Oral cancer is a type of head and neck cancer and
its main subtype is oral squamous cell carcinoma (OSCC) (1). OSCC occurs in the oral cavity, which
includes the tongue, floor of the mouth, buccal mucosa and alveolar
rim (2). OSCC presents a major
global health concern, with an estimated >300,000 cases per
annum (3) and ~1.8 million deaths
(4). Currently, the diagnosis and
treatment of OSCC represent a clinical challenge.
miRNAs are a class of small non-coding RNAs that are
involved in the regulation of a variety of physiological processes
by targeting specific mRNAs (5).
miRNAs have been reported to play an important regulatory role in
cancer occurrence and progression (6), and they may be of value as targets in
oral cancer treatment (7). Due to
their stability in human peripheral blood and body fluids and
disease-specific expression, an increasing number of studies
indicate that miRNAs may represent an ideal set of biomarkers
applied in early diagnosis and prognosis of cancers (8).
The focus of the present review was the detailed
regulatory role of miRNAs in oral cancer and their value in
diagnosis and treatment. The conclusions of this review may
contribute to the early diagnosis and targeted therapy of oral
cancer.
miRNAs are a family of short, single-stranded, small
non-coding RNAs, containing ~20–22 nucleotides (9). The miRNA expression profiles and
levels differ between patients with cancer and healthy individuals,
and they are implicated in human carcinogenesis (10,11).
According to their chemical and structural properties, circulating
miRNAs are stable in the serum, plasma and other body fluids
(8), and they may be considered as
potential clinical diagnostic and prognostic biomarkers.
The occurrence of oral cancer is multifactorial, and
is accompanied by genetic and epigenetic instability (12). With the widespread application of
next-generation sequencing, a growing number of studies have
demonstrated that certain miRNAs are differentially expressed in
oral cancer. In addition, the results of receiver operating
characteristic (ROC) curve and area under the curve (AUC) analysis
have indicated that the differentially expressed miRNAs may help
distinguish patients with oral cancer from healthy subjects
(13,14). As revealed in Table I (13–31),
35 miRNAs were screened, which were reported in the last 5 years
with a ROC (AUC) >0.500. For example, Momen-Heravi et al
reported that upregulated miR-27b and downregulated miR-136 derived
from the saliva were able to differentiate between patients with
oral cancer and healthy subjects (15). The ROC (AUC) of miR-21 extracted
from oral cytology and miR-99a from the serum were up to 0.910
(16) and 0.911 (17), respectively. Gombos et al
reported that miR-155 derived from tissues was able to distinguish
between patients with oral cancer and healthy individuals [ROC
(AUC)=0.925] (13). These data
revealed that miRNAs originating from different samples may be
considered as biomarkers for oral cancer diagnosis.
The expression profiles of certain miRNAs have
demonstrated a positive correlation with clinical stage, metastasis
and patient survival, indicating that these miRNAs may be
considered as prognostic indices in oral cancer. Lai et al
revealed that dysregulated miR-31-5p expression enhanced OSCC cell
migration and invasion and accelerated oral cancer progression
(32). In recent years, miRNAs have
been reported to be strongly correlated with the survival of oral
cancer patients. Chen et al indicated that patients with
high expression level of miR-99a displayed a better prognosis and
longer overall survival (17).
Supic et al revealed that patients with miR-183
overexpression had markedly shorter overall survival and higher
risk of poor outcome (23). Zheng
et al reported that miR-503-5p, miR-450b-5p, miR-27a-3p,
miR-181a-5p and miR-183-5p were overexpressed in patients with oral
cancer, and they were all highly associated with cancer cell
proliferation, advanced clinical stage and poor prognosis (33). Cheng et al revealed that the
expression level of miR-455-5p was associated with the nodal
status, stage and overall survival of the patients, suggesting that
miR-455-5p may be a promising prognostic marker for predicting the
outcome of patients with oral cancer (34). We herein also summarized other
miRNAs that may be of prognostic value in oral cancer (Table II). All the aforementioned data
indicated that miRNAs may be valuable prognostic indicators in oral
cancer.
miRNAs play a key role in regulating the translation
or degradation of mRNAs by interacting with the 3′ untranslated
region (3′UTR) or coding region of mRNAs and regulating the
expression level of their target genes (35). miRNAs are involved in the cellular
processes of cancer, such as inflammation, proliferation, stress
response, growth, apoptosis, survival and migration (10). Therefore, manipulating miRNA
expression in cancer has been attracting increasing attention as a
novel therapeutic strategy. It was reported that miR-24-3p,
miR-155-5p and miRNA-10a may significantly promote the
proliferation of oral cancer cells (18,36,37).
These findings suggested that silencing the expression of specific
miRNAs may prevent the progression of oral cancer. Conversely,
numerous miRNAs have been revealed to have an anticancer function.
miR-6887-5p, miR-34a-5p and miR-142-3p markedly suppressed the
proliferation of oral cancer cells (38–40).
In addition, certain miRNAs, such as miR-204-5p and miR-34a-5p,
exert their anticancer effects by inhibiting the aggressiveness and
metastasis of oral cancer cells (39,41).
The targets and functional roles of miRNAs are listed in Table II (17–19,23,32,33,36–54).
miRNAs belong to a family of non-coding RNAs that
play key roles in suppressing or promoting cancer by interacting
with their target mRNAs. Several studies have demonstrated that
miRNAs are involved in the occurrence, progression and metastasis
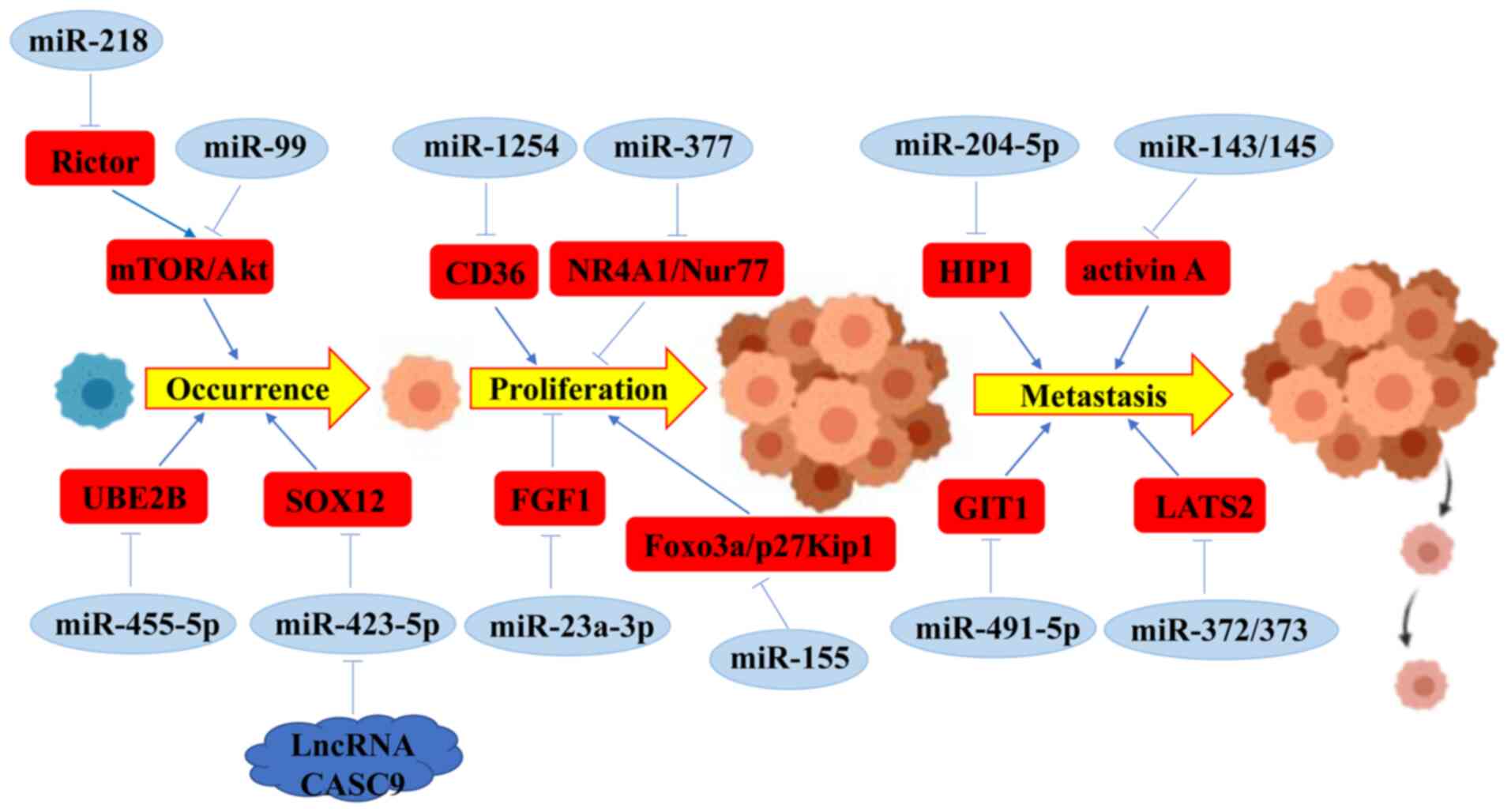
of oral cancer (Fig. 1) (53,55).
Accumulating evidence indicates that miRNAs play a
key role in the occurrence and progression of oral cancer. The
protein kinase B (AKT) and mammalian target of rapamycin (mTOR)
pathways are known to participate in the regulation of oral cancer
occurrence (56). Manikandan et
al revealed that >40 differentially expressed miRNAs in OSCC
may activate the AKT pathway (57).
miR-218 can inhibit the activation of mTOR/AKT pathway by targeting
Rictor, and then suppress oral carcinogenesis (58). In oral cancer, miR-99 has been
revealed to decreased the expression level of mTOR by directly
binding with mTOR mRNA, thereby promoting cancer cell growth and
increasing tumor size (59,60). The expression level of miR-455-5p
has been revealed to be regulated by the TGF-β-dependent pathway,
which subsequently promotes oral tumorigenesis by downregulating
ubiquitin-conjugating enzyme E2B (UBE2B) (34). Chen et al revealed that CD36
contributed to the proliferation and invasion of OSCC, and miR-1254
may inhibit the progression of OSCC by partially downregulating the
expression level of CD36 (61). In
addition, inhibiting the expression of miR-423-5p may rescue the
carcinogenic effect of lncRNA CASC9 by silencing the expression of
SRY-box transcription factor 12 (SOX12) (62). These data revealed that miRNAs
participate in oral carcinogenesis and progression by regulating
their target genes.
The proliferation and apoptosis of oral cancer cells
are regulated by multiple factors, with an increasing number of
studies demonstrating that miRNAs are involved in the regulation of
these cellular process. Rastogi et al reported that in
vitro restoration of miR-377 suppressed OSCC cell growth and
induced apoptosis by regulating HDAC9 and its pro-apoptotic target,
NR4A1/Nur77 (63). miR-23a-3p may
suppress proliferation and promote apoptosis of OSCC cells by
targeting fibroblast growth factor 2 (FGF1) (64). By contrast, it was reported that
miR-155 contributed to oral cancer cell proliferation, inhibited
cell apoptosis and reduced the sensitivity of oral cancer cells to
DDP by downregulating Foxo3a expression (65). Furthermore, miR-155 promoted cell
cycle progression and cell proliferation and suppressed apoptosis
by inhibiting p27Kip1 expression (66).
The migration, invasion and metastasis of oral
cancer cells are highly associated with the therapeutic strategy.
miRNAs are involved in the regulation of these processes in oral
cancer cells. Fang et al reported that miR-204-5p suppressed
oral cancer cell aggressiveness, viability and migration by
targeting and inhibiting: Huntingtin-interacting protein 1 (HIP1)
expression (41). Overexpression of
miR-143 and miR-145 in OSCC cells markedly suppressed the
expression of activin A, and suppressed the migration and invasion
of oral cancer cells, prevented lymph node metastasis, increased
tumor differentiation and prolonged the survival of the patients
(67). Huang et al revealed
that enhancing the expression of miR-491-5p significantly
downregulated the expression of GPCR kinase 2 interacting protein 1
(GIT1), thereby suppressing OSCC cell migration in vitro and
lung metastasis in vivo (68). Conversely, overexpression of miR-21
was associated with perineural invasion and worse prognosis in OSCC
patients (69). Tu et al
reported that overexpression of miR-372 and miR-373 were associated
with nodal metastasis, lymph vascular invasion and poor survival by
regulating the expression of large tumor suppressor kinase 2
(LATS2) in OSCC cells (70).
A large volume of evidence has demonstrated that
miRNAs play key roles in oral cancer occurrence and growth, cancer
cell migration and invasion, cancer progression and patient
prognosis. This evidence suggests that miRNAs may be considered as
novel therapeutic tools for oral cancer.
It was reported that upregulation of miR-375
markedly inhibited cell proliferation, induced cell cycle arrest in
the G0/G1 phase, promoted apoptosis and increased radiosensitivity
in OSCC cells, suggesting that miR-375 may be a potential
therapeutic target for OSCC patients (71). miR-494-3p was able to enhance the
radiosensitivity of OSCC cells by promoting cellular senescence
(72). Min et al revealed
that overexpression of miR-148a in cancer-associated fibroblasts
significantly decreased the migration and invasion abilities of
oral cancer cells by directly targeting WNT10B, suggesting that
miR-148a may be a novel promising target for the treatment of OSCC
(73). Chen et al reported
that miR-1254 may inhibit the progression of OSCC, and restoring
miR-1254 expression may represent an effective treatment strategy
for OSCC (61). Similarly, miR-377
and miR-23a-3p suppressed cell proliferation and promoted apoptosis
in OSCC, suggesting that both miR-377 and miR-23a-3p may prove to
be of value as therapeutic targets for OSCC in the future (63,64).
In summary, all the aforementioned data suggest that miRNAs may be
considered as effective anticancer targets, and they may be used to
develop novel treatment strategies for oral cancer.
Although great progress has been made in the
diagnosis of oral cancer, the methods for early diagnosis and
prognosis require further improvements. Early diagnosis may
markedly increase the effectiveness of treatment and prolong the
survival of patients with oral cancer. Therefore, it is urgent to
develop novel biomarkers with higher accuracy, sensitivity and
specificity, that are more convenient for clinical detection.
With the development of sequencing technology, an
increasing number of miRNAs have been revealed to be specifically
expressed in a variety of samples from oral cancer (15,16).
These miRNAs also play a key role in regulating cellular processes
and behaviors (66,68). miRNAs may represent optimal
biomarkers and new therapeutic tools for oral cancer. However,
although the miRNAs aforementioned appear to be promising
candidates as biomarkers, they require further investigation,
including biological study and clinical verification. Furthermore,
the source, formation and regulatory networks of miRNAs are quite
complex, and the expression level of miRNAs in different stages of
oral cancer is also different. Hence, it is necessary to elucidate
the mechanism of miRNA formation, which may contribute to early
diagnosis, targeted therapy and prognosis evaluation of oral cancer
patients. Notably, the therapeutic miRNAs that may be used to
develop novel targeted drugs require further research in terms of
suitable and effective in vivo delivery methods. Identifying
key targets and building a targeted delivery system (such as a
nano-miRNA system) will be our future research priorities.
Not applicable.
The present study was supported by the National
Science Foundation of China (grant no. 81372908).
All data generated or analyzed during this study are
included in this published article.
JW, RY, JY designed the review and edited the
manuscript. JW, NL, XL wrote the manuscript. JW, RY and ZC
collected and analyzed data. All authors read and approved the
final manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Santoro A, Pannone G, Papagerakis S,
Serpico R and Bufo P: Epigenetic profiling of oral cancer. InTech.
Mar 14–2012.(Epub ahead of print). doi: 10.5772/31386.
|
|
2
|
Tsantoulis PK, Kastrinakis NG, Tourvas AD,
Laskaris G and Gorgoulis VG: Advances in the biology of oral
cancer. Oral Oncol. 43:523–534. 2007. View Article : Google Scholar
|
|
3
|
Gupta B, Johnson NW and Kumar N: Global
epidemiology of head and neck cancers: A continuing challenge.
Oncology. 91:13–23. 2016. View Article : Google Scholar
|
|
4
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar
|
|
5
|
Huppi K, Volfovsky N, Mackiewicz M,
Runfola T, Jones TL, Martin SE, Stephens R and Caplen NJ: MicroRNAs
and genomic instability. Semin Cancer Biol. 17:65–73. 2007.
View Article : Google Scholar
|
|
6
|
Mitchell PS, Parkin RK, Kroh EM, Fritz BR,
Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant
KC, Allen A, et al: Circulating microRNAs as stable blood-based
markers for cancer detection. Proc Natl Acad Sci USA.
105:10513–10518. 2008. View Article : Google Scholar
|
|
7
|
Chawla JP, Iyer N, Soodan KS, Sharma A,
Khurana SK and Priyadarshni P: Role of miRNA in cancer diagnosis,
prognosis, therapy and regulation of its expression by Epstein-Barr
virus and human papillomaviruses: With special reference to oral
cancer. Oral Oncol. 51:731–737. 2015. View Article : Google Scholar
|
|
8
|
Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K,
Guo J, Zhang Y, Chen J, Guo X, et al: Characterization of microRNAs
in serum: A novel class of biomarkers for diagnosis of cancer and
other diseases. Cell Res. 18:997–1006. 2008. View Article : Google Scholar
|
|
9
|
Croce CM and Calin GA: miRNAs, cancer, and
stem cell division. Cell. 122:6–7. 2005. View Article : Google Scholar
|
|
10
|
Di Leva G, Garofalo M and Croce CM:
MicroRNAs in cancer. Annu Rev Pathol. 9:287–314. 2014. View Article : Google Scholar
|
|
11
|
Fabbri M: MicroRNAs and cancer: Towards a
personalized medicine. Curr Mol Med. 13:751–756. 2013. View Article : Google Scholar
|
|
12
|
Madhura MG, Rao RS, Patil S, Fageeh HN,
Alhazmi A and Awan KH: Advanced diagnostic aids for oral cancer.
Dis Mon. Jun 25–2020.(Epub ahead of print). doi:
10.1016/j.disamonth.2020.101034. View Article : Google Scholar
|
|
13
|
Gombos K, Horváth R, Szele E, Juhász K,
Gocze K, Somlai K, Pajkos G, Ember I and Olasz L: miRNA expression
profiles of oral squamous cell carcinomas. Anticancer Res.
33:1511–1517. 2013.
|
|
14
|
Maclellan SA, Lawson J, Baik J, Guillaud
M, Poh CF and Garnis C: Differential expression of miRNAs in the
serum of patients with high-risk oral lesions. Cancer Med.
1:268–274. 2012. View
Article : Google Scholar
|
|
15
|
Momen-Heravi F, Trachtenberg AJ, Kuo WP
and Cheng YS: Genomewide study of salivary MicroRNAs for detection
of oral cancer. J Dent Res. 93 (Suppl 7):86S–93S. 2014. View Article : Google Scholar
|
|
16
|
He Q, Chen Z, Cabay RJ, Zhang L, Luan X,
Chen D, Yu T, Wang A and Zhou X: microRNA-21 and microRNA-375 from
oral cytology as biomarkers for oral tongue cancer detection. Oral
Oncol. 57:15–20. 2016. View Article : Google Scholar
|
|
17
|
Chen L, Hu J, Pan L, Yin X, Wang Q and
Chen H: Diagnostic and prognostic value of serum miR-99a expression
in oral squamous cell carcinoma. Cancer Biomark. 23:333–339. 2018.
View Article : Google Scholar
|
|
18
|
He L, Ping F, Fan Z, Zhang C, Deng M,
Cheng B and Xia J: Salivary exosomal miR-24-3p serves as a
potential detective biomarker for oral squamous cell carcinoma
screening. Biomed Pharmacother. 121:1095532020. View Article : Google Scholar
|
|
19
|
Lu Z, He Q, Liang J, Li W, Su Q, Chen Z,
Wan Q, Zhou X, Cao L, Sun J, et al: miR-31-5p is a potential
circulating biomarker and therapeutic target for oral cancer. Mol
Ther Nucleic Acids. 16:471–480. 2019. View Article : Google Scholar
|
|
20
|
Gai C, Camussi F, Broccoletti R, Gambino
A, Cabras M, Molinaro L, Carossa S, Camussi G and Arduino PG:
Salivary extracellular vesicle-associated miRNAs as potential
biomarkers in oral squamous cell carcinoma. BMC Cancer. 18:4392018.
View Article : Google Scholar
|
|
21
|
Chang YA, Weng SL, Yang SF, Chou CH, Huang
WC, Tu SJ, Chang TH, Huang CN, Jong YJ and Huang HD: A
Three-MicroRNA signature as a potential biomarker for the early
detection of oral cancer. Int J Mol Sci. 19:7582018. View Article : Google Scholar
|
|
22
|
Berania I, Cardin GB, Clément I, Guertin
L, Ayad T, Bissada E, Nguyen-Tan PF, Filion E, Guilmette J, Gologan
O, et al: Four PTEN-targeting co-expressed miRNAs and
ACTN4-targeting miR-548b are independent prognostic biomarkers in
human squamous cell carcinoma of the oral tongue. Int J Cancer.
141:2318–2328. 2017. View Article : Google Scholar
|
|
23
|
Supic G, Zeljic K, Rankov AD, Kozomara R,
Nikolic A, Radojkovic D and Magic Z: miR-183 and miR-21 expression
as biomarkers of progression and survival in tongue carcinoma
patients. Clin Oral Investig. 22:401–409. 2018. View Article : Google Scholar
|
|
24
|
Xu H, Yang Y, Zhao H, Yang X, Luo Y, Ren
Y, Liu W and Li N: Serum miR-483-5p: A novel diagnostic and
prognostic biomarker for patients with oral squamous cell
carcinoma. Tumour Biol. 37:447–453. 2016. View Article : Google Scholar
|
|
25
|
Gu WL, Ye DX and Wu JJ: Expression and
clinical significance of plasma microRNA-125b level in patients
with oral squamous cell carcinoma. Shanghai Kou Qiang Yi Xue.
24:71–75. 2015.(In Chinese).
|
|
26
|
Zahran F, Ghalwash D, Shaker O, Al-Johani
K and Scully C: Salivary microRNAs in oral cancer. Oral Dis.
21:739–747. 2015. View Article : Google Scholar
|
|
27
|
Lu YC, Chang JT, Huang YC, Huang CC, Chen
WH, Lee LY, Huang BS, Chen YJ, Li HF and Cheng AJ: Combined
determination of circulating miR-196a and miR-196b levels produces
high sensitivity and specificity for early detection of oral
cancer. Clin Biochem. 48:115–121. 2015. View Article : Google Scholar
|
|
28
|
Ries J, Vairaktaris E, Kintopp R, Baran C,
Neukam FW and Nkenke E: Alterations in miRNA expression patterns in
whole blood of OSCC patients. In Vivo. 28:851–861. 2014.
|
|
29
|
Ries J, Baran C, Wehrhan F, Weber M, Motel
C, Kesting M and Nkenke E: The altered expression levels of
miR-186, miR-494 and miR-3651 in OSCC tissue vary from those of the
whole blood of OSCC patients. Cancer Biomark. 24:19–30. 2019.
View Article : Google Scholar
|
|
30
|
Wu N, Lu Y and Liang JZ: Expression and
correlation of survivin and hsa-miR-542-3p in patients with oral
squamous cell carcinoma. Shanghai Kou Qiang Yi Xue. 25:720–724.
2016.(In Chinese).
|
|
31
|
Duz MB, Karatas OF, Guzel E, Turgut NF,
Yilmaz M, Creighton CJ and Ozen M: Identification of miR-139-5p as
a saliva biomarker for tongue squamous cell carcinoma: A pilot
study. Cell Oncol (Dordr). 39:187–193. 2016. View Article : Google Scholar
|
|
32
|
Lai YH, Liu H, Chiang WF, Chen TW, Chu LJ,
Yu JS, Chen SJ, Chen HC and Tan BC: MiR-31-5p-ACOX1 Axis enhances
tumorigenic fitness in oral squamous cell carcinoma via the
promigratory prostaglandin E2. Theranostics. 8:486–504. 2018.
View Article : Google Scholar
|
|
33
|
Zheng X, Wu K, Liao S, Pan Y, Sun Y, Chen
X, Zhang Y, Xia S, Hu Y and Zhang J: MicroRNA-transcription factor
network analysis reveals miRNAs cooperatively suppress RORA in oral
squamous cell carcinoma. Oncogenesis. 7:792018. View Article : Google Scholar
|
|
34
|
Cheng CM, Shiah SG, Huang CC, Hsiao JR and
Chang JY: Up-regulation of miR-455-5p by the TGF-β-SMAD signalling
axis promotes the proliferation of oral squamous cancer cells by
targeting UBE2B. J Pathol. 240:38–49. 2016. View Article : Google Scholar
|
|
35
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
Elife. 4:e050052015. View Article : Google Scholar
|
|
36
|
Wu M, Duan Q, Liu X, Zhang P, Fu Y, Zhang
Z, Liu L, Cheng J and Jiang H: MiR-155-5p promotes oral cancer
progression by targeting chromatin remodeling gene ARID2. Biomed
Pharmacother. 122:1096962020. View Article : Google Scholar
|
|
37
|
Chen YH, Song Y, Yu YL, Cheng W and Tong
X: miRNA-10a promotes cancer cell proliferation in oral squamous
cell carcinoma by upregulating GLUT1 and promoting glucose
metabolism. Oncol Lett. 17:5441–5446. 2019.
|
|
38
|
Higaki M, Shintani T, Hamada A, Rosli SNZ
and Okamoto T: Eldecalcitol (ED-71)-induced exosomal miR-6887-5p
suppresses squamous cell carcinoma cell growth by targeting
heparin-binding protein 17/fibroblast growth factor-binding
protein-1 (HBp17/FGFBP-1). In vitro cellular & developmental
biology. In Vitro Cell Dev Biol Anim. 56:222–233. 2020. View Article : Google Scholar
|
|
39
|
Li YY, Tao YW, Gao S, Li P, Zheng JM,
Zhang SE, Liang J and Zhang Y: Cancer-associated fibroblasts
contribute to oral cancer cells proliferation and metastasis via
exosome-mediated paracrine miR-34a-5p. EBioMedicine. 36:209–220.
2018. View Article : Google Scholar
|
|
40
|
Dickman CT, Lawson J, Jabalee J, MacLellan
SA, LePard NE, Bennewith KL and Garnis C: Selective extracellular
vesicle exclusion of miR-142-3p by oral cancer cells promotes both
internal and extracellular malignant phenotypes. Oncotarget.
8:15252–15266. 2017. View Article : Google Scholar
|
|
41
|
Fang X, Tang Z, Zhang H and Quan H: Long
non-coding RNA DNM3OS/miR-204-5p/HIP1 axis modulates oral cancer
cell viability and migration. J Oral Pathol Med. 49:865–875. 2020.
View Article : Google Scholar
|
|
42
|
Jakob M, Mattes LM, Küffer S, Unger K,
Hess J, Bertlich M, Haubner F, Ihler F, Canis M, Weiss BG and Kitz
J: MicroRNA expression patterns in oral squamous cell carcinoma:
hsa-mir-99b-3p and hsa-mir-100-5p as novel prognostic markers for
oral cancer. Head Neck. 41:3499–3515. 2019. View Article : Google Scholar
|
|
43
|
Falzone L, Lupo G, La Rosa GR, Crimi S,
Anfuso CD, Salemi R, Rapisarda E, Libra M and Candido S:
Identification of Novel MicroRNAs and their diagnostic and
prognostic significance in oral cancer. Cancers. 11:6102019.
View Article : Google Scholar
|
|
44
|
Hsing EW, Shiah SG, Peng HY, Chen YW, Chuu
CP, Hsiao JR, Lyu PC and Chang JY: TNF-α-induced miR-450a mediates
TMEM182 expression to promote oral squamous cell carcinoma
motility. PLoS One. 14:e02134632019. View Article : Google Scholar
|
|
45
|
Kurihara-Shimomura M, Sasahira T,
Shimomura H, Nakashima C and Kirita T: miR-29b-1-5p the oncogenic
activity of induces the epithelial-mesenchymal transition in oral
squamous cell carcinoma. J Clin Med. 24:2732019. View Article : Google Scholar
|
|
46
|
Lu Y, Li Y, Wang Z, Xie S, Wang Q, Lei X,
Ruan Y and Li J: Downregulation of RGMA by HIF-1A/miR-210-3p axis
promotes cell proliferation in oral squamous cell carcinoma. Biomed
Pharmacother. 112:1086082019. View Article : Google Scholar
|
|
47
|
Yuan G, Wu H, Du Y and He F: Tumor
suppressor role of microRNA-545 in oral squamous cell carcinoma.
Oncol Lett. 17:2063–2068. 2019.
|
|
48
|
Qiu YF, Wang MX, Meng LN, Zhang R and Wang
W: MiR-21 regulates proliferation and apoptosis of oral cancer
cells through TNF-α. Eur Rev Med Pharmacol Sci. 22:7735–7741.
2018.
|
|
49
|
Zheng J, Wang J, Jia Y, Liu T, Duan Y,
Liang X and Liu L: microRNA-211 promotes proliferation, migration,
and invasion ability of oral squamous cell carcinoma cells via
targeting the bridging integrator 1 protein. J Cell Biochem.
120:4644–4653. 2019. View Article : Google Scholar
|
|
50
|
Peng SY, Tu HF, Yang CC, Wu CH, Liu CJ,
Chang KW and Lin SC: miR-134 targets PDCD7 to reduce E-cadherin
expression and enhance oral cancer progression. Int J Cancer.
143:2892–2904. 2018. View Article : Google Scholar
|
|
51
|
Kim JS, Choi DW, Kim CS, Yu SK, Kim HJ, Go
DS, Lee SA, Moon SM, Kim SG, Chun HS, et al: MicroRNA-203 induces
apoptosis by targeting in YD-38 oral cancer cells. Anticancer Res.
38:3477–3485. 2018. View Article : Google Scholar
|
|
52
|
Wang L, Wei Y, Yan Y, Wang H, Yang J,
Zheng Z, Zha J, Bo P, Tang Y, Guo X, et al: CircDOCK1 suppresses
cell apoptosis via inhibition of miR-196a-5p by targeting BIRC3 in
OSCC. Oncol Rep. 39:951–966. 2018.
|
|
53
|
Ruan P, Tao Z and Tan A: Low expression of
miR-30a-5p induced the proliferation and invasion of oral cancer
via promoting the expression of FAP. Biosci Rep.
38:BSR201710272018. View Article : Google Scholar
|
|
54
|
Sakha S, Muramatsu T, Ueda K and Inazawa
J: Exosomal microRNA miR-1246 induces cell motility and invasion
through the regulation of DENND2D in oral squamous cell carcinoma.
Sci Rep. 6:387502016. View Article : Google Scholar
|
|
55
|
Chuerduangphui J, Ekalaksananan T,
Chaiyarit P, Patarapadungkit N, Chotiyano A, Kongyingyoes B,
Promthet S and Pientong C: Effects of arecoline on proliferation of
oral squamous cell carcinoma cells by dysregulating c-Myc and
miR-22, directly targeting oncostatin M. PLoS One. 13:e01920092018.
View Article : Google Scholar
|
|
56
|
Harsha C, Banik K, Ang HL, Girisa S,
Vikkurthi R, Parama D, Rana V, Shabnam B, Khatoon E, Kumar AP and
Kunnumakkara AB: Targeting AKT/mTOR in oral cancer: Mechanisms and
advances in clinical trials. Int J Mol Sci. 21:32852020. View Article : Google Scholar
|
|
57
|
Manikandan M, Deva Magendhra Rao AK,
Arunkumar G, Manickavasagam M, Rajkumar KS, Rajaraman R and
Munirajan AK: Oral squamous cell carcinoma: microRNA expression
profiling and integrative analyses for elucidation of
tumourigenesis mechanism. Mol Cancer. 15:282016. View Article : Google Scholar
|
|
58
|
Uesugi A, Kozaki K, Tsuruta T, Furuta M,
Morita K, Imoto I, Omura K and Inazawa J: The tumor suppressive
microRNA miR-218 targets the mTOR component Rictor and inhibits AKT
phosphorylation in oral cancer. Cancer Res. 71:5765–5778. 2011.
View Article : Google Scholar
|
|
59
|
Moratin J, Hartmann S, Brands R, Brisam M,
Mutzbauer G, Scholz C, Seher A, Müller-Richter U, Kübler AC and
Linz C: Evaluation of miRNA-expression and clinical tumour
parameters in oral squamous cell carcinoma (OSCC). J
Craniomaxillofac Surg. 44:876–881. 2016. View Article : Google Scholar
|
|
60
|
Chen D, Chen Z, Jin Y, Dragas D, Zhang L,
Adjei BS, Wang A, Dai Y and Zhou X: MicroRNA-99 family members
suppress Homeobox A1 expression in epithelial cells. PLoS One.
8:e806252013. View Article : Google Scholar
|
|
61
|
Chen R, Zhang Y and Zhang X: MiR-1254
functions as a tumor suppressor in oral squamous cell carcinoma by
targeting CD36. Technol Cancer Res Treat. Aug 11–2019.doi:
10.1177/1533033819859447. View Article : Google Scholar
|
|
62
|
Chen X, Xu H, Sun G and Zhang Y: LncRNA
CASC9 affects cell proliferation, migration, and invasion of tongue
squamous cell carcinoma via regulating miR-423-5p/SOX12 Axes.
Cancer Manage Res. 12:277–287. 2020. View Article : Google Scholar
|
|
63
|
Rastogi B, Kumar A, Raut SK, Panda NK,
Rattan V, Joshi N and Khullar M: Downregulation of miR-377 promotes
oral squamous cell carcinoma growth and migration by targeting
HDAC9. Cancer Invest. 35:152–162. 2017. View Article : Google Scholar
|
|
64
|
Chen F, Qi S, Zhang X, Wu J, Yang X and
Wang R: miR-23a-3p suppresses cell proliferation in oral squamous
cell carcinomas by targeting FGF2 and correlates with a better
prognosis: miR-23a-3p inhibits OSCC growth by targeting FGF2.
Pathol Res Pract. 215:660–667. 2019. View Article : Google Scholar
|
|
65
|
Li X, Liu K, Zhou W and Jiang Z: MiR-155
targeting FoxO3a regulates oral cancer cell proliferation,
apoptosis, and DDP resistance through targeting FoxO3a. Cancer
Biomark. 27:105–111. 2020. View Article : Google Scholar
|
|
66
|
Fu S, Chen HH, Cheng P, Zhang CB and Wu Y:
MiR-155 regulates oral squamous cell carcinoma Tca8113 cell
proliferation, cycle, and apoptosis via regulating p27Kip1. Eur Rev
Med Pharmacol Sci. 21:937–944. 2017.
|
|
67
|
Bufalino A, Cervigne NK, de Oliveira CE,
Fonseca FP, Rodrigues PC, Macedo CC, Sobral LM, Miguel MC, Lopes
MA, Paes Leme AF, et al: Low miR-143/miR-145 cluster levels induce
activin A overexpression in oral squamous cell carcinomas, which
contributes to poor prognosis. PLoS One. 10:e01365992015.
View Article : Google Scholar
|
|
68
|
Huang WC, Chan SH, Jang TH, Chang JW, Ko
YC, Yen TC, Chiang SL, Chiang WF, Shieh TY, Liao CT, et al:
miRNA-491-5p and GIT1 serve as modulators and biomarkers for oral
squamous cell carcinoma invasion and metastasis. Cancer Res.
74:751–764. 2014. View Article : Google Scholar
|
|
69
|
Yu EH, Tu HF, Wu CH, Yang CC and Chang KW:
MicroRNA-21 promotes perineural invasion and impacts survival in
patients with oral carcinoma. J Chin Med Assoc. 80:383–388. 2017.
View Article : Google Scholar
|
|
70
|
Tu HF, Chang KW, Cheng HW and Liu CJ:
Upregulation of miR-372 and −373 associates with lymph node
metastasis and poor prognosis of oral carcinomas. Laryngoscope.
125:E365–E370. 2015. View Article : Google Scholar
|
|
71
|
Zhang B, Li Y, Hou D, Shi Q, Yang S and Li
Q: MicroRNA-375 inhibits growth and enhances radiosensitivity in
oral squamous cell carcinoma by targeting insulin like growth
factor 1 receptor. Cell Physiol Biochem. 42:2105–2117. 2017.
View Article : Google Scholar
|
|
72
|
Weng JH, Yu CC, Lee YC, Lin CW, Chang WW
and Kuo YL: miR-494-3p induces cellular senescence and enhances
radiosensitivity in human oral squamous carcinoma cells. Int J Mol
Sci. 17:10922016. View Article : Google Scholar
|
|
73
|
Min A, Zhu C, Peng S, Shuai C, Sun L, Han
Y, Qian Y, Gao S and Su T: Downregulation of Microrna-148a in
cancer-associated fibroblasts from oral cancer promotes cancer cell
migration and invasion by targeting Wnt10b. J Biochem Mol Toxicol.
30:186–191. 2016. View Article : Google Scholar
|