Introduction
Lung cancer remains one of the leading causes of
cancer-related deaths worldwide. As the most common and aggressive
type of lung cancer, the incidence of non-small cell lung cancer
(NSCLC) accounts for approximately 85% (1). Clinically, surgical resection and
postoperative adjuvant therapy, including chemotherapy and
radiotherapy, remain the primary choice of treatment for NSCLC
patients at early stages. However, a considerable proportion of
NSCLC patients are in advanced stages or the cancer has
metastasized at the time of diagnosis. Thereby, a great number of
patients with advanced NSCLC cannot benefit from surgical resection
(2). Hence, the 5-year survival
rate of NSCLC patients remains markedly poor despite the great
progress that has been made in the past few years in chemotherapy
and radiotherapy treatments (3).
Therefore, the elucidation of the mechanism involved in the
progression of NSCLC is essential to improve the prognosis of
patients.
Circular RNAs (circRNAs) are commonly expressed in
eukaryotic cells, and are associated with the progression of human
tumors (4,5). However, limited research has focused
on the underlying mechanism of circRNAs in regulating the
development of NSCLC. Through circRNA microarrays, a previous
research screened hsa_circ_0016760 as a tumor-specific circRNA
candidate towards NSCLC tissues and adjacent normal tissues
(6). Furthermore, Li et al
(7) revealed the enhanced
expression of hsa_circ_0016760 in NSCLC tissues and cells, and that
high expression of hsa_circ_0016760 was closely related to the poor
prognosis of patients as well as lymph node metastasis and advanced
TNM stages. The upregulated hsa_circ_0016760 could facilitate NSCLC
cell growth and metastatic properties in vitro. In terms of
mechanism, the carcinogenic properties of circ_0016760 were
attributed to the enhancement of GAGE1 expression via sponging
miR-1287. To date, more studies concerning the function of
circ_0016760 in NSCLC or other human tumors are scarce.
Hence, the present study was designed to explore the
effect of circ_0016760 in regulating NSCLC progression in order to
provide an effective theoretical basis for the application of
circ_0016760 in the targeted therapy of NSCLC. In terms of the
molecular mechanism network, it was observed from biological
software online analysis that miR-145-5p possessed binding sites
for circ_0016760 and FGF5. Previous studies have reported that
miR-145-5p and FGF5 are involved in the progression of lung cancer
(8,9). Thus, the present study researched the
mechanism of circ_0016760 in regulating NSCLC with the
miR-145-5p/FGF5 axis.
Materials and methods
Patients and tissues
NSCLC patients (n=80) who were treated at our
hospital (the Affiliated Changzhou No. 2 People's Hospital of
Nanjing Medical University, Changzhou, Jiangsu) from August 2013 to
October 2014 were enrolled in the present study. All patients were
diagnosed with NSCLC for the first time and had no previous history
of cancer-related treatment. The inclusion criteria were as
follows: i) Patients who voluntarily joined the study; ii) patients
who were diagnosed with NSCLC for the first time; iii) patients
without previous treatment history related to cancer diseases.
The exclusion criteria were as follows: i) Patients
who did not want to join this study; ii) patients with previous
treatment history of cancer-related diseases. Surgical resection
was performed on these patients. During surgery, tumor tissues and
adjacent normal tissues were collected and stored at −80°C. The
clinical characteristics of all patients are recorded and listed in
Table I. All patients were
followed-up for 60 months after surgery.
 | Table I.High hsa_circ_0016760 expression is
associated with poor prognosis of NSCLC patients. |
Table I.
High hsa_circ_0016760 expression is
associated with poor prognosis of NSCLC patients.
|
|
| Hsa_circ_0016760
expression |
|
|---|
|
|
|
|
|
|---|
| Characteristics | No. of patients | Low (<median) | High (≥median) | P-value |
|---|
| Number | 80 | 38 | 42 |
|
| Age (years) |
|
|
| 0.504 |
|
<65 | 39 | 19 | 20 |
|
|
≥65 | 41 | 19 | 22 |
|
| Sex |
|
|
| 0.420 |
|
Female | 38 | 19 | 19 |
|
|
Male | 42 | 19 | 23 |
|
| Smoking |
|
|
| 0.412 |
|
Yes | 40 | 18 | 22 |
|
| No | 40 | 20 | 20 |
|
| Tumor size |
|
|
| 0.039 |
| ≤3
cm | 37 | 22 | 15 |
|
| >3
cm | 43 | 16 | 27 |
|
|
Differentiation |
|
|
| 0.513 |
| Well
differentiated | 37 | 18 | 19 |
|
| Lowly
or undifferentiated | 43 | 20 | 23 |
|
| T
classification |
|
|
| 0.023 |
| T1 +
T2 | 38 | 23 | 15 |
|
| T3 +
T4 | 42 | 15 | 27 |
|
| N
classification |
|
|
| 0.037 |
| N0 +
N1 | 39 | 23 | 16 |
|
| N2 +
N3 | 41 | 15 | 26 |
|
| Distant
metastasis |
|
|
| 0.251 |
| M1 | 40 | 17 | 23 |
|
| M0 | 40 | 21 | 19 |
|
| Clinical stage |
|
|
| 0.013 |
|
I–II | 39 | 24 | 15 |
|
|
III–IV | 41 | 14 | 27 |
|
All patients volunteered to join the study and
written informed consent was signed by all subjects. The present
study complied with the Declaration of Helsinki and was approved by
the Ethics Committee of the Affiliated Changzhou No. 2 People's
Hospital of Nanjing Medical University.
Cell culture
Six NSCLC cell lines (NCI-H1395, A549, H460, H1975,
H1299 and Calu3) and normal human bronchial epithelial cell line
(NHBE) were obtained from the Institute of Biochemistry and Cell
Biology of the Chinese Academy of Sciences. Cells were grown in
Dulbecco's modified Eagle's medium (DMEM) supplemented with 10%
(v/v) fetal bovine serum (FBS) in a humidified incubator at 37°C
and 5% CO2.
Structural stability test of
hsa_circ_0016760
Tumor tissues collected from NSCLC patients were
ground into powder in liquid nitrogen. Total RNA in tumor tissues
were extracted with TRIzol reagent (Thermo Fisher Scientific,
Inc.). A total of 5.0 µg of total RNA sample was subjected to
incubation with 10 units RNase R for 30 min at 37°C (used as the
RNase R group). In addition, 5.0 µg of total RNA sample without any
treatment was collected and used as the Mock group. The expression
of hsa_circ_0016760 and glyceraldehyde-3-phosphate dehydrogenase
(GAPDH) in the two groups was monitored through reverse
transcription quantitative polymerase chain reaction (RT-qPCR). It
is well known that the structure of linear RNA is easily destroyed
by RNase R, whereas the circular structure of circular RNA is
difficult to be degraded by RNase R (10). Therefore, the circular structure
stability of hsa_circ_0016760 could be detected.
Cell transfection
In this experiment, Lipofectamine 2000 reagent
(Thermo Fisher Scientific, Inc.) was used for transfection. H1299
and Calu3 cells (1×106 cells/ml; 1 ml) cultured in
6-well plates with serum-free DMEM were transfected by small
interfering RNA (100 nM; siRNA-1: 5′-GUCUGGCAUGCAGAGGCAGAA-3′,
siRNA-2: 5′-CUGGCAUGCAGAGGCAGAAGA-3′ and siRNA-3:
5′-AUGCAGAGGCAGAAGAGGCCU-3′, respectively) targeting
hsa_circ_0016760 (named the si-hsa_circ_0016760-1 group,
si-hsa_circ_0016760-2 group and si-hsa_circ_0016760-3 group,
respectively). The transfection was performed at 37°C for 8 h.
H1299 and Calu3 cells transfected by siRNA negative control (100
nM; 5′-AGGAGGCUAGCUCUGCGACUU-3′) were used as the si-Ctrl groups.
The miR-145-5p mimic (100 nM; miR-145-5p mimic group, sequence:
5′-GUCCAGUUUUCCCAGGAAUCCCU-3′) and mimic negative control (100 nM;
miR-NC group, sequence: 5′-CGCGAGUUAACGGACCAUACGGU-3′) were used to
transfect H1299 and Calu3 cells. siRNA and corresponding negative
control, miR-145-5p mimic and mimic negative control were
synthesized by Shanghai GenePharma Co., Ltd.
pCDNA3.1 vector containing the full length of
hsa_circ_0016760 was provided by Shanghai Genechem Co., Ltd., and
was then transfected into A549 cells (1×106 cells/ml; 1
ml) cultured in 6-well plates with serum-free DMEM
(hsa_circ_0016760 group). A549 cells transfected by pCDNA3.1 empty
vector (100 nM) were set as the Ctrl group. The transfection was
performed at 37°C for 8 h. FGF5 siRNA (Shanghai GenePharma Co.,
Ltd.) was commercially obtained. A549 cells were co-transfected by
pCDNA3.1-hsa_circ_0016760 vector and FGF5 siRNA (hsa_circ_0016760 +
siFGF5 group). In addition, pCDNA3.1-hsa_circ_0016760 vector and
miR-145-5p mimic were both used to co-transfect A549 cells, and
served as the hsa_circ_0016760 + miR-145-5p mimic group.
All cells were incubated at 37°C and 5%
CO2 for 8 h after transfection. Subsequently, fresh DMEM
containing 10% FBS was used to culture cells for 48 h. RT-qPCR was
used for the transfection efficiency detection.
Cell Counting Kit-8 (CCK-8) assay
The proliferation ability of cells was evaluated by
CCK-8 assay. Cells were harvested and dispersed in DMEM (with 10%
FBS) to a density of 1×105 cells/ml. A total of 100 µl
of cell suspension was added into 96-well plates with 5 multiple
wells. Cells were maintained at 37°C and 5% CO2 for 24,
48 and 72 h. CCK-8 solution (10 µl) was added into each well and
the cells were incubated for 4 h at 37°C. The optical density (OD)
value was monitored using a microplate reader (Bio-Tek Instruments,
Inc.) at a wavelength of 450 nm.
5-Ethynyl-2′-deoxyuridine (EdU)
assay
EdU assays were conducted to assess the
proliferation ability of cells using an EdU assay kit (Beijing
Solarbio Science & Technology Co., Ltd.) strictly according to
the manufacturer's instructions. Briefly, cells were collected and
seeded into 6-well plates with 1×105 cells/well. EdU
buffer (50 µM) was used to incubate cells for 2 h at 37°C. Then, 4%
formaldehyde was used to fix cells for 30 min at 37°C, and 0.1%
Triton X-100 was used for 20 min of permeabilization at 37°C. EdU
solution was then applied to incubate cells for 20 min at 37°C and
Hoechst 33342 (5 µg/ml; Beyotime Institute of Biotechnology) was
used to stain the nucleus for 15 min at 37°C. Under a fluorescence
microscope, the EdU-positive cells were photographed and counted as
previously described (11).
Transwell assays
Cells were collected and dispersed in serum-free
DMEM. The Transwell inserts (8 µm pore size; Corning, Inc.) were
inserted into 6-well plates and Matrigel was pre-coated on the
upper chamber. Then 5×104 cells dispersed in 300 µl of
serum-free DMEM was added into the upper chamber, while 600 µl of
DMEM (with 10% FBS) was added into the lower chamber. Cells were
incubated at 37°C, 5% CO2 for 24 h. The chamber was
removed and the non-invading cells were removed with a cotton swab.
The invasive cells were fixed at room temperature for 20 min with
4% paraformaldehyde, after which, 0.1% crystal violet was used to
stain the cells for 10 min at room temperature. The number of
invasive cells was counted under a confocal microscope in five
random fields (magnification, ×200). Cell migration was performed
according to the aforementioned steps, without the use of Matrigel
coated on the upper chamber.
Luciferase reporter gene assay
Through biological software online analysis
[Circular RNA Interactome (http://circinteractome.nia.nih.gov/), miRDB
(http://www.mirdb.org/) and TargetScan (http://www.targetscan.org/vert_71/)], it was
revealed that hsa_circ_0016760 and FGF5 both had binding sites for
miR-145-5p. Therefore, H1299 cells were used for the luciferase
reporter gene assay to detect the relationship between miR-145-5p
and hsa_circ_0016760 (or FGF5). Fragments of hsa_circ_0016760
wild-type (WT) and mutant type (Mut) as well as FGF5 WT and Mut
were designed and synthesized by Shanghai GenePharma Co., Ltd..
These fragments were loaded into the pmirGLO luciferase vectors.
H1299 cells transfected by miR-145-5p mimic (miR-145-5p mimic
group, sequence: 5′-GUCCAGUUUUCCCAGGAAUCCCU-3′) and mimic negative
control (miR-NC group, sequence: 5′-CGCGAGUUAACGGACCAUACGGU-3′)
were seeded in 6-well plates with serum-free DMEM. Then these cells
underwent co-transfection with pmirGLO-hsa_circ_0016760-WT
luciferase vector, or pmirGLO-hsa_circ_0016760-Mut luciferase
vector, or pmirGLO-FGF5-WT luciferase vector or pmirGLO-FGF5-Mut
luciferase vector. Lipofectamine 2000 reagent (Thermo Fisher
Scientific, Inc.) was used for the transfection. After 8 h, the
serum-free DMEM in each well was replaced by DMEM containing 10%
FBS. After 48 h of culture at 37°C and 5% CO2, the cells
were harvested to detect the luciferase activity using Dual-Glo
luciferase assay kit (Promega Corporation) strictly according to
the manufacturer's instructions. The relative luciferase activity
was normalized to Renilla luciferase activity.
RNA immunoprecipitation (RIP)
assay
EZ-Magna RIP kit (EMD Millipore) was used for RIP
assays strictly according to the manufacturer's instructions. H1299
cells transfected by miR-145-5p mimic (miR-145-5p mimic group) and
mimic negative control (miR-NC group) was harvested and lysed in
RIP lysis buffer. Then 100 µl of the cell extract was collected for
6-h incubation at 4°C with RIP buffer containing magnetic beads
conjugated with antibodies against AGO2 (1:200; product code
ab5072) or IgG (1:200; product code ab109489; both from Abcam).
Wash buffer was used to wash the beads. Subsequently, Proteinase K
was applied to incubate the complexes for 30 min at 55°C. Finally,
the immunoprecipitated hsa_circ_0016760 was purified using phenol,
chloroform, salt solution I, salt solution II, precipitate enhancer
and absolute ethanol (no RNase), and was assessed with RT-qPCR.
Animal xenograft experiment
Animal experiments involved in the present study
were approved by the Animal Ethics Committee of the Affiliated
Changzhou No. 2 People's Hospital of Nanjing Medical University.
Nude mice (n=12; male, 4 weeks old, 18-20 g) were commercially
provided by Shanghai Experimental Animal Center, Chinese Academy of
Sciences. Mice were kept in a 12-h day/night cycle room with free
access to water and food. H1299 cells were transfected by
hsa_circ_0016760 shRNA (100 nM) and corresponding negative control
(100 nM) (Shanghai GenePharma Co., Ltd.). After 48 h of
transfection, H1299 cells were harvested and washed with
phosphate-buffered saline (PBS). A total of 1×106 cells
dispersed in 100 µl PBS was prepared for subcutaneous implantation.
Thereafter, 6 nude mice were randomly selected and subcutaneously
injected with H1299 cells transfected by hsa_circ_0016760 shRNA.
These mice were named the sh-hsa_circ_0016760 group. In addition,
the other 6 mice were subcutaneously implanted with H1299 cells
transfected by corresponding negative control, which were used as
sh-Ctrl group. Mice were maintained for 28 days with free access to
food and water. Every 7 days, the longitudinal diameter (L) and
latitudinal diameter (D) were measured using a Vernier caliper. The
tumor volume was calculated by the equation of V = 0.5 × L ×
D2. On the 28th day, all mice were sacrificed by rapid
neck dislocation. The xenograft tumors were stripped, weighed and
then stored at −80°C.
Immunohistochemical staining
The xenograft tumors were fixed with 4%
paraformaldehyde for 12 h at 4°C, embedded in paraffin and prepared
into paraffin tissue sections with a thickness of 4 µm. The
sections were then dewaxed and rehydrated. Sodium citrate buffer
was used for the antigen retrieval of the sections. Bovine serum
albumin (5%; Beijing Solarbio Science & Technology Co., Ltd.)
was applied to block the sections for 30 min at room temperature.
Thereafter, the sections were incubated with rabbit anti-Ki67
(product no. 9449; Cell Signaling Technology, Inc.) for 12 h at
4°C, and then with goat anti-rabbit HRP conjugated secondary
antibody (product no. 7074; Cell Signaling Technology, Inc.) for 1
h at room temperature. Diaminobenzidine (DAB) and hematoxylin were
used for the staining of the sections for 15 min at room
temperature. Under a confocal microscope, the Ki67-positive signals
(brown particles) were observed (magnification, ×200).
RT-qPCR
TRIzol reagent (Takara Bio, Inc.) was used to
isolate total RNA in tissues and cells. According to the
manufacturer's instructions, cDNA was reverse transcribed using
PrimeScript RT reagent Kit (Takara Bio, Inc.). qPCR was performed
in a 10-µl reaction system, including 2.0 µl of cDNA, 0.5 µl of
forward primer, 0.5 µl of reverse primer, 5.0 µl of 2X PCR Master
mix and 2.0 µl of H2O. The qPCR reaction was performed
with SYBR Green reagent (Takara Bio, Inc.) using the ABI 7500 Fast
Real-Time PCR System (Applied Biosystems; Thermo Fisher Scientific,
Inc.) according to the following conditions: 10 min initial
denaturation at 95°C, 40 cycles of amplification at 95°C for 10
sec, annealing at 56°C for 45 sec and extension at 60°C for 60 sec.
Primers used were as follows: hsa_circ_0016760 forward,
5′-TGCATTGGTGCTCAGAAGCG-3′ and reverse,
5′-TCTGTTCCTGGGTCTGTGTGC-3′; miR-145-5p forward,
5′-GTCCAGTTTTCCCAGGAATCCCT-3′ and reverse,
5′-GCTGTCAACATACGCTACGTAACG-3′; FGF5 forward,
5′-CCCGGATGGCAAAGTCAATGG-3′ and reverse,
5′-TTCAGGGCAACATACCACTCCCG-3′; GAPDH forward,
5′-AATGTGTCCGTCGTGGATCTG-3′ and reverse,
5′-CAACCTGGTCCTCAGTGTAGC-3′. GAPDH was set as the internal control
and the relative expression of hsa_circ_0016760, miR-145-5p and
FGF5 mRNA was calculated by 2−ΔΔCq method (12).
Western blotting
Total proteins in cells were extracted with RIPA
lysis buffer (Beyotime Institute of Biotechnology). Protease
inhibitors (Roche Diagnostics) were contained in the RIPA lysis
buffer. A BCA Protein Assay kit (Beyotime Institute of
Biotechnology) was applied for the determination of total protein
concentration. Then 10 µl of total protein (50 µg) extract was
subjected to separation with 10% sodium dodecyl
sulphate-polyacrylamide gel electrophoresis (SDS-PAGE). The
separated proteins were transferred onto polyvinylidene fluoride
(PVDF) membranes. Subsequently, the membranes were blocked for 1 h
with 5% non-fat milk at room temperature. Rabbit polyclonal FGF5
antibody (1:1,000; cat. sc-376264; Santa Cruz Biotechnology, Inc.)
was used for the incubation of the membrane for 12 h at 4°C.
Thereafter, goat anti-rabbit secondary antibody (1:5,000; cat. no.
PAB10821; Abnova) was applied to incubate the membrane for 1 h at
room temperature. The blots were captured by an enhanced
chemiluminescence (ECL) system (Pierce Biotechnology; Thermo Fisher
Scientific, Inc.). The intensity of the blots was quantified by
ImageJ software (version 1.51t; National Institutes of Health). The
internal control was GAPDH in this research.
Statistical analysis
The statistical analysis was processed through SPSS
19.0 software (IBM Corp.) The graphs were constructed using
GraphPad Prism 6 software (GraphPad Software, Inc.). Two-tailed
paired Student's t-test and one-way analysis of variance (ANOVA)
was respectively used for the comparison between two groups and
more than two groups. Tukey's post hoc test was used to validate
ANOVA for pairwise comparisons. The relationship between
hsa_circ_0016760 expression and the clinicopathological
characteristics of NSCLC patients was analyzed by χ2
test. Kaplan-Meier survival analysis and log-rank tests were
applied for the analysis of survival curves. Among
hsa_circ_0016760, miR-145-5p and FGF5, Pearson's correlation
analysis was used to assess the correlation between two genes.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Hsa_circ_0016760 is aberrantly
upregulated in NSCLC, which is associated with the poor prognosis
of patients
The present study firstly explored the expression of
hsa_circ_0016760 in 80 pairs of tumor tissues/adjacent normal
tissues. The results presented in Fig.
1A revealed thathsa_circ_0016760 expression was significantly
increased in tumor tissues than that in paired adjacent normal
tissues (P<0.01). Thereafter, the correlation between
hsa_circ_0016760 expression and the clinicopathological
characteristics of NSCLC patients was analyzed to evaluate the
significance of hsa_circ_0016760 upregulation in NSCLC. As a
result, high hsa_circ_0016760 expression was significantly
associated with poor 60-month survival (P=0.0151), large tumor size
(P=0.039), advanced T classification (P=0.023), N classification
(P=0.037) and clinical stage (P=0.013) (Fig. 1B; Table
I). The expression of hsa_circ_0016760 in NHBE cells and 6
NSCLC cell lines (NCL-H1395, A549, H460, H1975, H1299 and Calu3)
was further assessed. Notably, the 6 NSCLC cell lines were
expressed significantly higher hsa_circ_0016760 than the NHBE cells
(P<0.05 and P<0.01). Among the 6 NSCLC cell lines, H1299 and
Calu3 cell lines had higher hsa_circ_0016760 expression than the
other 4 NSCLC cell lines. Therefore, H1299 and Calu3 cell lines
were used in subsequent experiments (Fig. 1C). To demonstrate the expression
stability of hsa_circ_0016760, total RNA in tumor tissues of NSCLC
patients was collected and incubated with RNase R. As a result,
RNase R treatment significantly decreased GAPDH expression
(P<0.01), but had no obvious effect on the expression level of
hsa_circ_0016760 (Fig. 1D). Thus,
hsa_circ_0016760 possessed as table circular structure, which could
be stably expressed in NSCLC. Collectively it was revealed that the
high expression of hsa_circ_0016760 in NSCLC was associated with
the poor prognosis of patients.
Hsa_circ_0016760 silencing suppresses
NSCLC cell proliferation, migration and invasion in vitro
H1299 and Calu3 cells were transfected by
hsa_circ_0016760 siRNA. RT-qPCR was performed to monitor the
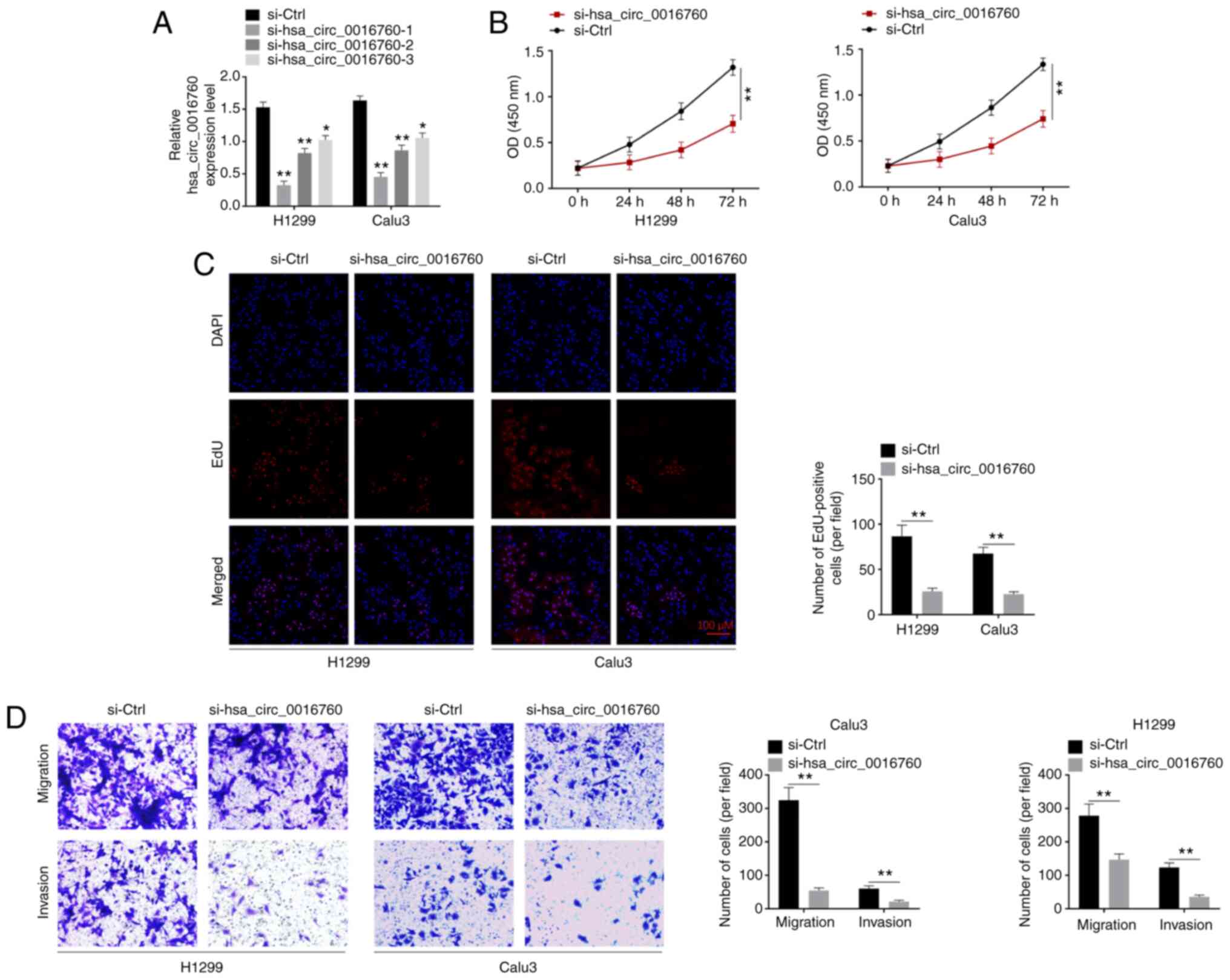
transfection efficiency. As revealed in Fig. 2A, the si-hsa_circ_0016760-1,
si-hsa_circ_0016760-2 and si-hsa_circ_0016760-3 groups of H1299 and
Calu3 cells exhibited significantly lower hsa_circ_0016760
expression than that of the si-Ctrl group (P<0.05 and
P<0.01). Thus, hsa_circ_0016760 expression in H1299 and Calu3
cells was successfully silenced via transfection. Notably, the
si-hsa_circ_0016760-1 group exhibited lower hsa_circ_0016760
expression than that of the si-hsa_circ_0016760-2 group and
si-hsa_circ_0016760-3 group of H1299 and Calu3 cells. Therefore,
the si-hsa_circ_0016760-1 group of H1299 and Calu3 cells was used
in subsequent experiments, and was renamed as si-hsa_circ_0016760
in the following studies.
Next, the phenotype of H1299 and Calu3 cells was
explored to research the effect of hsa_circ_0016760 on NSCLC
development in vitro. The proliferation of H1299 and Calu3
cells was analyzed by CCK-8 and EdU assays respectively. The
results revealed that the si-hsa_circ_0016760 group of H1299 and
Calu3 cells had a significantly lower OD value than that of the
si-Ctrl group (P<0.01) (Fig.
2B). In addition, a significantly less EdU-positive number of
cells was observed in the si-hsa_circ_0016760 group when compared
with si-Ctrl group of H1299 and Calu3 cells (P<0.01) (Fig. 2C). In addition, cell migration and
invasion abilities were investigated by Transwell assay. In
comparison with the siCtrl group, the number of migrated and
invasive cells of the si-hsa_circ_0016760 group of H1299 and Calu3
cells was significantly decreased (P<0.01) (Fig. 2D). These data revealed that
hsa_circ_0016760 silencing suppressed NSCLC cell proliferation,
migration and invasion in vitro.
Hsa_circ_0016760 facilitates FGF5
expression via sponging miR-145-5p
The transfection efficiency of H1299 and Calu3 cells
was assessed by RT-qPCR. Compared with the miR-NC group, the
miR-145-5p mimic group of H1299 and Calu3 cells had significantly
increased miR-145-5p expression (P<0.01) (Fig. 3A). Thus, H1299 and Calu3 cells were
successfully transfected by miR-145-5p mimic and mimic NC. The
exact target of hsa_circ_0016760 was investigated in order to
elucidate the mechanism of hsa_circ_0016760 in the promotion of the
malignant phenotype of NSCLC cells. According to online
bioinformatics databases (Circular RNA Interactome and miRDB),
miR-145-5p may be a target of hsa_circ_0016760, as it contained a
binding site for hsa_circ_0016760 (Fig.
3B). Luciferase reporter gene assay and RIP assay were then
performed to verify the relationship between hsa_circ_0016760 and
miR-145-5p. As revealed in Fig. 3C,
the miR-145-5p mimic group of H1299 cells exhibited significantly
decreased relative luciferase activity of hsa_circ_0016760-WT
reporter than the miR-NC group (P<0.01). However, the difference
in relative luciferase activity of hsa_circ_0016760-Mut reporter
between the miR-145-5p mimic group and miR-NC group was not
statistically significant. Furthermore, based on the results from
the RIP assay, it was observed that hsa_circ_0016760 could be
effectively precipitated by the AGO2 antibody. Notably,
hsa_circ_0016760 expression was significantly increased in the
miR-145-5p mimic group when compared with the miR-NC group
(P<0.01) (Fig. 3D). Thus,
miR-145-5p was confirmed as a target of hsa_circ_0016760. For the
si-hsa_circ_0016760 group, significantly higher miR-145-5p
expression was observed compared with the si-Ctrl group of H1299
and Calu3 cells (P<0.01) (Fig.
3E). In addition, in comparison with adjacent normal tissues,
significantly decreased miR-145-5p expression was observed in tumor
tissues of NSCLC patients (P<0.01) (Fig. 3F). In tumor tissues of NSCLC
patients, the expression level ofmiR-145-5p was negatively
correlated with the expression level of hsa_circ_0016760
(P<0.01) (Fig. 3G). Therefore,
it was confirmed that miR-145-5p was directly inhibited by
hsa_circ_0016760 in NSCLC according to these data.
 | Figure 3.Hsa_circ_0016760 facilitates FGF5
expression via sponging miR-145-5p. (A) RT-qPCR revealed that H1299
and Calu3 cells were successfully transfected by miR-145-5p mimic
and mimic NC. **P<0.01 when compared with miR-NC group. (B)
According to an online bioinformatics database, miR-145-5p
contained a binding site for hsa_circ_0016760. (C) A luciferase
reporter gene assay indicated that miR-145-5p was a target of
hsa_circ_0016760. **P<0.01 when compared with miR-NC group. (D)
A RIP assay revealed that, hsa_circ_0016760 could be effectively
precipitated by miR-145-5p, and miR-145-5p was further confirmed to
be a target of hsa_circ_0016760. **P<0.01 when compared with
miR-NC group. (E) Hsa_circ_0016760 silencing significantly
increased the expression of miR-145-5p in H1299 and Calu3 cells.
**P<0.01 when compared with the si-Crtl group. (F) miR-145-5p
expression was significantly reduced in tumor tissues compared with
adjacent normal tissues. (G) miR-145-5p expression was negatively
correlated with hsa_circ_0016760 in NSCLC tumor tissues. (H)
According to TargetScan online database, FGF5 maybe target of
miR-145-5p, since it possessed a binding site for miR-145-5p. (I) A
luciferase reporter gene assay revealed that FGF5 was a target of
miR-145-5p. **P<0.01 when compared with miR-NC group. (J) In
H1299 and Calu3 cells, miR-145-5p upregulation significantly
reduced FGF5 mRNA and protein expression. In addition,
hsa_circ_0016760 downregulation significantly reduced FGF5 mRNA and
protein expression. **P<0.01 when compared with miR-NC group or
si-Crtl group. (K) FGF5 expression was significantly increased in
tumor tissues compared with adjacent normal tissues. (L) Pearson's
correlation analysis indicated that, in NSCLC tumor tissues, the
expression level of FGF5was positively correlated with the
expression level of hsa_circ_0016760. Conversely, a negative
correlation was revealed between the expression levels of FGF5 and
miR-145-5p. NSCLC, non-small cell lung cancer; RT-qPCR, reverse
transcription-quantitative polymerase chain reaction; NC, negative
control; si-, small interfering; Ctrl, control; RIP, RNA
immunoprecipitation. |
According to TargetScan online database, FGF5 maybe
target of miR-145-5p, since it possessed a binding site for
miR-145-5p (Fig. 3H). A luciferase
reporter gene assay revealed that the miR-145-5p mimic group
exhibited significantly reduced relative luciferase activity of
FGF5-WT reporter compared with the miR-NC group of H1299 cells
(P<0.01), whereas no statistically significant difference was
revealed in the relative luciferase activity of the FGF5-Mut
reporter between the miR-NC group and miR-145-5p mimic group
(Fig. 3I). Hence, FGF5 was
identified as a target of miR-145-5p. In vitro studies
revealed that the miR-145-5p mimic group expressed significantly
decreased FGF5 mRNA and protein expression levels than those of the
miR-NC group of H1299 and Calu3 cells (P<0.01). Concurrently,
relative to the si-Ctrl group, the FGF5 mRNA and protein expression
levels of the si-hsa_circ_0016760 group of H1299 and Calu3 cells
were significantly decreased (P<0.01) (Fig. 3J). Moreover, for NSCLC patients,
significantly increased FGF5 expression was observed in tumor
tissues than that in adjacent normal tissues (P<0.01) (Fig. 3K). The correlation analysis of FGF5
and hsa_circ_0016760 (or miR-145-5p) in tumor tissues revealed
that, the expression level of FGF5 was positively correlated with
hsa_circ_0016760 (P<0.01). Conversely, a negative correlation
was revealed for FGF5 and miR-145-5p expression levels in tumor
tissues (P<0.01) (Fig. 3L). All
of these data demonstrated that hsa_circ_0016760 enhanced the
expression of FGF5 via sponging miR-145-5p.
Hsa_circ_0016760 exacerbates the
malignant development of NSCLC by sponging the miR-145-5p/FGF5
axis
The mechanism of hsa_circ_0016760 in promoting NSCLC
malignant development was verified by rescue experiments. A549
cells had relatively low hsa_circ_0016760 expression among the 6
NSCLC cell lines. Thus, hsa_circ_0016760 was overexpressed in A549
cells. As revealed in Fig. 4A,
compared with the Ctrl group, FGF5 protein expression in the
hsa_circ_0016760 group of A549 cells was significantly increased
(P<0.01). A549 cells were also subjected to co-transfection. The
results revealed that compared with the hsa_circ_0016760 group,
significantly decreased FGF5 protein expression was observed in the
hsa_circ_0016760 + miR-145-5p mimic group and hsa_circ_0016760 +
si-FGF5 group of A549 cells (P<0.01). In addition, the
expression of hsa_circ_0016760, miR-145-5p and FGF5 mRNA in A549
cells of the 4 groups was assessed. As revealed in Fig. 4B, compared with the Ctrl group, the
hsa_circ_0016760 group exhibited higher hsa_circ_0016760
expression, lower miR-145-5p expression and higher FGF5 mRNA
expression in A549 cells (P<0.01). However, in comparison with
the hsa_circ_0016760 group, the hsa_circ_0016760 + miR-145-5p mimic
group and hsa_circ_0016760 + si-FGF5 group presented significantly
decreased hsa_circ_0016760 expression, increased miR-145-5p
expression and decreased FGF5 mRNA expression (P<0.01).
Then, the malignant phenotype of A549 cells was
investigated in vitro. According to a CCK-8 assay, the
hsa_circ_0016760 group of A549 cells exhibited a significantly
increased OD value than that of the Ctrl group (P<0.01).
Conversely, compared with the hsa_circ_0016760 group, the OD values
of A549 cells in the hsa_circ_0016760 + miR-145-5p mimic group and
hsa_circ_0016760 + si-FGF5 group were significantly decreased
(P<0.01) (Fig. 4C). Similarly,
an EdU assay revealed that, more Edu-positive cells were observed
in the hsa_circ_0016760 group when compared with the Ctrl group of
A549 cells (P<0.01). However, the number of Edu-positiveA549
cells in thehsa_circ_0016760 + miR-145-5p mimic group and
hsa_circ_0016760 + si-FGF5 group were significantly decreased
compared with the hsa_circ_0016760 group (P<0.01) (Fig. 4D). Transwell assays revealed
significantly increased migration and invasion abilities of A549
cells in thehsa_circ_0016760 group compared with the Ctrl group
(P<0.01). Conversely, decreased migration and invasion abilities
of A549 cells were observed in the hsa_circ_0016760 + miR-145-5p
mimic and hsa_circ_0016760 + si-FGF5 groups in comparison with the
hsa_circ_0016760 group (P<0.01) (Fig. 4E). Hence, these results revealed
that hsa_circ_0016760 exacerbated the malignant development of
NSCLC by targeting the miR-145-5p/FGF5 axis.
Hsa_circ_0016760 silencing inhibits
NSCLC cell growth in nude mice
To explore the effect of hsa_circ_0016760 on NSCLC
development in vivo, H1299 cells were transfected with
hsa_circ_0016760 shRNA and corresponding NC and subcutaneously
implanted into nude mice. The results revealed that compared with
the sh-NC group, the tumor volume and weight of sh-hsa_circ_0016760
group were significantly reduced (P<0.01) (Fig. 5A and B). Thus, silencing of
hsa_circ_0016760 markedly slowed down the growth of xenograft
tumors in vivo. RT-qPCR revealed that, xenograft tumors of
sh-hsa_circ_0016760 group exhibited significantly decreased
hsa_circ_0016760 expression, increasedmiR-145-5p expression and
decreasedFGF5 expression compared with the sh-NC groups (P<0.01)
(Fig. 5C). Immunohistochemical
staining revealed decreased Ki67-positive signals in the xenograft
tumors of the sh-hsa_circ_0016760 group when compared with the
sh-NC group (Fig. 5D). Therefore,
hsa_circ_0016760 silencing inhibited NSCLC cell growth in
vivo.
Discussion
In recent years, the biological role of circRNAs in
human tumors has attracted increasing attention. Several functions
of circRNAs have been elucidated, including RNA transport,
regulation of translation and protein binding (13). Notably, circRNAs display the
potential of gene regulation and have been confirmed as efficient
sponges for microRNAs (14).
Currently, a number of circRNAs have been revealed to participate
in the regulation of NSCLC by indirectly regulating coding gene
expression via sponging microRNAs (15–17).
The discovery of these circRNAs and related molecular mechanisms in
regulating NSCLC progression provides wide selection for the
targeted treatment of patients. The discovery of more circRNAs is
clinically significant for the targeted treatment of NSCLC
patients. In the present research, hsa_circ_0016760 was identified
as a novel target for NSCLC, as well as an oncogene of NSCLC and
was associated with poor prognosis of patients. In terms of the
mechanism, it was reported that hsa_circ_0016760 exacerbated the
malignant development of NSCLC by enhancing FGF5 expression via
sponging of miR-145-5p.
In the present study, miR-145-5p was identified as a
tumor suppressor gene of NSCLC. Gan et al (18) revealed that miR-145-5p had clinical
value in the diagnosis and treatment of NSCLC. Through
meta-analysis and microRNA microarray analysis, it was determined
that, miR-145-5p expression was significantly decreased in NSCLC
tissues compared with that in adjacent non-tumor tissues and
decreased miR-145-5p expression was closely related to the positive
lymph node metastasis of NSCLC patients. Epithelial-mesenchymal
transition (EMT) is involved in the important biological process of
tumor migration and metastasis. Chang et al (8) demonstrated that miR-145-5p could
suppress the EMT in NSCLC by inhibiting the activity of the c-Jun
N-terminal kinase (JNK) signaling pathway via targeting
mitogen-activated protein kinase kinase kinase 1 (MAP3K1). Results
from the present study also identified that, as a tumor suppressor
gene, miR-145-5p expression was reduced in NSCLC samples. According
to existing literature, miR-145-5p in NSCLC has been reported to be
regulated by multiple long-chain non-coding RNAs, including SNHG1,
JPX and PVT1. These long-chain non-coding RNAs have been revealed
to exhibit cancer-promoting effects in NSCLC by enhancing the
expression of downstream coding genes via sponging miR-145-5p
(19–21). At present, studies on whether
miR-145-5p is regulated by circRNAs in NSCLC are scarce. The
present study reported for the first time, to the best of our
knowledge, that miR-145-5p was sponged by hsa_circ_0016760 in
NSCLC.
In the present study, FGF5 expression was revealed
to be upregulated in NSCLC and FGF5 was demonstrated as a potential
target gene of miR-145-5p. After overexpression of miR-145-5p, the
expression of FGF5 mRNA and protein levels were both decreased.
Thus, miR-145-5p may suppress FGF5 expression at transcription and
translation levels. Rescue experiments revealed that silencing of
FGF5 reversed the promoting effect of hsa_circ_0016760 on NSCLC
cell malignant phenotype, including proliferation, migration and
invasion. Thus, FGF5 was an oncogene in NSCLC. Zhao et al
(9) suggested that FGF5 was
overexpressed in lung adenocarcinoma. High FGF5 expression was an
independent prognostic factor for patients, and was associated with
worse overall survival and relapse-free survival in lung
adenocarcinoma patients. Zhou et al (22) revealed that FGF5 expression was
aberrantly increased in NSCLC tissues and cell lines. The
inhibition of FGF5 significantly suppressed NSCLC cell line
proliferation, migration and invasion. It is suggested that FGF5
may be a promising treatment strategy for NSCLC. Furthermore,
overexpression of FGF5 has also been revealed to contribute to
malignant development in several other human tumors, such as
osteosarcoma, melanoma and breast cancer (23–25).
Similarly, in the present study, FGF5 played a role in the
promotion of the malignant development of NSCLC.
There is a limitation in the present study. It was
observed that miR-145-5p overexpression reduced the expression
levels of FGF5 mRNA and protein. It was speculated that miR-145-5p
may suppress FGF5 expression at both the transcription and
translation levels. However, we could not conduct more experiments
to verify this theory. This point will be the focus of our future
research.
In summary, the present study reported the role and
mechanism of hsa_circ_0016760 in regulating NSCLC progression. It
was demonstrated that hsa_circ_0016760 was an oncogene in NSCLC,
and was related to the poor prognosis of patients. Hsa_circ_0016760
silencing could inhibit the malignant progression of NSCLC in
vitro and in vivo. With regard to the mechanism,
hsa_circ_0016760 exacerbated the malignant development of NSCLC by
enhancing FGF5 expression via sponging miR-145-5p. Therefore,
hsa_circ_0016760 was recommended as a novel potential target for
the treatment of NSCLC.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Major Science
and Technology Project of Changzhou Health Commission (grant no.
ZD201910), the ‘Six One Project’ Research Projects of High-level
Medical Personnel of Jiangsu Province (grant no. LGY2019025), the
Medical Scientific Research Foundation of Jiangsu Commission of
Health (grant no. H2018083), the High-level Talent Selection and
Training Project of The 16th Batch of ‘Six Talent Peak’ of Jiangsu
Province (grant no. WSN-245), the 333 High-Level Talent Training
Project (grant no. 2016, III-0719), the High-Level Medical Talents
Training Project (grant no. 2016CZBJ042) and the Jiangsu Provincial
Medical Youth Talent [Jiangsu Health Scientific Education (2017)
No. 3].
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
ZZ, KY and QW designed the study. ZZ, QW, MZ, and JT
performed the experiments. BZ and KY performed the data analysis.
ZZ and KY wrote the manuscript. All authors read and approved the
manuscript and agree to be accountable for all aspects of the
research in ensuring that the accuracy or integrity of any part of
the work are appropriately investigated and resolved.
Ethics approval and consent to
participate
The present study was approved (approval no.
H3511EC) by the Ethics Committee of the Affiliated Changzhou No. 2
People's Hospital of Nanjing Medical University (Changzhou, China).
All patients volunteered to join the study and written informed
consent was signed by all subjects. Animal experiments involved in
the present study were approved (approval no. 20140351AEC) by the
Animal Ethics Committee of the Affiliated Changzhou No. 2 People's
Hospital of Nanjing Medical University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Brahmer JR, Govindan R, Anders RA, Antonia
SJ, Sagorsky S, Davies MJ, Dubinett SM, Ferris A, Gandhi L, Garon
EB, et al: The society for immunotherapy of cancer consensus
statement on immunotherapy for the treatment of non-small cell lung
cancer (NSCLC). J Immunother Cancer. 6:752018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
He W, Zhang H, Wang Y, Zhou Y, Luo Y, Cui
Y, Jiang N, Jiang W, Wang H, Xu D, et al: CTHRC1 induces non-small
cell lung cancer (NSCLC) invasion through upregulating MMP-7/MMP-9.
BMC Cancer. 18:4002018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen S, Shi F, Zhang W, Zhou Y and Huang
J: miR-744-5p inhibits non-small cell lung cancer proliferation and
invasion by directly targeting PAX2. Technol Cancer Res Treat.
18:15330338198769132019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Nicolaou A, Northoff BH, Zhao Z, Kohlmaier
A, Sass K, Rose-John S, Steffens S, Weber C, Teupser D and Holdt
LM: The ADAM17 metalloproteinase maintains arterial elasticity.
Thromb Haemost. 118:210–213. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhang J, Hou L, Liang R, Chen X, Zhang R,
Chen W and Zhu J: CircDLST promotes the tumorigenesis and
metastasis of gastric cancer by sponging miR-502-5p and activating
the NRAS/MEK1/ERK1/2 signaling. Mol Cancer. 18:802019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang S, Zeng X, Ding T, Guo L, Li Y, Ou S
and Yuan H: Microarray profile of circular RNAs identifies
hsa_circ_0014130 as a new circular RNA biomarker in non-small cell
lung cancer. Sci Rep. 8:28782018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li Y, Hu J, Li L, Cai S, Zhang H, Zhu X,
Guan G and Dong X: Upregulated circular RNA circ_0016760 indicates
unfavorable prognosis in NSCLC and promotes cell progression
through miR-1287/GAGE1 axis. Biochem Biophys Res Commun.
503:2089–2094. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chang Y, Yan W, Sun C, Liu Q, Wang J and
Wang M: miR-145-5p inhibits epithelial-mesenchymal transition via
the JNK signaling pathway by targeting MAP3K1 in non-small cell
lung cancer cells. Oncol Lett. 14:6923–6928. 2017.PubMed/NCBI
|
|
9
|
Zhao T, Qian K and Zhang Y: High
expression of FGF5 is an independent prognostic factor for poor
overall survival and relapse-free survival in lung adenocarcinoma.
J Comput Biol. 27:948–957. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xiao MS and Wilusz JE: An improved method
for circular RNA purification using RNase R that efficiently
removes linear RNAs containing G-quadruplexes or structured 3′
ends. Nucleic Acids Res. 47:8755–8769. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen Y, Peng C, Chen J, Chen D, Yang B, He
B, Hu W, Zhang Y, Liu H, Dai L, et al: WTAP facilitates progression
of hepatocellular carcinoma via m6A-HuR-dependent epigenetic
silencing of ETS1. Mol Cancer. 18:1272019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang X, Yan Y, Lei X, Li A, Zhang H, Dai
Z, Li X, Chen W, Lin W, Chen F, et al: Circular RNA alterations are
involved in resistance to avian leukosis virus subgroup-J-induced
tumor formation in chickens. Oncotarget. 8:34961–34970. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shi P, Sun J, He B, Song H, Li Z, Kong W,
Wang J, Wang J and Xue H: Profiles of differentially expressed
circRNAs in esophageal and breast cancer. Cancer Manag Res.
10:2207–2221. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang Y, Li Y, He H and Wang F: Circular
RNA circ-PRMT5 facilitates non-small cell lung cancer proliferation
through upregulating EZH2 via sponging miR-377/382/498. Gene.
720:1440992019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang H, Wang X, Hu B, Zhang F, Wei H and
Li L: Circular RNA ZFR accelerates non-small cell lung cancer
progression by acting as a miR-101-3p sponge to enhance CUL4B
expression. Artif Cells Nanomed Biotechnol. 47:3410–3416. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chen T, Yang Z, Liu C, Wang L, Yang J,
Chen L and Li W: Circ_0078767 suppresses non-small-cell lung cancer
by protecting RASSF1A expression via sponging miR-330-3p. Cell
Prolif. 52:e125482019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gan TQ, Xie ZC, Tang RX, Zhang TT, Li DY,
Li ZY and Chen G: Clinical value of miR-145-5p in NSCLC and
potential molecular mechanism exploration: A retrospective study
based on GEO, qRT-PCR, and TCGA data. Tumour Biol.
39:10104283176916832017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lu Q, Shan S, Li Y, Zhu D, Jin W and Ren
T: Long noncoding RNA SNHG1 promotes non-small cell lung cancer
progression by up-regulating MTDH via sponging miR-145-5p. FASEB J.
32:3957–3967. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jin M, Ren J, Luo M, You Z, Fang Y, Han Y,
Li G and Liu H: Long non-coding RNA JPX correlates with poor
prognosis and tumor progression in non-small-cell lung cancer by
interacting with miR-145-5p and CCND2. Carcinogenesis. 41:634–645.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wei CM, Zhao XF, Qiu HB, Ming Z, Liu K and
Yan J: The long non-coding RNA PVT1/miR-145-5p/ITGB8 axis regulates
cell proliferation, apoptosis, migration and invasion in non-small
cell lung cancer cells. Neoplasma. 67:802–812. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhou Y, Yu Q, Chu Y, Zhu X, Deng J, Liu Q
and Wang Q: Downregulation of fibroblast growth factor 5 inhibits
cell growth and invasion of human nonsmall-cell lung cancer cells.
J Cell Biochem. Dec 5–2018.(Epub ahead of print).doi:
https://doi.org/10.1002/jcb.28107.
|
|
23
|
Han D, Wang M, Yu Z, Yin L, Liu C, Wang J,
Liu Y, Jiang S, Ren Z and Yin J: FGF5 promotes osteosarcoma cells
proliferation via activating MAPK signaling pathway. Cancer Manag
Res. 11:6457–6466. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ghassemi S, Vejdovszky K, Sahin E,
Ratzinger L, Schelch K, Mohr T, Peter-Vörösmarty B, Brankovic J,
Lackner A, Leopoldi A, et al: FGF5 is expressed in melanoma and
enhances malignancy in vitro and in vivo. Oncotarget.
8:87750–87762. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Huang Y, Wang H and Yang Y: Expression of
fibroblast growth factor 5 (FGF5) and its influence on survival of
breast cancer patients. Med Sci Monit. 24:3524–3530. 2018.
View Article : Google Scholar : PubMed/NCBI
|