majority of lncRNAs and target mRNAs in this
global map. Similarly, the present study selected four dysregulated
lncRNAs (upregulated ENST00000433108; downregulated
ENST00000598996, ENST00000524265 and ENST00000398461) and 93
targeted DE-mRNAs to build the ceRNA local-area network (Fig. 6). The miRNA hsa-miR-1276 was
associated with the most lncRNAs and target mRNAs in this localized
network. In the ceRNA network map, the preset study demonstrated
the top 5 cancer-associated miRNAs that potentially bind to the
lncRNAs and the five most likely target genes of each miRNA. Among
the 93 intersection genes, the present study also built a PPI
network based on the diverse modes of action (Fig. 7 and Data S1). The ceRNA and PPI networks were
established using Cytoscape 3.6.1 and String pool, respectively.
Although it provided us with a clear direction in the progression
of BC, the deeper functional mechanism of screened DE-lncRNAs and
their target genes involved in ceRNA and PPI networks still require
further research in the BC cell lines.
 | Figure 6.A magnified partial ceRNA network for
the selected four lncRNAs was constructed using Cytoscape 3.6.1
tool. There were 4 lncRNAs, 19 miRNAs, 93 mRNAs and 143 edges in
this network. The yellow arrowheads and purple rectangles indicate
miRNAs and pathways, respectively. The circles represent mRNAs,
hexagons represent lncRNAs, red indicates upregulation, and blue
indicates downregulation. ceRNA, competing endogenous RNA; lncRNA,
long non-coding RNA; miRNA, microRNA; mRNA, messenger RNA. |
 | Figure 7.The PPI analysis for 93 target genes
were constructed with the String tool. Spheres represent the genes,
and lines represent the protein interaction between the genes.
Colored nodes represent query proteins and first shell of
interactors while white nodes second shell of interactors. The
information inside the sphere describes the protein structure:
Empty nodes, protein of unknown 3D structure; filled nodes, some 3D
structure is known or predicted. Blue-green lines represent known
interactions from curated databases; purple lines, known
interactions from experimentally determined; green lines, predicted
interactions form the gene neighborhood; red lines, predicted
interactions from gene fusions; dark blue lines, predicted
interactions from gene co-occurrence; yellow lines, interactions
from textmining; black lines, interactions from co-expression;
light blue lines, interactions form protein homology. PPI,
protein-protein interaction. |
Clinical impact of selected lncRNAs in
patients with BC
According to the validation results, ENST00000598996
and ENST00000524265 were the most significantly decreased in BC
specimens compared with those in normal specimens. Subsequently,
the present study assessed the correlation between the expression
of these four lncRNAs and various clinicopathological
characteristics of patients with BC. As presented in Table IV, a decreased expression level of
ENST00000598996 and ENST00000524265 was markedly associated with
histological stage, tumor pathological stage and lymphatic
metastasis, rather than the sex and age of the patients.
Subsequently, the ROC curve analysis was conducted in order to
estimate the diagnostic performance of these two lncRNAs in
patients with BC. The AUC was 0.867 [95% confidence interval (CI):
0.795–0.939; P<0.0001] for ENST00000598996, 0.815 (95% CI:
0.732–0.899, P<0.0001) for ENST00000524265 (Fig. 8A).
 | Figure 8.Clinical significance of lnc8996 and
lnc4265 in patients with BC. (A) ROC curves of lnc8996 and lnc4265
differentiating BC samples from adjacent normal samples
(P<0.0001). (B and C) Kaplan-Meier curves for OS based on
lnc8996 and lnc4265 expression in a group of 64 patients with BC
(P<0.001). Horizontal axis, OS time (months); vertical axis,
survival function. (D and E) Kaplan-Meier curves for PFS based on
lnc8996 and lnc4265 expression in a group of 64 patients with BC
(P<0.001). Horizontal axis, PFS time (months); vertical axis,
survival function. lnc-8996, ENST00000598996; lnc-4265,
ENST00000524265; BC, bladder cancer; ROC, receiver operating
characteristic; AUC, area under the curve; OS, overall survival;
PFS, progression-free survival. |
 | Table IV.Correlation analysis between the
expression of selected four lncRNAs and clinicopathological
characteristics in 64 BC patients. |
Table IV.
Correlation analysis between the
expression of selected four lncRNAs and clinicopathological
characteristics in 64 BC patients.
|
| lnc-8996
expression |
| lnc-4265
expression |
| lnc-3108
expression |
| lnc-8461
expression |
|
|---|
|
|
|
|
|
|
|
|
|
|
|---|
| Variables
(total) | Low n=56 | High n=8 | P-value | Low n=53 | High n=11 | P-value | Low n=32 | High n=32 | P-value | Low n=55 | High n=9 | P-value |
|---|
| Sex |
|
|
|
|
|
|
|
|
|
|
|
|
| Female
(7) | 7 | 0 | 0.582 | 5 | 2 | 0.594 | 2 | 5 | 0.426 | 5 | 2 | 0.253 |
| Male
(57) | 49 | 8 |
| 48 | 9 |
| 30 | 27 |
| 50 | 7 |
|
| Age |
|
|
|
|
|
|
|
|
|
|
|
|
| <65
(23) | 20 | 3 | >0.9 | 17 | 6 | 0.182 | 10 | 13 | 0.603 | 19 | 4 | 0.711 |
| ≥65
(41) | 36 | 5 |
| 36 | 5 |
| 22 | 19 |
| 36 | 5 |
|
| Tumor
sizea |
|
|
|
|
|
|
|
|
|
|
|
|
| <3
cm (32) | 26 | 6 | 0.257 | 27 | 5 | >0.9 | 19 | 13 | 0.211 | 26 | 6 | 0.474 |
| ≥3 cm
(32) | 30 | 2 |
| 26 | 6 |
| 13 | 19 |
| 29 | 3 |
|
| Multiplicity |
|
|
|
|
|
|
|
|
|
|
|
|
| Soli.
(31) | 26 | 5 | 0.468 | 24 | 7 | 0.331 | 19 | 12 | 0.133 | 27 | 4 | >0.9 |
| Mult.
(33) | 30 | 3 |
| 29 | 4 |
| 13 | 20 |
| 28 | 5 |
|
| Pathological
stage |
|
|
|
|
|
|
|
|
|
|
|
|
| T1-T2
(36) | 29 | 7 | 0.07 | 26 | 10 | 0.017b | 19 | 17 | 0.801 | 28 | 8 | 0.066 |
| T3-T4
(28) | 27 | 1 |
| 27 | 1 |
| 13 | 15 |
| 27 | 1 |
|
| Histologic
grade |
|
|
|
|
|
|
|
|
|
|
|
|
| Low
(25) | 19 | 6 | 0.048b | 17 | 8 | 0.018b | 13 | 12 | >0.9 | 18 | 7 | 0.022b |
| High
(39) | 37 | 2 |
| 36 | 3 |
| 19 | 20 |
| 37 | 2 |
|
| Lymphatic
metastasis |
|
|
|
|
|
|
|
|
|
|
|
|
| No
(38) | 30 | 8 | 0.017c | 27 | 11 | 0.002c | 19 | 19 | >0.9 | 29 | 9 | 0.008c |
| Yes
(26) | 26 | 0 |
| 26 | 0 |
| 13 | 13 |
| 26 | 0 |
|
In addition, the present study investigated the
prognostic value of ENST00000598996 and ENST00000524265 expression
in BC by performing a Kaplan-Meier survival analysis of OS and PFS.
The results indicated that patients with a lower expression level
of ENST00000598996 and ENST00000524265 had shorter postoperative OS
and PFS time than patients with higher expression of
ENST00000598996 and ENST00000524265 (Fig. 8B-E). Furthermore, univariate and
multivariate Cox regression analyses revealed that ENST00000598996
and ENST00000524265 expression levels were potential prognostic
predictors for OS and PFS of patients with BC (Table V).
 | Table V.Univariate and multivariate analysis
of prognostic factors for OS and PFS in patients with BC. |
Table V.
Univariate and multivariate analysis
of prognostic factors for OS and PFS in patients with BC.
|
| Univariate analysis
(OS) | Multivariate
analysis (OS) | Univariate analysis
(PFS) | Multivariate
analysis (PFS) |
|---|
|
|
|
|
|
|
|---|
| Factors | HR (95% CI) | P-value | HR (95% CI) | P-value | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Age
(≧65/<65) | 1.508
(0.52–4.38) | 0.45 | – | – | 1.162
(0.54–2.52) | 0.703 | – | – |
| Sex
(Male/Female) | 2.114
(0.28–16.24) | 0.472 | – | – | 1.058
(0.32–3.56) | 0.927 | – | – |
| Tu. size (≧3
cm/<3 cm) | 1.088
(0.40–2.97) | 0.87 | – | – | 0.885
(0.42–1.85) | 0.746 | – | – |
| Mul.
(Multiple/Single) | 1.699
(0.62–4.68) | 0.306 | – | – | 1.541
(0.69–3.43) | 0.289 | – | – |
| Lym. no. Met.
(Yes/No) | 6.782
(2.13–21.64) | 0.001 | 0.748
(0.13–4.41) | 0.748 | 4.483
(2.02–9.97) | <0.001 | 1.023
(0.27–3.81) | 0.973 |
| T stage
(T2-T4/Ta-T1) | 6.176
(1.92–19.87) | 0.002 | 2.073
(0.34–12.64) | 0.429 | 6.192
(2.55–15.02) | <0.001 | 1.564
(0.37–6.55) | 0.54 |
| Histo. gr.
(High/Low) | 8.161
(1.80–36.91) | 0.006 | 1.937
(0.19–19.79) | 0.577 | 5.462
(2.02–14.75) | 0.001 | 3.199
(0.67–15.36) | 0.146 |
| lnc8996
(High/Low) | 0.153
(0.04–0.55) | 0.004 | 0.203
(0.05–0.86) | 0.031 | 0.275
(0.12–0.61) | 0.002 | 0.281
(0.12–0.65) | 0.003 |
| lnc4265
(High/Low) | 0.136
(0.04–0.49) | 0.002 | 0.15
(0.03–0.73) | 0.018 | 0.232
(0.10–0.54) | 0.001 | 0.316
(0.13–0.79) | 0.013 |
These results demonstrated an association between
downregulation of ENST00000598996 and ENST00000524265 and
aggressive tumor characteristics of BC, and may serve as tumor
suppressor genes in BC. These results indicated the potential
diagnostic and prognostic value of these two lncRNAs in patients
with BC.
Both ENST00000598996 and
ENST00000524265 play a tumor-suppressive role in BC cells
Next, the biological roles of ENST00000598996 (named
lnc8996) and ENST00000524265 (lnc4265) was investigated in BC
cells. To achieve this, the BC cell lines EJ-1 and HTB-9 cells were
transfected with lnc-8996 or lnc-4265 overexpressed plasmid with
PCDNA3.1 frame or negative control (NC), respectively. After 48 h
of treatment, the expression levels of lnc-8996 and lnc-4265 were
significantly increased compared to NC (Fig. 9A). As presented in Fig. 9B, the proliferation curves detected
by CCK-8 experiments demonstrated that cell growth ability was
attenuated followed by upregulation of lnc-8996 or lnc-4265
compared with the vector group (NC) in EJ-1 and HTB-9 cells. The
present study demonstrated that enhanced expression of lnc-8996 or
lnc-4265 weakened the clonogenic capacity of EJ-1 and HTB-9 cells
using a colony formation assay (Fig.
9C). To research other biological behaviors of lnc-8996 and
lnc-4265 in BC, migration and invasion, scratch wound assays and
Transwell assays were carried out in the EJ-1 and HTB-9 cells. Both
lnc-8996 and lnc-4265 significantly inhibited the wound healing
ability of BC cells (Fig. 9D).
Furthermore, as presented in Fig.
9E, both migration potential and invasive ability of BC cells
was significantly impaired by lnc-8996 and lnc-4265 overexpression.
Collectively, these data clearly revealed that lnc-8996 or lnc-4265
overexpression strongly inhibited the proliferation, migration and
invasion of BC cells in vitro.
Discussion
Recently, an increasing number of studies have
confirmed that lncRNAs are involved in the molecular biological
processes including pathogenesis, invasion, metastasis and survival
prognosis, of a number of different types of cancer (16,25,35).
However, information regarding advanced BC and lncRNAs remains
limited.
Hence, the present study performed high-throughput
total transcriptome sequencing to identify DE lncRNAs and mRNAs
between five matched BC tissues and adjacent normal tissues. Then,
the present study obtained 37,684 lncRNAs and 20,266 mRNAs from the
two groups. Among them, compared with controls, the present study
detected 35 lncRNAs that were significantly upregulated and 68 that
were significantly downregulated in the BC specimens. To date, a
considerable proportion of these DE-lncRNAs have been reported in
multiple carcinomas. For example, lncRNA XIST (ENST00000429829)
(36,37), TINCR (ENST00000448587) (38,39),
MEG3 (ENST00000398460, ENST00000398461 and ENST00000452120)
(40,41), FENDRR (ENST00000593604 and
ENST00000598996) (42,43), and ADAMTS9-AS2 (ENST00000460833,
ENST00000474768 and ENST00000481312) (44,45)
were observed in gastric, lung and breast cancers; LINC01419
(ENST00000522365) in esophageal (46) and ovarian (47) carcinomas;
JHDM1D-AS1(ENST00000566699) in pancreatic cancer (48); MIR205HG (ENST00000433108) in
cervical cancer (49); MBNL1-AS1
(ENST00000445466) (50) and PART1
(ENST00000515734) (51) in
non-small cell lung cancer. MAGI2-AS3 (ENST00000452320) was
observed in hepatocellular carcinoma (52). Notably, it has been reported that
lncRNA XIST (53), MEG3 (54), MAGI2-AS3 (55), FENDRR and ADAMTS9-AS2 (56) act as prognosis markers of patients
with BC or exert significant roles in BC progression through
different cancer-associated pathways. For instance, Kouhsar et
al (56) revealed that based on
the analysis of the ceRNA networks in the early stage of BC, the
lncRNAs FENDRR, ADAMTS9-AS2 and CARMN may regulate MEG3 in NMIBC
via harboring some critical miRNAs such as miR-143-3p, miR-34a-3p
and miR-106a-5p.
In order to identify the potential functional
effects of these dysregulated lncRNAs in BC, the 236 DEmRNAs
revealed in the global ceRNA network were used for the GO function
and KEGG pathway analyses. As revealed, the significant GO items of
these dysregulated genes were ‘gene regulation of apoptotic
process’ (GO:0042981), ‘regulation of cell cycle’ (GO:0051726),
‘positive regulation of gene expression’ (GO:0010628) and ‘positive
regulation of cell proliferation’ (GO:0008284) in BP. Furthermore,
the present study revealed that several cancer-associated pathways
were significantly enriched, including ‘pathways in cancer’, ‘cell
cycle’, ‘apoptosis’, ‘MAPK signaling pathway’, ‘Focal adhesion’,
‘PI3K-Akt signaling pathway’, ‘transcriptional misregulation in
cancer’, ‘cAMP signaling pathway’, ‘p53 signaling pathway’ and
‘NF-κB signaling pathway’. Hence, it is reasonable to assume that
these dysregulated lncRNAs in BC act principally by modulating the
cell cycle, cell proliferation and the aforementioned pivotal
pathways, which then impact cancer cell aggression. Zhang et
al revealed that when compared with paired non-cancerous liver
tissues, LINC01419 was significantly upregulated in HCC and may be
associated with initiation of HCC via regulating cell cycle genes
(57). Wang et al revealed
that EPIC1 regulated the cell cycle and promoted breast cancer
proliferation (13), and Su et
al reported that BLACAT1 affected colorectal cancer cell
proliferation by silencing p15 (15). It has also been revealed that
downregulation of lncRNA ADAMTS9-AS2 contributed to gastric cancer
development through activation of the PI3K/Akt pathway (44). However, although numerous miRNAs,
genes and pathways indicated in this ceRNA network have been
reported in types of cancer, the detailed molecular mechanism
involved in BC development and progression requires further
investigation.
Notably, according to their nearby genes, the
present study was also able to identify a few tumor-associated
lncRNAs altered in BC. ENST00000562848, ENST00000587187,
ENST00000398878, ENST00000421206, ENST00000443576 and
ENST00000562393 are spliced from the neighboring areas of HOXC6
(58), UHRF1 (59), SULT1A3 (60), GRK5 (61), STARD13 (62) and BDKRB2 (63), which plays a critical role in
malignant proliferation, migration and metastasis. Overall, these
findings indicated that these lncRNAs are associated with a diverse
range of different types of cancer, thereby emphasizing the
accuracy of the sequencing data in the present study, and
indicating the potential critical role of these lncRNAs in the
development of BC as well. Therefore, efforts should be made to
ascertain their biological function in BC. These four lncRNAs
(upregulated, lnc-3108; downregulated, lnc-8996, lnc-4265 and
lnc-8461) were selected and then investigated in another 64 newly
diagnosed patients with BC and two lncRNAs (downregulated, lnc-8996
and lnc-4265) in four BC cells by RT-qPCR. These verification
results were nearly accordant with the RNA-seq results.
An increasing amount evidence has demonstrated a
correlation between lncRNA expression and the clinicopathological
characteristics of different types of cancer, such as World Health
Organization grade and KPS score in glioma (64), lymph node metastasis and
Tumor-Node-Metastasis classification in HCC (65), and invasion depth, tumor stage and
lymphatic metastasis in gastric carcinoma (43). In the present study, according to
aforementioned validation data, lnc-8996 and lnc-4265 were found to
be expressed at significantly lower levels in BC tissues than in
the normal controls, and associated with advanced T stage,
histological grade or lymph node metastasis in patients with BC.
Furthermore, overexpression of lnc-8996 and lnc-4265 suppressed BC
cell line proliferation, migration and invasion as demonstrated by
the in vitro experiments. For lnc-8996, similar results have
also been demonstrated in lung adenocarcinoma (43,66).
Notably, ectopic expression of lnc-8996 and lnc-4265 affected the
migration and invasion abilities of BC cells to a greater extent
than the proliferation ability. A potential complex biological
regulatory mechanism of lnc-8996 or lnc-4265 may be mainly
responsible for the discrepancies. With regard to this phenomenon,
similar results were also revealed in numerous studies (67,68).
Furthermore, prior studies suggested that lncRNAs
function as diagnostic or prognostic biomarkers and therapeutic
targets for malignancies, such as LINC00261 and lncRNA FOXD2-AS1
for gastric cancer (18,35) and ZEB1-AS1 for hepatocellular
carcinoma (8,16). In accordance with the results of
these previous studies, by applying Kaplan-Meier analysis and
log-rank tests, the present study indicated that patients with BC
with low lnc-8996 or lnc-4265 expression levels had poorer OS and
PFS compared with those patients with higher levels. Furthermore,
after the univariate and multivariate Cox regression analyses,
lnc-8996 and lnc-4265 could serve as potential prognostic markers
for both OS and PFS. In the multivariate Cox regression analysis,
only lnc-8996 and lnc-4265, rather than T stage and lymph
metastasis were statistically significant. The most likely reason
for this result was a multicollinearity problem. With regard to
this phenomenon, similar results were also found in numerous tumor
studies (69–71). Next, the prediction power of each of
the two lncRNAs was estimated on the basis of the ROC curve, and
the results revealed that lnc-8996 and lnc-4265 had better
diagnostic prediction power (AUC equals 0.867 and 0.815,
respectively). These results indicated that decreased expression of
lnc-8996 and lnc-4265 may act as significant biomarkers for the
diagnosis and prognosis of BC.
LncRNAs have been reported to exert multifaceted
roles by functioning as cytoplasmic miRNA sponges, combined with
RNA-binding proteins or other functional proteins, and acting as
nuclear transcriptional regulators (13,17,72).
Among these roles, functioning as an miRNA sponge represents the
most classic function, which has been confirmed in various
diseases, such as bladder carcinoma (23), colorectal cancer (14) and lung cancer (37). Accordingly, by employing
bioinformatics analysis, the present study first revealed that
these four lncRNAs may interact with 19 types of predicted crucial
miRNAs and therefore, the presumptive 93 key DE-genes stemmed from
the sequencing results that were used to form ceRNA and PPI
networks. Notably, in the ceRNA network, it was revealed that
miR-661 (73) and miR-326 (74) exhibited high and low expression in
cancer tissues, respectively. Li et al also revealed that
upregulated miR-558 promoted cell migration, invasion and
angiogenesis by targeting HPSE in T24T and UMUC3 cells (75). CCNB1, the G2/M transition member of
the cell cycle, was significantly upregulated in BC tumor tissues,
which was consistent with a previous study (76). Notably, these studies also reflect
the accuracy of the predicted ceRNA network in the present
study.
In conclusion, the present study presented general
perspectives of DElncRNAs and DEmRNAs in BC samples vs. adjacent
non-BC samples using RNA high-throughput sequencing. Four lncRNAs
(lnc-3108, lnc-8996, lnc-4265 and lnc-8461) were further validated
in both BC tissues and cell lines via RT-qPCR. The results revealed
that lnc-8996 and lnc-4265 could be potential diagnostic biomarkers
for BC. It was also revealed that lnc-8996 and lnc-4265 expression
levels were markedly downregulated and closely associated with
unfavorable clinicopathological parameters and poor survival
prognoses. In addition, through bioinformatics, the present study
predicted a number of potential gene regulatory networks and
signaling pathways associated with these DE-lncRNAs. Ultimately,
lnc-8996 and lnc-4265 overexpression retarded bladder cancer cell
proliferation, migration and invasion in vitro. All these
results indicated that lnc-8996 and lnc-4265 may be potential
biomarkers for diagnosis and prognosis, as well as promising
therapeutic targets in patients with BC. However, larger clinical
samples and deeper insights into the regulatory mechanisms of these
lncRNAs are required to confirm the present data.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
We would like to thank Cloud-Seq Biotech Co., Ltd.
(Shanghai, China) for the high-throughput RNA-Seq service.
Funding
The present study received financial support from
the Key Project of Health Industry in Tianjin (grant no. 16KG119),
the Natural Science Foundation Project of Tianjin (grant no.
18PTLCSY00010), the Tianjin Urological Key Laboratory Foundation
(grant no. 2017ZDSYS13) and the Youth Fund of Tianjin Medical
University Second Hospital (grant no. 2018ydey09).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
HH and CS conceived and designed the study. DT and
ZW collected the clinical specimens. CS, YW, LD and SG performed
the experiments and analyzed the results. CS wrote the manuscript.
HH, LX, YQ, YW and ZZ edited the manuscript and provided critical
comments. All authors read and approved the final version of this
manuscript and agree to be accountable for all aspects of the
research in ensuring that the accuracy or integrity of any part of
the work are appropriately investigated and resolved.
Ethics approval and consent to
participate
The protocol of the present study was conducted
based on the Declaration of Helsinki and approved by the Medical
Review Committee of the Second Hospital of Tianjin Medical
University. Written informed consent was obtained from all patients
for the use of their tissue in this study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2019. CA Cancer J Clin. 69:7–34. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Di Meo A, Bartlett J, Cheng Y, Pasic MD
and Yousef GM: Liquid biopsy: A step forward towards precision
medicine in urologic malignancies. Mol Cancer. 16:802017.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Singh R, Ansari JA, Maurya N, Mandhani A,
Agrawal V and Garg M: Epithelial-to-mesenchymal transition and its
correlation with clinicopathologic features in patients with
urothelial carcinoma of the bladder. Clin Genitourin Cancer.
15:e187–e197. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cao W, Zhao Y, Wang L and Huang X:
Circ0001429 regulates progression of bladder cancer through binding
miR-205-5p and promoting VEGFA expression. Cancer Biomark.
25:101–113. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Li B, Xie F, Zheng FX, Jiang GS, Zeng FQ
and Xiao XY: Overexpression of CircRNA BCRC4 regulates cell
apoptosis and MicroRNA-101/EZH2 signaling in bladder cancer. J
Huazhong Univ Sci Technolog Med Sci. 37:886–890. 2017.PubMed/NCBI
|
|
7
|
Antoni S, Ferlay J, Soerjomataram I, Znaor
A, Jemal A and Bray F: Bladder cancer incidence and mortality: A
global overview and recent trends. Eur Urol. 71:96–108. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Qiu L, Tang Q, Li G and Chen K: Long
non-coding RNAs as biomarkers and therapeutic targets: Recent
insights into hepatocellular carcinoma. Life Sci. 191:273–282.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tang Y, Cheung BB, Atmadibrata B, Marshall
GM, Dinger ME, Liu PY and Liu T: The regulatory role of long
noncoding RNAs in cancer. Cancer Lett. 391:12–19. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Vishnubalaji R, Shaath H, Elkord E and
Alajez NM: Long non-coding RNA (lncRNA) transcriptional landscape
in breast cancer identifies LINC01614 as non-favorable prognostic
biomarker regulated by TGFβ and focal adhesion kinase (FAK)
signaling. Cell Death Discov. 5:1092019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yang H, Chen T, Xu S, Zhang S and Zhang M:
Long noncoding RNA FOXC2-AS1 predicts poor survival in breast
cancer patients and promotes cell proliferation. Oncol Res.
27:219–226. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen C, He W, Huang J, Wang B, Li H, Cai
Q, Su F, Bi J, Liu H, Zhang B, et al: LNMAT1 promotes lymphatic
metastasis of bladder cancer via CCL2 dependent macrophage
recruitment. Nat Commun. 9:38262018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang Z, Yang B, Zhang M, Guo W, Wu Z, Wang
Y, Jia L, Li S, Xie W and Yang D; Cancer Genome Atlas Research
Network, : lncRNA epigenetic landscape analysis identifies EPIC1 as
an oncogenic lncRNA that interacts with MYC and promotes cell-cycle
progression in cancer. Cancer Cell. 33:706–720. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Xu M, Chen X, Lin K, Zeng K, Liu X, Pan B,
Xu X, Xu T, Hu X, Sun L, et al: The long noncoding RNA SNHG1
regulates colorectal cancer cell growth through interactions with
EZH2 and miR-154-5p. Mol Cancer. 17:1412018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Su J, Zhang E, Han L, Yin D, Liu Z, He X,
Zhang Y, Lin F, Lin Q, Mao P, et al: Long noncoding RNA BLACAT1
indicates a poor prognosis of colorectal cancer and affects cell
proliferation by epigenetically silencing of p15. Cell Death Dis.
8:e26652017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jin L, He Y, Tang S and Huang S: LncRNA
GHET1 predicts poor prognosis in hepatocellular carcinoma and
promotes cell proliferation by silencing KLF2. J Cell Physiol.
233:4726–4734. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang Y, Yang L, Chen T, Liu X, Guo Y, Zhu
Q, Tong X, Yang W, Xu Q, Huang D and Tu K: A novel lncRNA
MCM3AP-AS1 promotes the growth of hepatocellular carcinoma by
targeting miR-194-5p/FOXA1 axis. Mol Cancer. 18:282019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Fan Y, Wang YF, Su HF, Fang N, Zou C, Li
WF and Fei ZH: Decreased expression of the long noncoding RNA
LINC00261 indicate poor prognosis in gastric cancer and suppress
gastric cancer metastasis by affecting the epithelial-mesenchymal
transition. J Hematol Oncol. 9:572016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li X, Xiao X, Chang R and Zhang C:
Comprehensive bioinformatics analysis identifies lncRNA HCG22 as a
migration inhibitor in esophageal squamous cell carcinoma. J Cell
Biochem. 121:468–481. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen W, Hang Y, Xu W, Wu J, Chen L, Chen
J, Mao Y, Song J, Song J and Wang H: BLACAT1 predicts poor
prognosis and serves as oncogenic lncRNA in small-cell lung cancer.
J Cell Biochem. 120:2540–2546. 2019. View Article : Google Scholar
|
|
21
|
Fang C, He W, Xu T, Dai J, Xu L and Sun F:
Upregulation of lncRNA DGCR5 correlates with better prognosis and
inhibits bladder cancer progression via transcriptionally
facilitating P21 expression. J Cell Physiol. 234:6254–6262. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shan H, Yang Y, Zhu X, Han X, Zhang P and
Zhang X: FAM83H-AS1 is associated with clinical progression and
modulates cell proliferation, migration, and invasion in bladder
cancer. J Cell Biochem. 120:4687–4693. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Rui X, Wang L, Pan H, Gu T, Shao S and
Leng J: LncRNA GAS6-AS2 promotes bladder cancer proliferation and
metastasis via GAS6-AS2/miR-298/CDK9 axis. J Cell Mol Med.
23:865–876. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Xu Z, Huang B, Zhang Q, He X, Wei H and
Zhang D: NOTCH1 regulates the proliferation and migration of
bladder cancer cells by cooperating with long non-coding RNA HCG18
and microRNA-34c-5p. J Cell Biochem. 120:6596–6604. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Cao X, Xu J and Yue D: LncRNA-SNHG16
predicts poor prognosis and promotes tumor proliferation through
epigenetically silencing p21 in bladder cancer. Cancer Gene Ther.
25:10–17. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhai X and Xu W: Long noncoding RNA ATB
promotes proliferation, migration, and invasion in bladder cancer
by suppressing MicroRNA-126. Oncol Res. 26:1063–1072. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tuo Z, Zhang J and Xue W: LncRNA TP73-AS1
predicts the prognosis of bladder cancer patients and functions as
a suppressor for bladder cancer by EMT pathway. Biochem Biophys Res
Commun. 499:875–881. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Paraskevopoulou MD, Vlachos IS, Karagkouni
D, Georgakilas G, Kanellos I, Vergoulis T, Zaggana K, Tsanakas P,
Floros E, Dalamagas T and Hatzigeorgiou AG: DIANA-LncBase v2:
Indexing microRNA targets on non-coding transcripts. Nucleic Acids
Res. 44:D231–D238. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chou CH, Shrestha S, Yang CD, Chang NW,
Lin YL, Liao KW, Huang WC, Sun TH, Tu SJ, Lee WH, et al: MiRtarBase
update 2018: A resource for experimentally validated
microRNA-target interactions. Nucleic Acids Res. 46:D296–D302.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
Elife. 4:e050052015. View Article : Google Scholar
|
|
33
|
Zhuang L, Yang Z and Meng Z: Upregulation
of BUB1B, CCNB1, CDC7, CDC20, and MCM3 in tumor tissues predicted
worse overall survival and disease-free survival in hepatocellular
carcinoma patients. Biomed Res Int. 2018:78973462018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yao H, Lu F and Shao Y: The E2F family as
potential biomarkers and therapeutic targets in colon cancer. Peer
J. 8:e85622020. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Xu TP, Wang WY, Ma P, Shuai Y, Zhao K,
Wang YF, Li W, Xia R, Chen WM, Zhang EB and Shu YQ: Upregulation of
the long noncoding RNA FOXD2-AS1 promotes carcinogenesis by
epigenetically silencing EphB3 through EZH2 and LSD1, and predicts
poor prognosis in gastric cancer. Oncogene. 37:5020–5036. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Xing F, Liu Y, Wu SY, Wu K, Sharma S, Mo
YY, Feng J, Sanders S, Jin G, Singh R, et al: Loss of XIST in
breast cancer activates MSN-c-Met and reprograms microglia via
exosomal miRNA to promote brain metastasis. Cancer Res.
78:4316–4330. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Li C, Wan L, Liu Z, Xu G, Wang S, Su Z,
Zhang Y, Zhang C, Liu X, Lei Z and Zhang HT: Long non-coding RNA
XIST promotes TGF-β-induced epithelial-mesenchymal transition by
regulating miR-367/141-ZEB2 axis in non-small-cell lung cancer.
Cancer Lett. 418:185–195. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Gao YW, Ma F, Xie YC, Ding MG, Luo LH,
Jiang S, Rao L and Liu XL: Sp1-induced upregulation of the long
noncoding RNA TINCR inhibits cell migration and invasion by
regulating miR-107/miR-1286 in lung adenocarcinoma. Am J Transl
Res. 11:4761–4775. 2019.PubMed/NCBI
|
|
39
|
Chen Z, Liu H, Yang H, Gao Y, Zhang G and
Hu J: The long noncoding RNA, TINCR, functions as a competing
endogenous RNA to regulate PDK1 expression by sponging miR-375 in
gastric cancer. Onco Targets Ther. 10:3353–3362. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Terashima M, Tange S, Ishimura A and
Suzuki T: MEG3 long noncoding RNA contributes to the epigenetic
regulation of epithelial-mesenchymal transition in lung cancer cell
lines. J Biol Chem. 292:82–99. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhang Y, Wu J, Jing H, Huang G, Sun Z and
Xu S: Long noncoding RNA MEG3 inhibits breast cancer growth via
upregulating endoplasmic reticulum stress and activating NF-κB and
p53. J Cell Biochem. 120:6789–6797. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Gong F, Dong D, Zhang T and Xu W: Long
non-coding RNA FENDRR attenuates the stemness of non-small cell
lung cancer cells via decreasing multidrug resistance gene 1 (MDR1)
expression through competitively binding with RNA binding protein
hur. Eur J Pharmacol. 853:345–352. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Xu TP, Huang MD, Xia R, Liu XX, Sun M, Yin
L, Chen WM, Han L, Zhang EB, Kong R, et al: Decreased expression of
the long non-coding RNA FENDRR is associated with poor prognosis in
gastric cancer and FENDRR regulates gastric cancer cell metastasis
by affecting fibronectin1 expression. J Hematol Oncol. 7:632014.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Cao B, Liu C and Yang G: Down-regulation
of lncRNA ADAMTS9-AS2 contributes to gastric cancer development via
activation of PI3K/Akt pathway. Biomed Pharmacother. 107:185–193.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Deva Magendhra Rao AK, Patel K, Korivi
Jyothiraj S, Meenakumari B, Sundersingh S, Sridevi V, Rajkumar T,
Pandey A, Chatterjee A, Gowda H and Mani S: Identification of
lncRNAs associated with early-stage breast cancer and their
prognostic implications. Mol Oncol. 13:1342–1355. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Chen JL, Lin ZX, Qin YS, She YQ, Chen Y,
Chen C, Qiu GD, Zheng JT, Chen ZL and Zhang SY: Overexpression of
long noncoding RNA LINC01419 in esophageal squamous cell carcinoma
and its relation to the sensitivity to 5-fluorouracil by mediating
GSTP1 methylation. Ther Adv Med Oncol. 11:17588359198389582019.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Chen Y, Bi F, An Y and Yang Q:
Identification of pathological grade and prognosis-associated
lncRNA for ovarian cancer. J Cell Biochem. 120:14444–14454. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Kondo A, Nonaka A, Shimamura T, Yamamoto
S, Yoshida T, Kodama T, Aburatani H and Osawa T: Long noncoding RNA
JHDM1D-AS1 promotes tumor growth by regulating angiogenesis in
response to nutrient starvation. Mol Cell Biol. 37:e00125–e00117.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Li Y, Wang H and Huang H: Long non-coding
RNA MIR205HG function as a ceRNA to accelerate tumor growth and
progression via sponging miR-122-5p in cervical cancer. Biochem
Biophys Res Commun. 514:78–85. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Li P, Xing W, Xu J, Yuan D, Liang G, Liu B
and Ma H: MicroRNA-301b-3p downregulation underlies a novel
inhibitory role of long non-coding RNA MBNL1-AS1 in non-small cell
lung cancer. Stem Cell Res Ther. 10:1442019. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Zhu D, Yu Y, Wang W, Wu K, Liu D, Yang Y,
Zhang C, Qi Y and Zhao S: Long noncoding RNA PART1 promotes
progression of non-small cell lung cancer cells via JAK-STAT
signaling pathway. Cancer Med. 8:6064–6081. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Yin Z, Ma T, Yan J, Shi N, Zhang C, Lu X,
Hou B and Jian Z: LncRNA MAGI2-AS3 inhibits hepatocellular
carcinoma cell proliferation and migration by targeting the
miR-374b-5p/SMG1 signaling pathway. J Cell Physiol.
234:18825–18836. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Hu B, Shi G, Li Q, Li W and Zhou H: Long
noncoding RNA XIST participates in bladder cancer by downregulating
p53 via binding to TET1. J Cell Biochem. 120:6330–6338. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Shan G, Tang T, Xia Y and Qian HJ: MEG3
interacted with miR-494 to repress bladder cancer progression
through targeting PTEN. J Cell Physiol. 235:1120–1128. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Wang F, Zu Y, Zhu S, Yang Y, Huang W, Xie
H and Li G: Long noncoding RNA MAGI2-AS3 regulates CCDC19
expression by sponging miR-15b-5p and suppresses bladder cancer
progression. Biochem Biophys Res Commun. 507:231–235. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Kouhsar M, Azimzadeh Jamalkandi S, Moeini
A and Masoudi-Nejad A: Detection of novel biomarkers for early
detection of non-muscle-invasive bladder cancer using competing
endogenous RNA network analysis. Sci Rep. 9:84342019. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Zhang H, Zhu C, Zhao Y, Li M, Wu L, Yang
X, Wan X, Wang A, Zhang MQ, Sang X and Zhao H: Long non-coding RNA
expression profiles of hepatitis C virus-related dysplasia and
hepatocellular carcinoma. Oncotarget. 6:43770–43778. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Li PD, Chen P, Peng X, Ma C, Zhang WJ and
Dai XF: HOXC6 predicts invasion and poor survival in hepatocellular
carcinoma by driving epithelial-mesenchymal transition. Aging
(Albany NY). 10:115–130. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Hu Q, Qin Y, Ji S, Xu W, Liu W, Sun Q,
Zhang Z, Liu M, Ni Q, Yu X and Xu X: UHRF1 promotes aerobic
glycolysis and proliferation via suppression of SIRT4 in pancreatic
cancer. Cancer Lett. 452:226–236. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Zou J, Li H, Huang Q, Liu X, Qi X, Wang Y,
Lu L and Liu Z: Dopamine-induced SULT1A3/4 promotes EMT and cancer
stemness in hepatocellular carcinoma. Tumour Biol.
39:10104283177192722017. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Jiang LP, Fan SQ, Xiong QX, Zhou YC, Yang
ZZ, Li GF, Huang YC, Wu MG, Shen QS, Liu K, et al: GRK5 functions
as an oncogenic factor in non-small-cell lung cancer. Cell Death
Dis. 9:2952018. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Chen L, Hu W, Li G, Guo Y, Wan Z and Yu J:
Inhibition of miR-9-5p suppresses prostate cancer progress by
targeting starD13. Cell Mol Biol Lett. 24:202019. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Zhou Y, Wang W, Wei R, Jiang G, Li F, Chen
X, Wang X, Long S, Ma D and Xi L: Serum bradykinin levels as a
diagnostic marker in cervical cancer with a potential mechanism to
promote VEGF expression via BDKRB2. Int J Oncol. 55:131–141.
2019.PubMed/NCBI
|
|
64
|
Chen XD, Zhu MX and Wang SJ: Expression of
long non-coding RNA MAGI2-AS3 in human gliomas and its prognostic
significance. Eur Rev Med Pharmacol Sci. 23:3455–3460.
2019.PubMed/NCBI
|
|
65
|
Ni W, Zhang Y, Zhan Z, Ye F, Liang Y,
Huang J, Chen K, Chen L and Ding Y: A novel lncRNA uc.134 represses
hepatocellular carcinoma progression by inhibiting CUL4A-mediated
ubiquitination of LATS1. J Hematol Oncol. 10:912017. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Zhang L, Li S, Choi YL, Lee J, Gong Z, Liu
X, Pei Y, Jiang A, Ye M, Mao M, et al: Systematic identification of
cancer-related long noncoding RNAs and aberrant alternative
splicing of quintuple-negative lung adenocarcinoma through RNA-Seq.
Lung Cancer. 109:21–27. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Jiang MM, Mai ZT, Wan SZ, Chi YM, Zhang X,
Sun BH and Di QG: Microarray profiles reveal that circular RNA
hsa_circ_0007385 functions as an oncogene in non-small cell lung
cancer tumorigenesis. J Cancer Res Clin Oncol. 144:667–674. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Chen Y, Yang Z, Deng B, Wu D, Quan Y and
Min Z: Interleukin 1β/1RA axis in colorectal cancer regulates tumor
invasion, proliferation and apoptosis via autophagy. Oncol Rep.
43:908–918. 2020.PubMed/NCBI
|
|
69
|
Zhao HQ, Dong BL, Zhang M, Dong XH, He Y,
Chen SY, Wu B and Yang XJ: Increased KIF21B expression is a
potential prognostic biomarker in hepatocellular carcinoma. World J
Gastrointest Oncol. 12:276–288. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Wolf D, Fiegl H, Zeimet AG, Wieser V,
Marth C, Sprung S, Sopper S, Hartmann G, Reimer D and Boesch M:
High RIG-I expression in ovarian cancer associates with an
immune-escape signature and poor clinical outcome. Int J Cancer.
146:2007–2018. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Shiseki M, Ishii M, Miyazaki M, Osanai S,
Wang YH, Yoshinaga K, Mori N and Tanaka J: Reduced PLCG1 expression
is associated with inferior survival for myelodysplastic syndromes.
Cancer Med. 9:460–468. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Sun Q, Hao Q and Prasanth KV: Nuclear long
noncoding RNAs: Key regulators of gene expression. Trends Genet.
34:142–157. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Liu F, Cai Y, Rong X, Chen J, Zheng D,
Chen L, Zhang J, Luo R, Zhao P and Ruan J: MiR-661 promotes tumor
invasion and metastasis by directly inhibiting RB1 in non small
cell lung cancer. Mol Cancer. 16:1222017. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Liang X, Li Z, Men Q, Li Y, Li H and Chong
T: MiR-326 functions as a tumor suppressor in human prostatic
carcinoma by targeting mucin1. Biomed Pharmacother. 108:574–583.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Li Y, Zheng F, Xiao X, Xie F, Tao D, Huang
C, Liu D, Wang M, Wang L, Zeng F and Jiang G: CircHIPK3 sponges
miR-558 to suppress heparanase expression in bladder cancer cells.
EMBO Rep. 18:1646–1659. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Li D, Hao X and Song Y: An integrated
analysis of key microRNAs, regulatory pathways and clinical
relevance in bladder cancer. Onco Targets Ther. 11:3075–3085. 2018.
View Article : Google Scholar : PubMed/NCBI
|