Introduction
Ovarian cancer (OC) is one of the most lethal types
of malignant gynecological cancer in the world (1). In 2018 in the United States, there
were ~22,240 new cases of OC and 14,070 OC-associated mortalities
(2). In China, the latest reported
number of OC-associated mortalities was 25,000 in 2015 (3). Although surgery, chemotherapy and
radiotherapy have made marked progresses in the treatment of OC,
the treatment of patients with advanced OC with distant metastasis
or recurrence remains a challenge, and the median overall survival
(OS) time of patients with metastatic disease has not improved over
the past decades (4). Therefore,
the elucidation of the underlying mechanisms of OC and the
identification of novel therapeutic targets to improve the clinical
outcome of patients suffering from OC is of utmost importance.
MicroRNAs (miRNAs or miRs) are a class of RNAs
containing ~22 nucleotides that cannot be translated into proteins
(5). miRNAs can act as suppressors
of gene expression by binding to the 3-untranslated region (3′-UTR)
of target mRNAs (5,6). Previous studies have demonstrated that
miRNAs serve vital roles in the progression of various types of
cancer, such as ovarian, lung and breast cancer, by regulating the
expression of genes involved in cell proliferation, apoptosis,
differentiation and migration (7).
For example, miR-15a-3p suppresses the proliferation and invasion
of ovarian cancer cells by targeting Twist1 (8). This suggests the possible application
of miRNAs in cancer diagnosis, treatment or prognosis (9,10).
Increasing evidence has indicated that miR-134 acts as a tumor
suppressor by blocking the proliferation of various cancer cells
through different signaling pathways (11,12).
For instance, miR-134 can regulate the activation of the PI3K
signaling pathway to inhibit the proliferative and invasive
activities of glioma cells and facilitate their apoptosis (13). Furthermore, miR-134 is mediated by
interferon regulatory factor 1 to suppress the tumorigenesis and
progression of osteosarcoma by targeting VEGFA and MYCN (14).
Flap structure-specific endonuclease 1 (FEN1) is a
member of the Rad2 structure-specific nuclease family, involved in
a number of DNA processing pathways to maintain genome stability
(15,16). For instance, through the interaction
with proliferating cell nuclear antigen (PCNA), FEN1 helps
coordinate Okazaki fragment maturation by removing RNA-DNA primers
(17). FEN1 expression is
upregulated in a number of types of cancer, such as cancer of the
testes, lung and brain (18). In
addition, accumulating evidence has indicated that miRNAs can
inhibit FEN1 expression by directly binding to its 3′-UTR, leading
to impaired DNA repair and repressed cancer progression (19,20).
The present study aimed to reveal the role of
miR-134-3p in OC by measuring the expression levels of miR-134-3p
in OC cell lines. Cell Counting Kit (CCK)-8, TUNEL, flow cytometry,
colony formation, wound healing and Transwell assays were used to
assess the inhibition of miR-134-3p on OC cell proliferation,
migration and invasion.
Materials and methods
Cell lines and cell culture
The human OC SKOV-3, A2780 and OVCAR-3 cell lines,
and the normal ovarian epithelial ISOE-80 cell line were purchased
from the American Type Culture Collection. A2780 and OVCAR-3 cells
were maintained in DMEM supplemented with 10% fetal bovine serum
(FBS) (both Gibco; Thermo Fisher Scientific, Inc.), 100 U/ml
penicillin and 100 mg/ml streptomycin (HyClone; Cytiva). ISOE-80
cells were maintained in RPMI-1640 medium (Gibco; Thermo Fisher
Scientific, Inc.) supplemented with 10% FBS, 100 U/ml penicillin
and 100 mg/ml streptomycin. SKOV-3 cells were cultured in McCoys 5A
medium (Gibco; Thermo Fisher Scientific, Inc.) supplemented with
10% FBS, 100 U/ml penicillin and 100 mg/ml streptomycin. All cells
were cultured at 37°C with 5% CO2.
Cell transfection
The chemically synthesized miR-134-3p mimic
(miRBase, MIMAT0026481; 5′-CCUGUGGGCCACCUAGUCACCAA-3′),
non-targeting mimic negative control (NC;
5′-UUCUCCGAACGUGUCACGUTT-3′), Lv-FEN1 and Lv-NC (expression vector,
pcDNA3.1) were designed and obtained from Shanghai GenePharma Co.,
Ltd. Cell transfection was performed in SKOV-3 and OVCAR-3 cells
using the cell electroporation system operator H1 (Etta Biotech Co.
Ltd.) according to the manufacturers protocol. Briefly, SKOV-3 and
OVCAR-3 cells were collected and re-suspended in the
electroporation buffer. The cell concentration was adjusted to
6×105 cells/ml and mixed with miRNA mimics (125 nM)
and/or plasmid (1 mg/ml) prior to electroporation. A total of 100
µl cell suspension mixed with RNAs was then added into a 96-well
plate, and used for gene transfection using the operator H1. The
electroporation device was operated at DC square wave with 200 V
voltage, 100 µsec duration, 2-sec interval and 6 pulses. After
electroporation, the cells were diluted to appropriate
concentrations and seeded into appropriate cell culture plates for
use in further assays.
Reverse transcription-quantitative PCR
(RT-qPCR)
TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) was used for the isolation of total RNA
from cells. Reverse transcription was performed using the 4X
Reverse Transcription Master Mix kit (EZBioscience) according to
the manufacturers protocol. qPCR was performed on cDNA using
FastStart Universal SYBR-Green Master Mix (Roche Diagnostics GmbH).
Briefly, the reaction mixture contained 25 µl SYBR-Green Master
Mix, 0.5 µl each primer (30 µmol/l), 5 µl DNA template (10 ng/µl)
and molecular-grade H2O to a final reaction volume of 45
µl. The amplification protocol consisted of one cycle of initial
denaturation at 95°C for 5 min, 40 cycles of denaturation at 95°C
for 15 sec and annealing/extension at 60°C for 1 min. Gene
expression levels were quantified using the 2−∆∆Cq
method (21). U6 and β-actin were
used to normalize miR-134-3p or FEN1 expression, respectively. The
PCR primer sequences for the selected genes were as follows: FEN1
forward, 5′-GTGAAGGCTGGCAAAGTCTA-3′ and reverse,
5′-GTGAAGGCTGGCAAAGTCTA-3′; β-actin forward,
5′-GCTCGTCGTCGACAACGGCTC-3′ and reverse,
5′-CAAACATGATCTGGGTCATCTTCTC-3′; miR-134-3p forward,
5′-CTGTGGGCCACCTAGTCACCAA-3′ and reverse,
5′-GCTGTCAACGATACGCTACCTA-3′; U6 forward, 5′-CTCGCTTCGGCAGCACA-3′
and reverse, 5′-AACGCTTCACGAATTTGCGT-3′.
Cell proliferation assay
The transfected SKOV-3 and OVCAR-3 cells were seeded
in 96-well plates at a density of 5×103 cells/well. The
CCK-8 (Beyotime Institute of Biotechnology) colorimetric assay was
performed to measure cell proliferation according to the
manufacturers protocol. Briefly, the supernatant was removed after
24, 48 and 72 h of culture at 37°C and 5% CO2.
Subsequently, 100 µl of respective medium containing 10 µl CCK-8
reagent was added to each well for 3 h at 37°C. The absorbance was
measured at 450 nm using a plate reader (Thermo Multiskan MK3
spectrophotometer; Thermo Fisher Scientific, Inc.). The optical
density value was determined and used to construct a growth curve
to assess cell proliferation.
Colony formation assay
SKOV-3 and OVCAR-3 transfected cells were seeded in
10-cm dishes (1×103 cells/plate) and treated as follows:
First, the cells were cultured in complete culture medium for ~21
days; subsequently, they were fixed in 4% paraformaldehyde for 30
min at 4°C and stained with Giemsa dye for 20 min at 4°C. Images of
cells were captured using a light microscope (magnification, ×40),
and the number of colonies, consisting of ≥3 cells, was
calculated.
TUNEL assay
The transfected SKOV-3 and OVCAR-3 cells were
cultured for ~48 h at 37°C and 5% CO2. The apoptosis of
the transfected cells was then detected by TUNEL assay as
recommended in the ApopTag® Plus Peroxidase in
Situ Apoptosis kit (Sigma-Aldrich; Merck KGaA). Briefly, the
cells were fixed with 4% paraformaldehyde for 30 min at room
temperature, and permeabilized with 0.1% Triton X-100 in 0.1%
sodium citrate for 2 min at 4°C. Subsequently, the cells were
incubated with 1% TdT enzyme in a humidified atmosphere at 37°C for
90 min. Subsequently, the cells were stained with DAPI
(Sigma-Aldrich; Merck KGaA) at 4°C for 10 min. Finally, the cells
were observed under a fluorescence microscope (magnification,
×200). An average of 10 random fields with 100–200 nuclei per field
was analyzed. The number of TUNEL-positive nuclei (green
fluorescence) was expressed as the percentage of total nuclei (blue
fluorescence).
Flow cytometry
The effects of miR-134-3p on the cell cycle were
measured by flow cytometry as previously described (10). Briefly, the transfected cells were
harvested, washed and suspended in 250 µl of ice-cold PBS.
Subsequently, 750 µl of 100% ice-cold ethanol were added and slowly
mixed with the cell suspensions for ~8 h for fixation on ice.
Subsequently, the cells were washed with PBS and incubated with
RNase (50 µg/ml) and propidium iodide (50 µg/ml, Thermo Fisher
Scientific, Inc.) for 30 min on ice. Finally, the cell cycle was
detected using a Flow Cytometry System (BD Accuri C6; BD
Biosciences) and the relative ratios of G0/G1, S and
G2/M phases were analyzed using FlowJo VX software
(v10.0.7; FlowJo LLC).
Wound healing assay
Wound healing assay was performed to examine the
migration of the cells as previously described (22). Briefly, serum-starved SKOV-3 and
OVCAR-3 cells were seeded in each well of a 6-well plate, and a
wound was created using a 100-µl pipette tip. Following culture for
a further 48 h at 37°C, the wound recovery area was evaluated using
a light microscope (Nikon Corporation; magnification, ×200).
Transwell chamber assay
To explore the migration and invasion of SKOV-3 and
OVCAR-3 cells, the Transwell chamber assay was performed as
follows. The transfected SKOV-3 and OVCAR-3 cells
(3×104) were seeded into the upper chamber of a Matrigel
pre-coated Transwell chamber and cultured in their respective
serum-free medium. Complete culture medium was added to the lower
chamber. Following 48 h of culture at 37°C, the cells in the lower
chamber were fixed with 4% paraformaldehyde for 30 min at 4°C and
stained with 5% crystal violet at 4°C for 30 min. The number of
cells passing through the Matrigel matrix were counted and
photographed under a light microscope (magnification, ×200).
Dual-Luciferase reporter assay
Firstly, the starBase database (http://starbase.sysu.edu.cn/) was used for prediction
of miR-134-3p target genes. R<-0.1 and P<0.05 were set as the
cut-off criteria for identifying the significant miRNA/gene pairs.
One of the identified target genes was FEN1. Subsequently,
FEN1-wild-type (WT) or FEN1-mutant (MUT) reporter plasmids were
synthesized from Shanghai GenePharma Co., Ltd. Briefly, SKOV-3 and
OVCAR-3 cells were co-transfected with 0.24 µg of the FEN1-WT or
FEN1-MUT reporter plasmids together with 40 nM of miR-134-3p mimic
or NC mimic using the cell electroporation system operator H1, as
aforementioned. Additionally, 0.05 µg of Renilla luciferase
expression plasmid (Promega Corporation) was transfected into the
cells as a reference control. The transfected cells were seeded
into 24-well plates for 36 h of culture at 37°C. Finally, firefly
and Renilla luciferase activities in the cells were measured
using the Dual-Luciferase Reporter Assay System (Promega
Corporation). The ratio of firefly and Renilla luciferase
activities was calculated as the relative luciferase activity.
Western blot analysis
Cells were harvested using RIPA lysis buffer, Total
proteins (40 µg/lane) were separated via 12% SDS-PAGE and were
transferred onto PVDF membranes (EMD Millipore) using a MiniGenie
blotting system (Bio-Rad Laboratories, Inc.). The membranes were
then blocked with TBS-Tween (TBST; 0.1% Tween-20) containing 1%
skim milk powder at room temperature for 1 h, and then incubated
with rabbit monoclonal primary antibodies against human p21
(1:1,000; cat. no. 2947S), cyclooxygenase-2 (Cox-2; 1:1,000; cat.
no. 12282T), matrix metalloproteinase (MMP)2 (1:1,000; cat. no.
40994S), MMP9 (1:1,000; cat. no. 13667S), cyclin D1 (1:1,000; cat.
no. 55506S), CDK2 (1:1,000; cat. no. 2546S), Bax (1:1,000; cat. no.
5023S), cleaved caspase-3 (1:1,000; cat. no. 9654S), cleaved
caspase-9 (1:1,000; cat. no. 20750S), Bcl-2 (1:1,000; cat. no.
4223S), β-actin (1:1,000; cat. no. 4970T) and FEN1 (1:2,000; cat.
no. 82354S) (all from Cell Signaling Technology, Inc.) at 4°C
overnight. After washing with TBST, the membranes were incubated
with goat anti-rabbit secondary antibodies (1:10,000; cat. no.
14708S; Cell Signaling Technology, Inc.) for 1 h at room
temperature, followed by visualization with an enhanced
chemiluminescence system (BeyoECL Plus; Beyotime Institute of
Biotechnology). The protein bands were quantified using ImageJ
software (v1.48u; National Institutes of Health).
Statistical analysis
All experiments were performed in triplicate
independently. Quantitative data are presented as the mean ± SD.
GraphPad Prism (v6.01; GraphPad Software, Inc.) was used to perform
the statistical analyses. The unpaired Students t-test was used to
compare differences between two groups, while one-way ANOVA
followed Bonferronis post-hoc test was used when comparing >2
groups. P<0.05 was considered to indicate a statistically
significant difference.
Results
miR-134-3p inhibits the proliferation
of OC cells
To confirm whether miR-134-3p served a role in the
progression of OC, its expression levels were compared between
human ovarian cancer cell lines (SKOV-3, A2780 and OVCAR-3) and a
normal ovarian epithelial cell line (ISOE-80) by RT-qPCR. The
results revealed that the expression levels of miR-134-3p in the
human OC cell lines were significantly lower than those in the
normal ovarian cell line (P<0.01; Fig. 1A). As miR-134-3p expression was
lowest in the SKOV-3 and OVCAR-3 cells, these two cell lines were
selected for further analyses. Subsequently, to evaluate the
effects of miR-134-3p on the progression of OC, SKOV-3 and OVCAR-3
cells were transfected with miR-134-3p mimic or NC mimic. The
RT-qPCR results revealed that miR-134-3p mimic significantly
upregulated miR-134-3p expression in SKOV-3 and OVCAR-3 cells
(P<0.01; Fig. 1B). In addition,
the CCK-8 assay revealed that miR-134-3p mimic significantly
decreased the viability of SKOV-3 and OVCAR-3 cells after 72 h
(P<0.05; Fig. 1C). Colony
formation assay demonstrated that miR-134-3p mimic significantly
decreased the proliferation of SKOV-3 and OVCAR-3 cells (P<0.01;
Fig. 1D). Overall, these data
demonstrated that miR-134-3p inhibited the proliferation of OC
cells.
miR-134-3p facilitates cell apoptosis
and induces cell cycle arrest at the G0/G1
phase in SKOV-3 and OVCAR-3 cells
The regulation of apoptosis and cell cycle arrest
serves an important role in cell proliferation (12,13).
In the present study, to reveal the mechanisms of miR-134-3p in OC
cell proliferation, TUNEL and flow cytometry assays were performed
to examine the effects of miR-134-3p overexpression on cell
apoptosis and the cell cycle. First, the results of the TUNEL assay
indicated that the miR-134-3p mimic significantly facilitated
SKOV-3 and OVCAR-3 cell apoptosis (P<0.01; Fig. 2A). Furthermore, the analysis of cell
apoptosis-associated proteins by western blot analysis revealed
that the miR-134-3p mimic significantly increased the expression
levels of Bax, cleaved caspase-3 and cleaved caspase-9 (P<0.01),
and significantly decreased those of Bcl-2 in SKOV-3 and OVCAR-3
cells (P<0.05; Fig. 2B).
Subsequently, the data from the flow cytometry indicated that the
miR-134-3p mimic significantly increased the percentage of cells at
the G0/G1 phase, while decreasing the
percentage of cells at the S and G2/M phases in both
SKOV-3 and OVCAR-3 cells (P<0.01; Fig. 2C). Finally, western blot analysis
was performed to assess the expression levels of cell
cycle-associated proteins. The results revealed that miR-134-3p
mimic significantly decreased the protein expression levels of
cyclin D1 and CDK2, while increasing the protein expression levels
of p21 (P<0.05 and P<0.01, Fig.
2D). Overall, these results indicated miR-134-3p inhibited cell
proliferation by facilitating cell apoptosis and inducing cell
cycle arrest at the G0/G1 phase in OC.
miR-134-3p inhibits the migration of
SKOV-3 and OVCAR-3 cells
To further investigate the effects of miR-134-3p on
the migration and invasion of OC cells, wound healing and Transwell
assays were performed. The results of the wound healing assay
indicated that the miR-134-3p mimic significantly decreased the
wound closure ability of SKOV-3 and OVCAR-3 cells (P<0.01;
Fig. 3A), suggesting that
miR-134-3p inhibited cell migration. Similarly, Transwell assays
demonstrated that the miR-134-3p mimic significantly inhibited the
migration and invasion of SKOV-3 and OVCAR-3 cells (P<0.01;
Fig. 3B). At the molecular level,
western blot analysis revealed that the miR-134-3p mimic
significantly decreased the expression levels of the
migration-associated proteins MMP2, MMP9 and Cox-2 (P<0.01;
Fig. 3C). Collectively, these
results demonstrated that miR-134-3p decreased the migratory and
invasive abilities of OC cells.
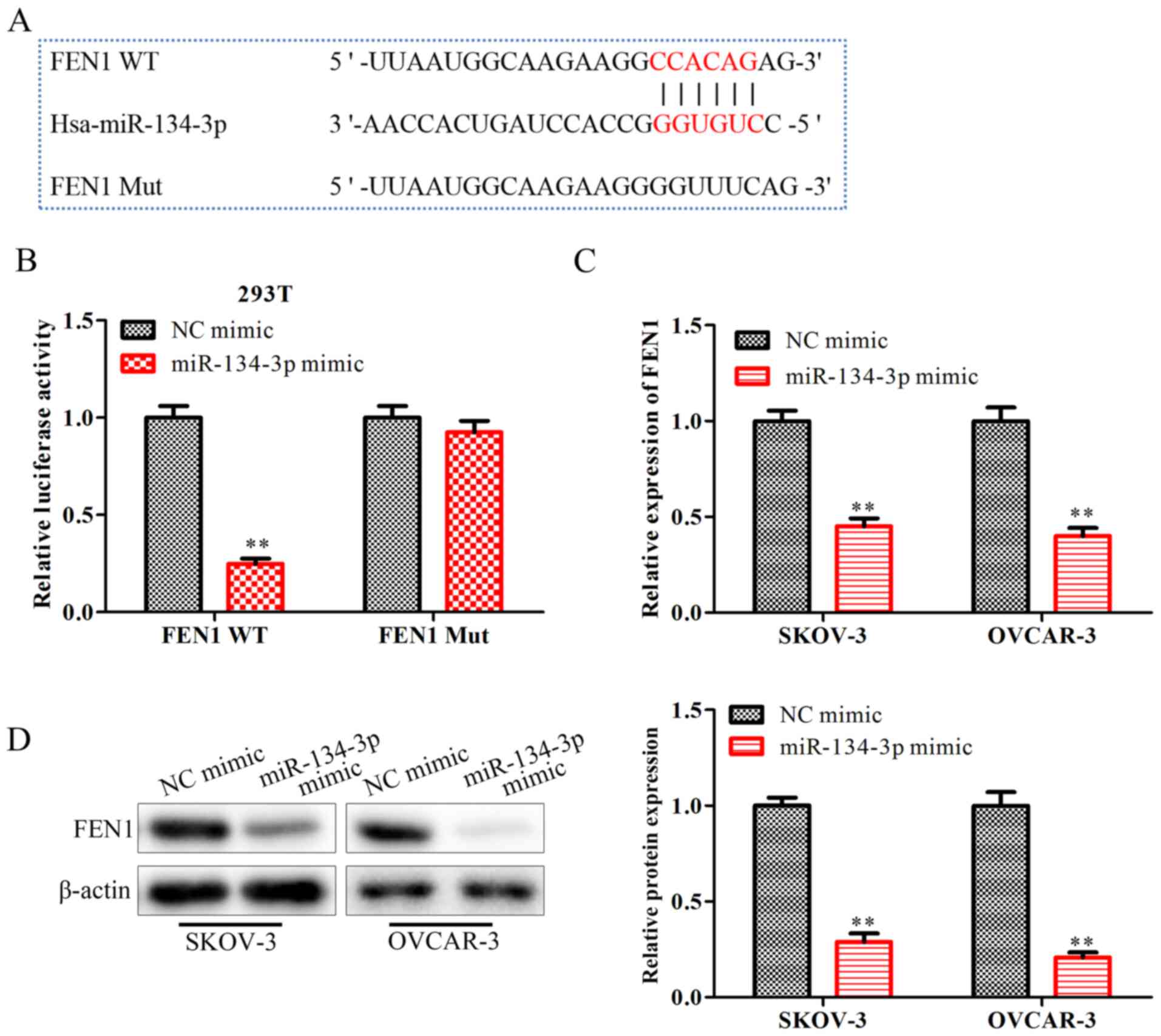
miR-134-3p inhibits FEN1 expression by
directly binding to the 3′-UTR of FEN1
To explore the detailed mechanisms responsible for
the regulatory effects of miR-134-3p on OC progression, starBase
was used to screen for miR-134-3p target genes, revealing that FEN1
may be one of the target genes of miR-134-3p (Fig. 4A). Subsequently, a Dual-Luciferase
reporter assay was performed to verify this interaction. The
results indicated that the miR-134-3p mimic significantly
downregulated the luciferase activity of FEN1-WT, but not that of
FEN1-MUT (P<0.01; Fig. 4B). The
RT-qPCR results (Fig. 4C) and
western blot analysis (Fig. 4D)
demonstrated that the miR-134-3p mimic significantly downregulated
the mRNA and protein expression levels of FEN1 (P<0.01). These
data confirmed that miR-134-3p inhibited the expression levels of
the FEN1 gene by directly binding to the 3′-UTR of FEN1 in OC
cells.
Overexpression of FEN1 reverses the
effects of miR-134-3p on SKOV-3 and OVCAR-3 cells
To confirm the association between miR-134-3p and
FEN1, miR-134-3p mimic or NC mimic were co-transfected with Lv-FEN1
or Lv-NC into SKOV-3 and OVCAR-3 cells. RT-qPCR revealed that
Lv-FEN1 significantly increased the expression levels of FEN1 in
SKOV-3 and OVCAR-3 cells (P<0.01; Fig. 5A). To examine the mediatory effects
of FEN1 on OC cells, a CCK-8 assay was conducted, revealing that
co-transfection with the miR-134-3p mimic + Lv-FEN1 or miR-134-3p
mimic + Lv-NC significantly decreased the viability of SKOV-3 and
OVCAR-3 cells compared with that of the cells co-transfected with
NC mimic + Lv-NC (P<0.05; Fig.
5B). Additionally, the viability of SKOV-3 and OVCAR-3 cells
co-transfected with miR-134-3p mimic + Lv-FEN1 was significantly
higher than that of cells transfected with miR-134-3p mimic + Lv-NC
(Fig. 5B). Colony formation assay
revealed that, compared with Lv-NC, Lv-FEN1 significantly promoted
the colony formation ability of SKOV-3 and OVCAR-3 cells
transfected with miR-134-3p mimic (P<0.05; Fig. 5C). Moreover, TUNEL assay indicated
that Lv-FEN1 significantly inhibited the apoptosis of miR-134-3p
mimic-transfected SKOV-3 and OVCAR-3 cells compared with Lv-NC
(P<0.01; Fig. 5D). Finally,
Transwell assay revealed that Lv-FEN1 significantly increased the
invasion of miR-134-3p mimic-transfected SKOV-3 and OVCAR-3 cells
compared with Lv-NC (P<0.01; Fig.
5E). Overall, these results suggested that the overexpression
of FEN1 partially reverses the effects of miR-134-3p overexpression
on OC.
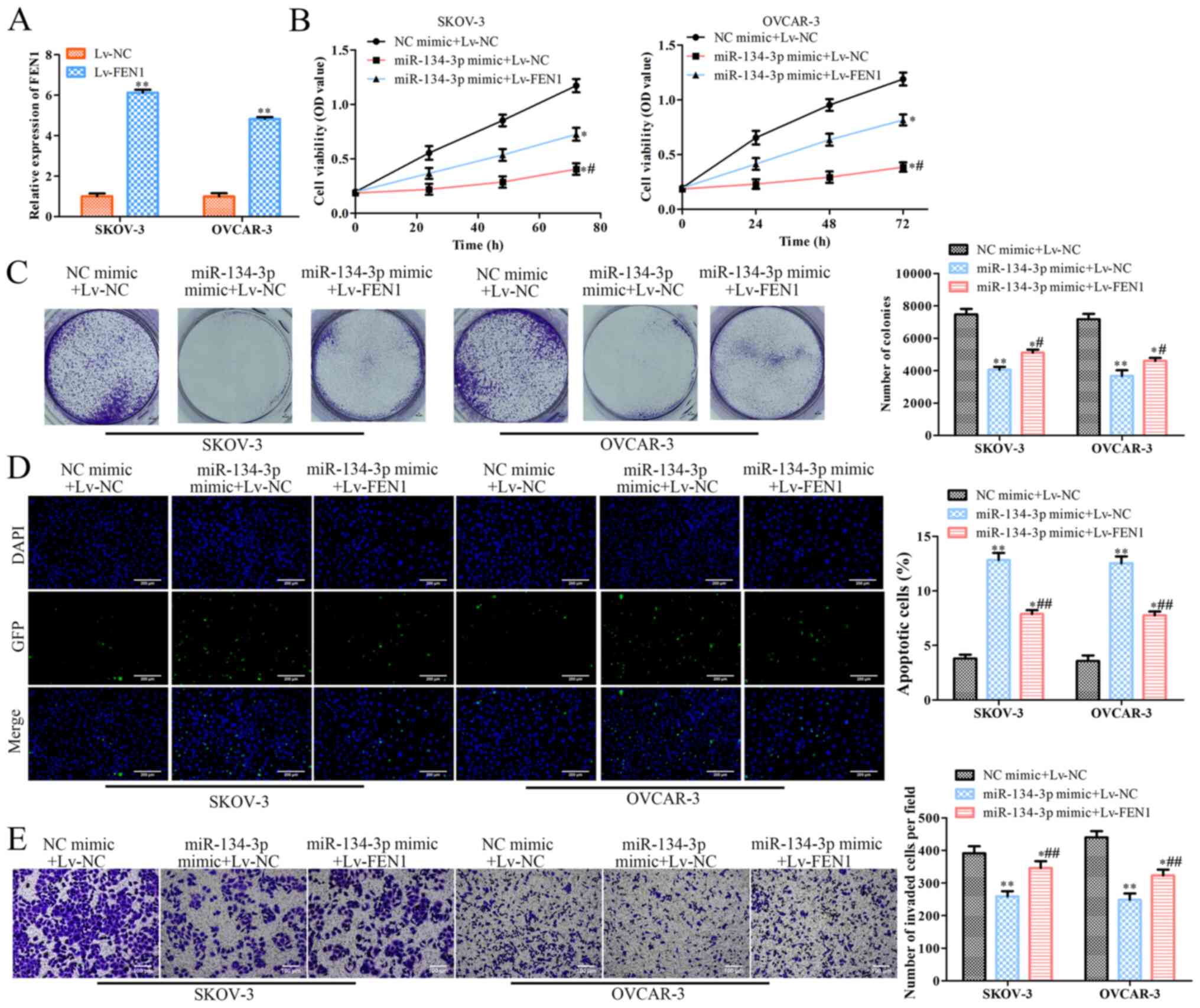 | Figure 5.Overexpression of FEN1 reverses the
effects of miR-134-3p on SKOV-3 and OVCAR-3 cells. SKOV-3 and
OVCAR-3 cells were co-transfected with miR-134-3p mimic or NC mimic
along with Lv-FEN1 or Lv-NC, respectively. (A) Reverse
transcription-quantitative PCR was performed to assess FEN1
expression. (B) Cell Counting Kit-8 was performed to assess cell
viability at 0, 24, 48 and 72 h. (C) Colony formation was performed
to assess colony formation of the cells (magnification, ×40). (D)
TUNEL (scale bar, 200 µm) and (E) Transwell assays (scale bar, 100
µm) were performed to assess the apoptosis and invasion of the
cells, respectively. The data are presented as the mean ± SD (n=3).
*P<0.05 and **P<0.01 vs. NC mimic+Lv-NC.
#P<0.05 and ##P<0.01 vs. miR-134-3p
mimic+Lv-NC. miR, microRNA; NC, negative control; FEN1, flap
structure-specific endonuclease 1; OD, optical density; Lv,
lentiviral expression vector (pcDNA3.1). |
Discussion
This study revealed that the expression levels of
miR-134-3p were decreased in OC cells compared with in normal
ovarian cells. Functional and mechanical experiments further
revealed that miR-134-3p inhibited the proliferation, migration and
invasion of SKOV-3 and OVCAR-3 cells by downregulating FEN1
expression. These preliminary experiments indicated that miR-134-3p
may be a promising candidate for use in OC therapy.
A previous study has demonstrated that miR-134
serves crucial roles in the onset, progression and metastasis of
various types of human cancer, such as lung cancer, glioma, breast
cancer and colorectal cancer, by inhibiting the translation of
target mRNAs (23). Furthermore,
miR-134 has been reported to suppress the migration and invasion of
non-small cell lung cancer by targeting integrin subunit β 1
(24). miR-134 has been identified
to target programmed cell death protein 7 to decrease E-cadherin
expression and enhance oral cancer progression (25). In addition, miR-134 has been
reported to function as a novel potential inhibitor of human OC
cell proliferation and cell cycle progression (26). The present study demonstrated that
upregulating miR-134-3p expression inhibited the proliferation of
SKOV-3 and OVCAR-3 cells by facilitating cell apoptosis and
arresting the cell cycle. The current results are in accordance
with those of a previous study demonstrating that downregulated
miR-134 expression contributes to paclitaxel resistance in human OC
cells (27). Additionally, the
present western blot analysis results indicated that the
overexpression of miR-134-3p decreased the protein expression
levels of cyclin D1, CDK2 and PCNA. The expression levels of Cox-2,
MMP2 and MMP9 were also decreased by the miR-134-3p mimic.
The prediction results by starBase revealed that
FEN1 was the direct target of miR-134-3p. Furthermore, this
interaction was validated by luciferase reporter assays, and it was
demonstrated that miR-134-3p interacted directly with the 3′-UTR of
FEN1. FEN1, also known as DNase IV, is the mammalian counterpart of
the distinct 59 nuclease domain of Escherichia coli DNA polymerase
I (28). FEN1 is directly involved
in DNA replication and repair, interacting with the interdomain
connector loop of PCNA (29). In
cancer progression, FEN1 mediates osteosarcoma cell autophagy
through the disruption of DNA damage repair processes (20). It has been demonstrated that FEN1
mediates the effects of miR-200a and promotes breast cancer cell
proliferation via MET and EGFR signaling (30). Moreover, miR-140 impedes DNA repair
by targeting FEN1 and sensitizes breast cancer cells to the
chemotherapeutic drug doxorubicin (19). It has been reported that FEN1
overexpression is associated with a high grade, high tumor stage
and a poor survival in patients with ovarian epithelial cancer
(31). The results of the present
study indicated that miR-134-3p decreased the expression levels of
FEN1 in SKOV-3 and OVCAR-3 cells. The overexpression of FEN1
reversed the effects of miR-134-3p on the proliferation, migration
and invasion of SKOV-3 and OVCAR-3 cells.
The present study has some limitations. No
comprehensive analysis of miR-134-3p expression in OC and normal
ovarian tissues was performed. The current results only
demonstrated the effects of miR-134-3p in OC cells in vitro.
Future studies should focus on the expression levels of miR-134-3p
in patients with different stages of OC. In addition, the effects
of miR-134-3p/FEN1 in OC in vivo should be further
investigated in future research.
In conclusion, the present study suggested that
miR-134-3p may modulate FEN1 expression to inhibit the progression
of OC. The present study provided the molecular mechanisms for
understanding the role of miR-134-3p in the regulation of the
proliferation and metastasis of OC.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Central South
University Post-Graduate Independent Exploration and Innovation
Project (grant no. 2017zzts382), the Central South University
Graduate excellent course (grant no. 2014jpkc003), the Hunan
Provincial Natural Science Foundation of China (grant no.
2015JJ2165) and the Central South University Fundamental Research
Funds Special Funding (grant no. 165611031).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors contributions
YY and MZ conceived and designed the study. MZ, HJ
and QF performed the experiments. QC and YZ analyzed the data. MZ
drafted the manuscript. YY and YZ edited and revised the
manuscript. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar
|
|
2
|
Torre LA, Trabert B, DeSantis CE, Miller
KD, Samimi G, Runowicz CD, Gaudet MM, Jemal A and Siegel RL:
Ovarian cancer statistics, 2018. CA Cancer J Clin. 68:284–296.
2018. View Article : Google Scholar
|
|
3
|
Zhang ML, Peng P, Wu CX, Gong YM, Zhang
SW, Chen WQ and Bao PP: [Report of breast cancer incidence and
mortality in China registry regions, 2008–2012]. Zhonghua Zhong Liu
Za Zhi. 41:315–320. 2019.(In Chinese).
|
|
4
|
Marth C, Reimer D and Zeimet AG:
Front-line therapy of advanced epithelial ovarian cancer: standard
treatment. Ann Oncol. 28 (Suppl 8):pp. viii36–viii39. 2019,
simplehttps://doi.org/10.1093/annonc/mdx450 View Article : Google Scholar
|
|
5
|
Bieg D, Sypniewski D, Nowak E and Bednarek
I: MiR-424-3p suppresses galectin-3 expression and sensitizes
ovarian cancer cells to cisplatin. Arch Gynecol Obstet.
299:1077–1087. 2019. View Article : Google Scholar
|
|
6
|
Yang B, Sun L and Liang L: MiRNA-802
suppresses proliferation and migration of epithelial ovarian cancer
cells by targeting YWHAZ. J Ovarian Res. 12:1002019. View Article : Google Scholar
|
|
7
|
Ding Y, Fang Q, Li Y and Wang Y:
Amplification of lncRNA PVT1 promotes ovarian cancer proliferation
by binding to miR-140. Mamm Genome. 30:217–225. 2019. View Article : Google Scholar
|
|
8
|
Fan B, Chen LP, Yuan YH, Xiao HN, Lv XS
and Xia ZY: MiR-15a-3p suppresses the growth and metastasis of
ovarian cancer cell by targeting Twist1. Eur Rev Med Pharmacol Sci.
23:1934–1946. 2019.
|
|
9
|
Zuo Y, Zheng W, Liu J, Tang Q, Wang SS and
Yang XS: MiR-34a-5p/PD-L1 axis regulates cisplatin chemoresistance
of ovarian cancer cells. Neoplasma. 67:93–101. 2020. View Article : Google Scholar
|
|
10
|
Zhang X, Xin G and Sun D: Serum exosomal
miR-328, miR-575, miR-134 and miR-671-5p as potential biomarkers
for the diagnosis of Kawasaki disease and the prediction of
therapeutic outcomes of intravenous immunoglobulin therapy. Exp
Ther Med. 16:2420–2432. 2018.
|
|
11
|
Chen CL, Zhang L, Jiao YR, Zhou Y, Ge QF,
Li PC, Sun XJ and Lv Z: miR-134 inhibits osteosarcoma cell invasion
and metastasis through targeting MMP1 and MMP3 in vitro and in
vivo. FEBS Lett. 593:1089–1101. 2019. View Article : Google Scholar
|
|
12
|
Qin Q, Wei F, Zhang J, Wang X and Li B:
miR-134 inhibits non-small cell lung cancer growth by targeting the
epidermal growth factor receptor. J Cell Mol Med. 20:1974–1983.
2016. View Article : Google Scholar
|
|
13
|
Qi A, Han J, Jia F and Liu C: miR-3175 and
miR-134 affect proliferation, invasion and apoptosis of glioma
cells through PI3K/AKT signaling pathway. J BUON. 24:2465–2474.
2019.
|
|
14
|
Ma Z, Li K, Chen P, Pan Q, Li X and Zhao
G: MiR-134, mediated by IRF1, suppresses tumorigenesis and
progression by targeting VEGFA and MYCN in osteosarcoma. Anticancer
Agents Med Chem. 20:1197–1208. 2020. View Article : Google Scholar
|
|
15
|
Greene AL, Snipe JR, Gordenin DA and
Resnick MA: Functional analysis of human FEN1 in Saccharomyces
cerevisiae and its role in genome stability. Hum Mol Genet.
8:2263–2273. 1999. View Article : Google Scholar
|
|
16
|
Gary R, Park MS, Nolan JP, Cornelius HL,
Kozyreva OG, Tran HT, Lobachev KS, Resnick MA and Gordenin DA: A
novel role in DNA metabolism for the binding of Fen1/Rad27 to PCNA
and implications for genetic risk. Mol Cell Biol. 19:5373–5382.
1999. View Article : Google Scholar
|
|
17
|
Gomes XV and Burgers PM: Two modes of FEN1
binding to PCNA regulated by DNA. EMBO J. 19:3811–3821. 2000.
View Article : Google Scholar
|
|
18
|
Nikolova T, Christmann M and Kaina B: FEN1
is overexpressed in testis, lung and brain tumors. Anticancer Res.
29:2453–2459. 2009.
|
|
19
|
Lu X, Liu R, Wang M, Kumar AK, Pan F, He
L, Hu Z and Guo Z: MicroRNA-140 impedes DNA repair by targeting
FEN1 and enhances chemotherapeutic response in breast cancer.
Oncogene. 39:234–247. 2020. View Article : Google Scholar
|
|
20
|
20. Dong S, Xiao Y, Ma X, He W, Kang J,
Peng Z, Wang L and Li Z: miR-193b Increases the chemosensitivity of
osteosarcoma cells by promoting FEN1-mediated autophagy.
OncoTargets Ther. 12:10089–10098. 2019. View Article : Google Scholar
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
22
|
Zhang YY, Li P, Zhu MZ, Guo Y and Yang J:
LINC01308 accelerates the malignant progression of ovarian cancer
by binding to miRNA-506. Eur Rev Med Pharmacol Sci. 23:3253–3260.
2019.
|
|
23
|
Pan JY, Zhang F, Sun CC, Li SJ, Li G, Gong
FY, Bo T, He J, Hua RX, Hu WD, et al: miR-134: A human cancer
suppressor? Mol Ther Nucleic Acids. 6:140–149. 2017. View Article : Google Scholar
|
|
24
|
Qin Q, Wei F, Zhang J and Li B: miR-134
suppresses the migration and invasion of non-small cell lung cancer
by targeting ITGB1. Oncol Rep. 37:823–830. 2017. View Article : Google Scholar
|
|
25
|
Peng SY, Tu HF, Yang CC, Wu CH, Liu CJ,
Chang KW and Lin SC: miR-134 targets PDCD7 to reduce E-cadherin
expression and enhance oral cancer progression. Int J Cancer.
143:2892–2904. 2018. View Article : Google Scholar
|
|
26
|
Chang C, Liu T, Huang Y, Qin W, Yang H and
Chen J: MicroRNA-134-3p is a novel potential inhibitor of human
ovarian cancer stem cells by targeting RAB27A. Gene. 605:99–107.
2017. View Article : Google Scholar
|
|
27
|
Shuang T, Wang M, Shi C, Zhou Y and Wang
D: Down-regulated expression of miR-134 contributes to paclitaxel
resistance in human ovarian cancer cells. FEBS Lett. 589 (20 Pt
B):3154–3164. 2015. View Article : Google Scholar
|
|
28
|
Prasad R, Dianov GL, Bohr VA and Wilson
SH: FEN1 stimulation of DNA polymerase beta mediates an excision
step in mammalian long patch base excision repair. J Biol Chem.
275:4460–4466. 2000. View Article : Google Scholar
|
|
29
|
Storici F, Henneke G, Ferrari E, Gordenin
DA, Hübscher U and Resnick MA: The flexible loop of human FEN1
endonuclease is required for flap cleavage during DNA replication
and repair. EMBO J. 21:5930–5942. 2002. View Article : Google Scholar
|
|
30
|
Zeng X, Qu X, Zhao C, Xu L, Hou K, Liu Y,
Zhang N, Feng J, Shi S, Zhang L, et al: FEN1 mediates miR-200a
methylation and promotes breast cancer cell growth via MET and EGFR
signaling. FASEB J. 33:10717–10730. 2019. View Article : Google Scholar
|
|
31
|
Abdel-Fatah TM, Russell R, Albarakati N,
Maloney DJ, Dorjsuren D, Rueda OM, Moseley P, Mohan V, Sun H,
Abbotts R, et al: Genomic and protein expression analysis reveals
flap endonuclease 1 (FEN1) as a key biomarker in breast and ovarian
cancer. Mol Oncol. 8:1326–1338. 2014. View Article : Google Scholar
|