Introduction
Lung cancer is the leading cause of
cancer-associated mortality globally, with a 5-year survival rate
<20% (1). Non-small cell lung
cancer (NSCLC) accounts for ~85% of all lung cancer cases and it is
usually diagnosed at an advanced stage (2). Molecularly targeted drugs, including
gefitinib and erlotinib, have been approved for clinical treatment
of NSCLC and have improved the survival and quality of life of
patients with NSCLC (3). However,
the problems of recurrence, metastasis and drug resistance have not
been resolved, resulting in poor outcomes in patients (4). Therefore, it is crucial to identify
new molecular targets for clinical diagnosis and therapy of
NSCLC.
Nucleolar and spindle-associated protein 1 (NUSAP1)
is a microtubule-binding protein that serves an important role in
mitotic progression, spindle assembly and stability (5,6). High
NUSAP1 expression has been identified in various types of cancer,
such as in metastatic prostate cancer (7–14). In
addition, high NUSAP1 expression has been associated with worse
clinical outcomes in several types of tumor, including estrogen
receptor-positive breast cancer, prostate cancer, cervical
carcinoma and hepatocellular carcinoma (HCC) (11–14).
Moreover, NUSAP1 expression is upregulated in cervical carcinoma
and contributes to metastasis by enhancing cancer stem cell traits
and epithelial-to-mesenchymal transition (EMT) (13). In HCC, NUSAP1-knockdown inhibits
cell proliferation and increases apoptotic cell death (14). These findings suggest that NUSAP1 is
involved in the initiation and progression of human cancer.
Notably, Bidkhori et al (15) reported that NUSAP1 expression is
upregulated in lung cancer, and Xu et al (16) demonstrated that NUSAP1-knockdown
inhibits NSCLC cell proliferation, migration and invasion. However,
the role and mechanism of action of NUSAP1 in NSCLC has not been
fully elucidated.
The present study aimed to analyze NUSAP1 expression
in NSCLC tissues and to analyze the association between its
expression and clinicopathological characteristics in patients with
NSCLC. Additionally, the present study aimed to investigate the
biological function and underlying molecular mechanisms of NUSAP1
in NSCLC. NSCLC cell lines stably inhibiting NUSAP1 were
established to investigate its effects on cell proliferation,
colony formation and invasion, and on in vivo
tumorigenicity. Furthermore, the upstream and downstream mechanisms
of NUSAP1 in regulating NSCLC progression were investigated.
Materials and methods
Patients and tissue specimens
A total of 115 NSCLC tissue specimens were collected
from patients who underwent surgical resection at Chongqing General
Hospital (Chongqing, China) between February 2009 and May 2011. The
age of the patients ranged between 37 and 65 years, with a median
age of 52 years. Among the 115 tissues, 35 pairs of human NSCLC and
adjacent normal tissues (2 cm from the tumor) were selected from
these patients and stored at −80°C. All patients provided written
informed consent, and the study protocol was approved by the Ethics
Committee of Chongqing General Hospital (approval no. 2015082104).
None of the patients had received radiotherapy or neoadjuvant
chemotherapy prior to surgery. All the samples used were examined,
and diagnosis was confirmed by two pathologists.
Clinicopathological data, including age, sex, smoking status, tumor
size, histological type, TNM stage (17) and lymph node metastasis, were
available for the patients diagnosed with NSCLC. The patient
samples were categorized into two groups according to the median
NUSAP1 expression (high vs. low expression). The clinical outcomes
of patients with NSCLC were recorded until January 2018. The
clinical information of these patients is summarized in Table I.
 | Table I.Associations between NUSAP1
expression and clinicopathological characteristics of patients with
non-small cell lung cancer. |
Table I.
Associations between NUSAP1
expression and clinicopathological characteristics of patients with
non-small cell lung cancer.
|
| NUSAP1
expression |
|
|---|
|
|
|
|
|---|
| Clinical
factors | Low (n=51) | High (n=64) | P-value |
|---|
| Age, years |
|
| 0.454 |
|
<60 | 30 | 42 |
|
|
≥60 | 21 | 22 |
|
| Sex |
|
| 0.849 |
|
Male | 23 | 30 |
|
|
Female | 28 | 34 |
|
| Smoking status |
|
| 0.573 |
| Never
smoke | 25 | 28 |
|
|
Smoke | 26 | 36 |
|
| Tumor size, cm |
|
| 0.005a |
|
<3 | 35 | 27 |
|
| ≥3 | 16 | 37 |
|
| Histological
type |
|
| 0.187 |
|
Squamous cell carcinoma | 20 | 33 |
|
|
Adenocarcinoma | 31 | 31 |
|
| TNM stage |
|
| 0.004a |
|
I+II | 33 | 24 |
|
|
III+IV | 18 | 40 |
|
| Lymph node
metastasis |
|
| 0.026b |
| No | 36 | 32 |
|
|
Yes | 15 | 32 |
|
Immunohistochemistry (IHC)
IHC examination of NUSAP1 expression was performed
as described previously (18).
NSCLC tissues were fixed in 10% (v/v) formaldehyde at room
temperature for 24 h, embedded in paraffin and cut into 4-µm-thick
sections. Subsequently, the sections were dewaxed using xylene.
Following rehydration in a descending alcohol series (100, 95, 80
and 70% ethanol) at room temperature, antigen retrieval was
performed by microwave treatment with citrate buffer (pH 6.0) for
15 min at 750 W, followed by the addition of 10% goat non-immune
serum (Boster Biological Technology) for blocking at room
temperature for 1 h. The sections were then incubated with a rabbit
anti-NUSAP1 polyclonal antibody (1:400; cat. no. 12024-1-AP;
ProteinTech Group, Inc.) at 4°C overnight, followed by incubation
with a biotin-labeled secondary antibody (1:500; cat. no. ab7089;
Abcam) at 37°C for 30 min. Finally, freshly prepared
3,3′-diaminobenzidine from a DAB Substrate kit (Abcam) was added
for color development.
Images were obtained using a light microscope
(magnification, ×100), and NUSAP1 expression was scored according
to the percentage of positive cells and the staining intensity
(19). The percentage of positive
cells was scored as follows: 0, 0; 1, <5; 2, 5–50; and 3,
>50% positive cells. The staining intensity was determined in
10–20 areas at ×400 magnification by visual assessment, regardless
of the percentage of staining, and the intensity of staining was
classified as follows: 0, negative (no positive cells); 1, weak; 2,
moderate; and 3, strong (20,21).
The final staining scores of NUSAP1 expression in NSCLC tissues
were calculated by multiplying the staining intensity score and the
percentage score. If the final score was ≥4, the protein expression
was considered as high, whereas if the score was ≤3, the protein
expression was considered as low.
Cell culture
Four NSCLC cell lines (A549, H1299, HCC827 and H358)
and one normal human lung epithelial cell line (BEAS-2B) were
obtained from the Cell Bank of Type Culture Collection of the
Chinese Academy of Sciences. H1299, HCC827 and H358 cells were
cultured in RPMI-1640 medium supplemented with 10% FBS (Gibco;
Thermo Fisher Scientific, Inc.) and 1% penicillin/streptomycin
(Invitrogen; Thermo Fisher Scientific, Inc.), and A549 cells were
cultured in F12K medium (Sigma-Aldrich; Merck KGaA) supplemented
with 10% FBS. BEAS-2B cells were cultured in medium from a
Bronchial Epithelial Cell Growth Medium Bullet kit (Clonetics
Corporation). All cell lines were cultured in an incubator with 5%
CO2 at 37°C.
Cell transfection
Short hairpin RNA (shRNA) fragments of NUSAP1,
MEF2D, zinc finger E-box binding homeobox 1 (ZEB1) and E2F
transcription factor 1 (E2F1) were synthesized and constructed into
the U6-shRNA-CMV-puromycin vector by SunBio, Inc. The shRNA
sequences are listed in Table II,
including a universal non-targeting control used as the negative
control (NC). Cell transfection was performed using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions.
H1299 and A549 cells were seeded into 6-well plates at
3×105 cells/well. When cells reached 70–90% confluence,
2.5 µg NUSAP1 shRNA, MEF2D shRNA or NC with 8 µl Lipofectamine 2000
in 250 µl Opti-MEM® (Gibco; Thermo Fisher Scientific,
Inc.) was added into each well. A549 cells were seeded into 6-well
plates at 3×105 cells/well. When cells reached 70–90%
confluence, 2.5 µg ZEB1 shRNA, E2F1 shRNA or NC with 8 µl
Lipofectamine 2000 in 250 µl Opti-MEM was added into each well.
After 72 h of transfection at 37°C, H1299 and A549 cells were
selected with 2 or 1 µg/ml puromycin, respectively, for 14 days.
Finally, a fluorescence microscope (MicroPublisher™ 3.3RTV; Olympus
Corporation; magnification, ×100) was used to evaluate transfection
efficiency. The transfection efficiency was calculated using the
percentage of RFP-positive cells in total cells. Three images at
×100 magnification per sample were used for quantification. The
mean percentage was obtained from three images, and three samples
were used per group. For NUSAP1 and MEF2D overexpression, the
coding sequences of NUSAP1 and MEF2D were subcloned into a pcDNA3.1
vector carrying a neomycin resistance gene by SunBio, Inc. An empty
vector was used as the NC. Cell transfection was performed using
Lipofectamine 2000 according to the manufacturer's instructions.
HCC827 cells were seeded into 6-well plates at 3×105
cells/well. When cells reached 70–90% confluence, 2.5 µg pcDNA3.1
or MEF2D overexpression vectors with 8 µl Lipofectamine 2000 in 250
µl Opti-MEM was added into each well. H1299 and A549 cells
pretreated with NC and MEF2D shRNA were seeded into 6-well plates
at 3×105 cells/well. When cells reached 70–90%
confluence, 2.5 µg pcDNA3.1 or NUSAP1 overexpression vectors with 8
µl Lipofectamine 2000 in 250 µl Opti-MEM was added into each well.
After 72 h of transfection at 37°C, H1299, A549 and HCC827 cells
were selected with 200, 500 or 400 µg/ml G418, respectively, for 14
days. Finally, a fluorescence microscope (MicroPublisher™ 3.3RTV;
Olympus Corporation; magnification, ×100) was used to evaluate
transfection efficiency. The transfection efficiency was calculated
using the percentage of EGFP-positive cells in total cells. Three
images at ×100 magnification per sample were used for
quantification. The mean percentage was obtained from three images,
and three samples were used per group.
 | Table II.Sequences of shRNAs and primers used
for reverse transcription-quantitative PCR. |
Table II.
Sequences of shRNAs and primers used
for reverse transcription-quantitative PCR.
| Name | Sequences |
|---|
| NUSAP1 forward |
5′-TTCTGCTGCTGTTATTAC-3′ |
| NUSAP1 reverse |
5′-GTGTGGTTCATAGTTGAG-3′ |
| MEF2D forward |
5′-CAGCAGCCAGCACTACAGAG-3′ |
| MEF2D reverse |
5′-GGCAGGGATGACCTTGTTTA-3′ |
| GAPDH forward |
5′-AGAAGGCTGGGGCTCATTTG-3′ |
| GAPDH reverse |
5′-AGGGGCCATCCACAGTCTTC-3′ |
| NUSAP1 shRNA
sense |
5′-CCGGCCTCAGGTAACAGAGATTCAACTCGAGTTGAATCTCTGTTACCTGAGGTTTTTTG-3′ |
| NUSAP1 shRNA
antisense |
5′-AATTCAAAAAACCTCAGGTAACAGAGATTCAACTCGAGTTGAATCTCTGTTACCTGAGG-3′ |
| MEF2D shRNA
sense |
5′-CCGGCAACAGCCTAAACAAGGTCATCTCGAGATGACCTTGTTTAGGCTGTTGTTTTTTG-3′ |
| MEF2D shRNA
antisense |
5′-AATTCAAAAAACAACAGCCTAAACAAGGTCACTCGAGATGACCTTGTTTAGGCTGTTG-3′ |
| ZEB1 shRNA
sense |
5′-CCGGTGTCTCCCATAAGTATCAATTCTCGAGAATTGATACTTATGGGAGACATTTTTTG-3′ |
| ZEB1 shRNA
antisense |
5′-AATTCAAAAAATGTCTCCCATAAGTATCAATTCTCGAGAATTGATACTTATGGGAGACA-3′ |
| E2F1 shRNA
sense |
5′-CCGGCAGGATGGATATGAGATGGGACTCGAGTCCCATCTCATATCCATCCTGTTTTTTG-3′ |
| E2F1 shRNA
antisense |
5′-AATTCAAAAAACAGGATGGATATGAGATGGGACTCGAGTCCCATCTCATATCCATCCTG-3′ |
| shNC sense |
5′-CCGGTTCTCCGAACGTGTCACGTTTCAAGAGAACGTGACACGTTCGGAGAATTTTTG-3′ |
| shNC antisense |
5′-AATTCAAAAATTCTCCGAACGTGTCACGTTCTCTTGAAACGTGACACGTTCGGAGAA-3′ |
Western blotting
Western blotting was performed as described
previously (19). Four NSCLC cell
lines (A549, H1299, HCC827 and H358) and one normal human lung
epithelial cell line (BEAS-2B) after transfection were harvested
and lysed using RIPA lysis and extraction buffer (cat. no. 89900;
Thermo Fisher Scientific, Inc.). The Nuclear and Cytoplasmic
Extraction Reagent kit (cat. no. 78835; Thermo Fisher Scientific,
Inc.) was used to isolate the nuclear and cytoplasmic fractions
according to the manufacturer's instructions. The BCA method was
used to determine the protein concentration. Equal amounts of total
protein (30 µg/lane) from each cell lysate were subjected to 10%
SDS-PAGE and transferred onto PVDF membranes. The membranes were
incubated with 5% skimmed milk at room temperature for 1 h.
Subsequently, the membranes were incubated overnight at 4°C with
the following primary antibodies: Rabbit anti-NUSAP1 polyclonal
antibody (1:100; cat. no. 12024-1-AP; ProteinTech Group, Inc.),
rabbit anti-MEF2D polyclonal antibody (1:800; cat. no. ab32845;
Abcam), mouse anti-GAPDH monoclonal antibody (1:20,000; cat. no.
60004-1-Ig; ProteinTech Group, Inc.), rabbit anti-β-catenin
monoclonal antibody (1:9,000; cat. no. ab32572; Abcam), rabbit
anti-c-Myc monoclonal antibody (1:600; cat. no. ab32072; Abcam),
mouse anti-cyclin D1 monoclonal antibody (1:6,000; cat. no.
60186-1-Ig; ProteinTech Group, Inc.), rabbit anti-matrix
metallopeptidase (MMP7) polyclonal antibody (1:500; cat. no.
10374-2-AP; ProteinTech Group, Inc.), rabbit anti-histone H3
polyclonal antibody (1:4,000; cat. no. 17168-1-AP; ProteinTech
Group, Inc.), rabbit anti-ZEB1 polyclonal antibody (1:1,000; cat.
no. 21544-1-AP; ProteinTech Group, Inc.) and rabbit anti-E2F1
polyclonal antibody (1:800; cat. no. 12171-1-AP; ProteinTech Group,
Inc.). After washing with TBS-Tween (0.1% Tween-20), the membranes
were incubated with HRP-conjugated Affinipure goat anti-rabbit
(1:10,000; cat. no. SA00001-2; ProteinTech Group, Inc.) or goat
anti-mouse (1:8,000; cat. no. SA00001-1; ProteinTech Group, Inc.)
at room temperature for 1 h. Finally, the Pierce ECL Western
Blotting Substrate (cat. no. 32106; Thermo Fisher Scientific, Inc.)
was used for visualization. GAPDH and Histone H3 were used as the
internal controls. ImageJ software (version 1.52q; National
Institutes of Health) was used for densitometric analysis, and grey
values were normalized to GAPDH or Histone H3, and expressed as
relative densities.
Reverse transcription-quantitative PCR
(RT-qPCR) analysis
Total RNA was extracted from H1299, A549 and HCC827
cells after transfection using TRIzol® reagent (cat. no.
15596026; Invitrogen; Thermo Fisher Scientific, Inc.) according to
the manufacturer's instructions. Subsequently, cDNA was synthesized
at 42°C for 1 h and 70°C for 10 min using a PrimeScript RT reagent
kit (cat. no. RR047A; Takara Biotechnology Co., Ltd.). qPCR
analysis was performed on an MxPro Mx3000P Sequence Detection
system (Stratagene; Agilent Technologies, Inc.) using a SYBR Prime
Script RT-PCR kit (cat. no. RR066A; Takara Biotechnology Co.,
Ltd.). The thermocycling conditions for PCR were as follows:
Pre-incubation at 95°C for 15 sec, followed by 45 cycles at 95°C
for 5 sec and 60°C for 30 sec. GAPDH was used as the internal
control, and the 2−ΔΔCq analysis method was used to
evaluate NUSAP1 expression (22).
The primer sequences are listed in Table II.
Cell Counting Kit-8 (CCK-8) assay
CCK-8 assay (cat. no. 96992; Sigma-Aldrich; Merck
KGaA) was used to determine NSCLC cell proliferation. H1299 and
A549 cells were seeded in 96-well plates at a density of
1×104 cells/well. Subsequently, 10 µl CCK-8 solution was
added to each well of the plate after 0, 1, 2, 3, 4 or 5 days, and
further incubated for 2 h at 37°C. The absorbance of each well of
the plate was measured at 450 nm using a microculture plate reader
(BioTek Instruments, Inc.).
Transwell assay
The invasion ability of NSCLC cells was analyzed
using a 24-well Transwell chamber with 8.0-µm pore membranes (cat.
no. 3577; Costar; Corning, Inc.). Matrigel® (100 µl;
cat. no. 356234; BD Biosciences) was added to the upper chambers of
the Transwell system for 15 min at 37°C. Subsequently,
1×105 H1299 and A549 cells in 100 µl serum-free medium
(RPMI-1640 or F12K medium) were seeded into the upper Transwell
chambers. The bottom chambers were filled with 500 µl RPMI-1640 or
F12K medium supplemented with 10% FBS. After culturing for 48 h at
37°C, the cells in the upper surface of the filter were removed
with a cotton swab, and the cells on the lower surface of the
filter were fixed with 4% paraformaldehyde at room temperature for
15 min and stained with 0.1% crystal violet for 5 min at room
temperature. Finally, the tumor cells were counted under a light
microscope (IX71; Olympus Corporation) in five random fields at a
magnification of ×200.
Colony formation assay
To analyze the cell colony-forming ability, H1299
and A549 cells were seeded at a concentration of 500 cells/well,
and cultured for 2 weeks at 37°C. Subsequently, the cells were
fixed with 4% paraformaldehyde at room temperature for 15 min and
stained with 0.1% crystal violet for 20 min at room temperature.
The number of colonies containing >50 cells was counted under a
light microscope (IX71; Olympus Corporation; magnification,
×400).
Xenograft model in nude mice
A total of 8 female BALB/c nude mice (age, 5 weeks;
weight, 14–19 g) were purchased from the Shanghai SLAC Laboratory
Animal Co., Ltd., and kept in an animal room at a constant
temperature of 23±1°C and humidity of 50±1% with a 12-h artificial
light/dark cycle and free access to food and water. Animal health
and behavior were monitored every day. All animal experiments were
approved by the Animal Care Committee of Youjiang Medical
University for Nationalities (approval no. 2018092703; Guangxi,
China) and complied with the recommendations of the Chinese
Guidelines for the Care and Use of Laboratory Animals (23). Mice were anesthetized with 3%
isoflurane. A549 cells (1×107 cells in 100 µl PBS)
stably expressing NUSAP1 shRNA (NUSAP1 RNAi) or a universal shNC
were injected subcutaneously into the flanks of BALB/c nude mice
(four in each group). The tumor volume was measured weekly. If the
tumor burden was >10% of the body weight in each mouse, or
tumors became ulcerated, infected or necrotic, or the longest tumor
diameter was >2 cm, euthanasia was used to halt the experiment.
At 5 weeks after injection, all mice were euthanized by
intraperitoneal injection of pentobarbital sodium (200 mg/kg;
ChemicalBook, Inc.), and the animal death was verified through a
combination of criteria, including lack of pulse, breathing,
corneal reflex, response to firm toe pinch, graying of the mucous
membranes and rigor mortis. After euthanasia, the tumors were
resected and the tumor volume was calculated as follows:
Volume=(length × width xwidth)/2. None of the mice presented
multiple tumors before euthanasia.
Luciferase assay
For luciferase reporter assays, the NUSAP1 promoter
containing the MEF2D-binding site (from 1,519 to 1,508 bp) was
amplified from A549 cells by PCR and cloned into the pGL3-Basic
luciferase reporter vector by SunBio, Inc. A mutated NUSAP1
promoter (pNUSAP1-mut) with deletion of the MEF2D-binding site was
also generated by SunBio, Inc. H1299 and A549 cells were
co-transfected with a universal shNC or MEF2D shRNA (MEF2D RNAi)
and wild-type NUSAP1 luciferase reporter vector (pNUSAP1-luc). In
addition, HCC827 cells were co-transfected with a MEF2D pcDNA3.1
vector (MEF2D) or a pcDNA3.1 control vector (pcDNA3.1) and
pNUSAP1-luc or pNUSAP1-mut. After 48 h of transfection using
Lipofectamine 2000 at 37°C, the cells were lysed in lysis buffer,
and the luciferase activity was determined using a Dual-luciferase
reporter assay system (cat. no. E1910; Promega Corporation)
according to the manufacturer's instructions. The firefly
luciferase activity was normalized to the Renilla luciferase
activity.
Bioinformatics and data analysis
The expression levels of NUSAP1 in NSCLC tissues
were investigated using the Gene Expression database of Normal and
Tumor tissues (GENT; http://medical-genome.kribb.re.kr/GENT/) (24). NUSAP1 mRNA and survival data in
NSCLC were obtained from Kaplan-Meier plotter (http://kmplot.com/analysis/). The promoter region of
NUSAP1 was scanned for known transcription factors using the JASPAR
database (http://jaspar.genereg.net/collection/core/) and
GeneCard (https://www.genecards.org/).
Statistical analysis
All data were analyzed using SPSS 20.0 (IBM Corp.)
and are presented as the mean ± SD from three independent
experiments. The correlation between NUSAP1 and MEF2D expression in
tissue specimens was analyzed using Pearson's correlation. The
expression levels of NUSAP1 and MEF2D mRNA in paired tissue
specimens was assessed using paired Student's t-tests. In
functional experiments, the differences between two groups were
assessed using unpaired Student's t-tests, while one-way ANOVA
followed by Tukey's post hoc test was conducted for comparisons of
multiple groups. The associations between NUSAP1 and the
clinicopathological characteristics of patients with NSCLC were
analyzed using Pearson's χ2 test. The Kaplan-Meier
method was used to construct survival curves, and the significance
was analyzed using the log-rank test. Multivariate analysis was
performed using the Cox proportional hazards model. P<0.05 was
considered to indicate a statistically significant difference.
Results
NUSAP1 expression is upregulated in
NSCLC tissues
The expression levels of NUSAP1 in NSCLC tissues
were investigated using the GENT database. The results demonstrated
that NUSAP1 expression was significantly higher in NSCLC tissues
compared with that in normal tissues (Fig. 1A). In addition, the mRNA expression
levels of NUSAP1 in 35 pairs of human NSCLC and adjacent normal
tissues were determined via RT-qPCR. As shown in Fig. 1B, NUSAP1 mRNA expression was
significantly higher in NSCLC tissues compared with that in normal
tissues. Furthermore, results from the Kaplan-Meier plotter
revealed that patients with high NUSAP1 expression exhibited a
shorter survival compared with patients with low expression
(Fig. 1C). Subsequently, IHC was
performed to confirm NUSAP1 protein expression in NSCLC tissues.
NUSAP1 expression was scored according to the percentage of
positive cells and the staining intensity (Fig. S1). The results revealed that NUSAP1
expression in lung adenocarcinoma and lung squamous cell carcinoma
tissues was markedly upregulated compared with that in normal lung
tissues (3.37±2.40 vs. 1.89±2.00; P=0.001; Fig. 1D). Moreover, high NUSAP1 expression
was significantly associated with large tumor size (P=0.005),
advanced TNM stage (P=0.004) and lymph node metastasis (P=0.026),
but not with age (P=0.454), sex (P=0.849), smoking status (P=0.573)
or histological type (P=0.287) (Table
I). In addition, the results of the Kaplan-Meier survival
analysis demonstrated that patients with NSCLC with high NUSAP1
expression had a significantly shorter overall survival than
patients with low expression (P<0.001; Fig. 1E). Furthermore, Cox regression
analysis revealed that high NUSAP1 expression, tumor size and lymph
node metastasis were independent risk factors for poor prognosis in
patients with NSCLC (Table III).
Next, NUSAP1 protein expression was measured in A549, H1299,
HCC827, H358 and BEAS-2B cells. The results demonstrated that the
expression levels of NUSAP1 were significantly higher in A549,
H1299, HCC827 and H358 cells compared with those in BEAS-2B cells
(Fig. 1F).
 | Table III.Cox proportional hazard model
analysis for prognostic factors. |
Table III.
Cox proportional hazard model
analysis for prognostic factors.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Variables | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Age (≥60 vs. <60
years) | 1.153
(0.744–1.786) | 0.524 |
|
|
| Sex (Female vs.
male) | 0.936
(0.597–1.466) | 0.771 |
|
|
| Smoking status
(Smoke vs. never smoke) | 1.516
(0.980–2.346) | 0.062 |
|
|
| Tumor size (≥3 vs.
<3 cm) | 1.600
(1.031–2.483) | 0.036a | 1.680
(1.094–2.580) | 0.018a |
| Histological type
(Adenocarcinoma vs. squamous cell carcinoma) | 1.121
(0.736–1.709) | 0.594 |
|
|
| TNM stage (III+IV
vs. I+II) | 2.022
(0.919–4.452) | 0.080 |
|
|
| Lymph node
metastasis (Yes vs. no) | 2.370
(1.075–5.223) | 0.032a | 3.870
(2.368–6.324) |
<0.001b |
| NUSAP1 (High vs.
now) | 1.824
(1.156–2.877) | 0.010a | 1.868
(1.186–2.940) | 0.007b |
NUSAP1 inhibition suppresses NSCLC
cell proliferation, colony formation and invasion
To evaluate the role of NUSAP1 in NSCLC cell
progression, H1299 and A549 cells were transfected with NUSAP1 RNAi
and a universal NC. As shown in Fig.
S2A, H1299 and A549 cells were successfully transfected with
NUSAP1 RNAi and a universal NC. Western blotting assays
demonstrated that H1299 and A549 cells transfected with NUSAP1 RNAi
exhibited significantly decreased NUSAP1 protein expression
compared with cells transfected with the NC (Fig. 2A). The cell function assays revealed
that NUSAP1 downregulation significantly suppressed the
proliferative (Fig. 2B),
colony-forming (Fig. 2C) and
invasive (Fig. 2D) abilities of
H1299 and A549 cells.
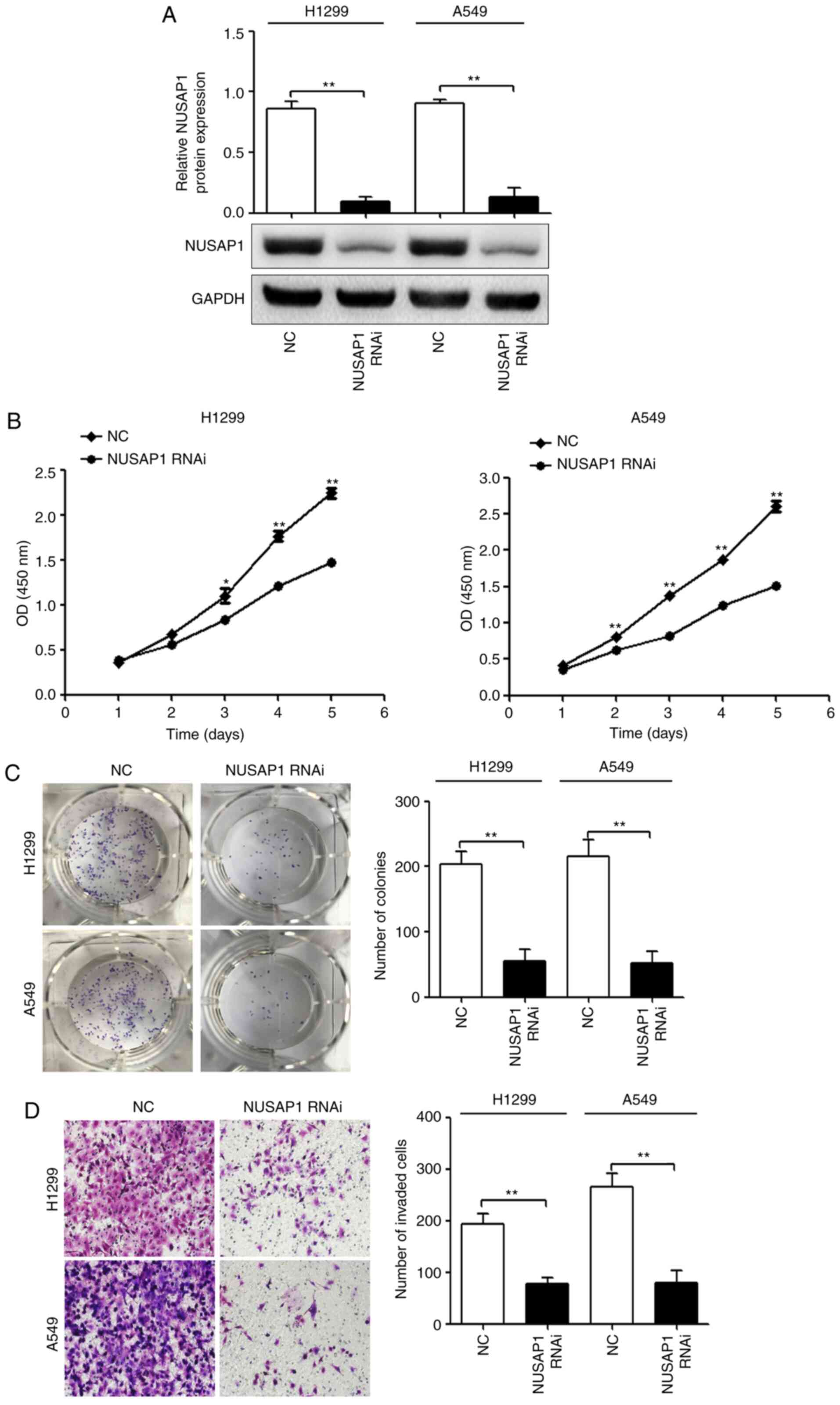 | Figure 2.NUSAP1 acts as an oncogene in NSCLC
cells. H1299 and A549 cells were transfected with a NUSAP1 shRNA
plasmid (NUSAP1 RNAi) or a universal NC plasmid. (A) NUSAP1 protein
expression was determined by western blotting. Proliferation,
colony formation and invasion of H1299 and A549 cells transfected
with NUSAP1 RNAi or a universal NC were determined by (B) Cell
Counting Kit-8, (C) colony formation and (D) Transwell assays,
respectively (magnification, ×200). *P<0.05; **P<0.01 vs.
NUSAP1 RNAi. NC, negative control; NSCLC, non-small cell lung
cancer; NUSAP1, nucleolar and spindle-associated protein 1; shRNA,
short hairpin RNA; RNAi, RNA interference; OD, optical density. |
NUSAP1-knockdown inhibits the
tumorigenic ability of NSCLC cells
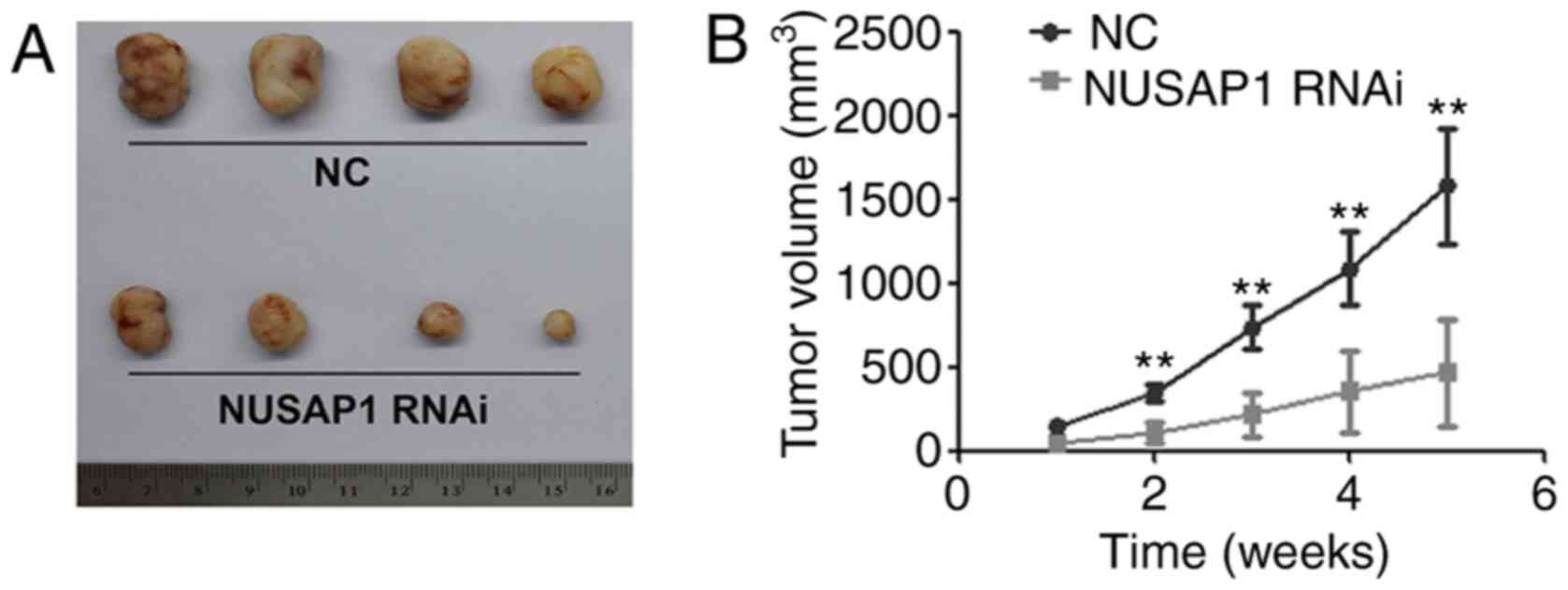
To confirm the effect of NUSAP1 on NSCLC growth
in vivo, A549 cells transfected with NUSAP1 RNAi and a
universal NC were subcutaneously injected into 5-week-old nude
mice. The results demonstrated that the mean tumor volume in the
group injected with NUSAP1-knockdown cells was significantly
smaller compared with that in the NC group (Fig. 3).
MEF2D transcriptionally regulates
NUSAP1 expression
To investigate the molecular mechanisms that control
NUSAP1 expression in NSCLC, the promoter region of NUSAP1 was
scanned for known transcription factors using the JASPAR database
and GeneCard. Several putative transcription factors, including
MEF2D, ZEB1 and E2F1, were selected. To determine whether MEF2D,
ZEB1 and E2F1 affected NUSAP1 expression, A549 cells were
transfected with shRNA plasmids against MEF2D (MEF2D RNAi), ZEB1
(ZEB1 RNAi) or E2F1 (E2F1 RNAi), or a universal NC plasmid. The
protein expression levels of MEF2D, ZEB1 and E2F1 were determined
by western blotting. The present results revealed that the protein
expression levels of MEF2D, ZEB1 and E2F1 were significantly
inhibited by MEF2D RNAi, ZEB1 RNAi and E2F1 RNAi, respectively
(Fig. S3A). As shown in Fig. S3B, MEF2D-knockdown significantly
inhibited the mRNA expression levels of NUSAP1; however, neither
ZEB1-nor E2F1-knockdown repressed the mRNA expression levels of
NUSAP1. In addition, the results revealed that the promoter region
(from 1,519 to 1,508 bp) of NUSAP1 had a putative MEF2D-binding
site (Fig. 4A). Therefore, it was
hypothesized that MEF2D, as an important transcription factor, may
transcriptionally regulate NUSAP1 expression, which may be one of
the key regulatory mechanisms underlying the overexpression of
NUSAP1 in NSCLC. To determine whether NUSAP1 was a direct target of
MEF2D, MEF2D-knockdown and overexpressing cell lines were
constructed using H1299, A549 and HCC827 cells. As shown in
Fig. S2A and B, H1299 and A549
cells were successfully transfected with a cell transfection
efficiency >95%. Western blot assays demonstrated that H1299 and
A549 cells transfected with MEF2D RNAi exhibited significantly
decreased MEF2D protein expression compared with cells transfected
with the NC, while HCC827 cells transfected with MEF2D
overexpression vectors exhibited significantly increased MEF2D
protein expression compared with control cells (Fig. 4B). In addition, RT-qPCR and western
blot analyses revealed that MEF2D-knockdown significantly inhibited
NUSAP1 mRNA and protein expression in H1299 and A549 cells
(Fig. 4C and D). On the other hand,
MEF2D overexpression in HCC827 cells significantly increased NUSAP1
mRNA and protein expression (Fig. 4C
and D). Subsequently, to determine whether NUSAP1 is a direct
target of MEF2D, wild-type and mutant NUSAP1 promoter luciferase
reporter vectors were constructed. As shown in Fig. 4E, MEF2D downregulation significantly
decreased the promoter activity of NUSAP1 in H1299 and A549 cells.
In addition, MEF2D upregulation induced the promoter activity of
NUSAP1 in HCC827 cells, while the mutant NUSAP1 promoter abolished
the promotional effect of MEF2D upregulation on luciferase activity
(Fig. 4F). These results supported
the hypothesis that MEF2D regulated NUSAP1 transcription by binding
to the NUSAP1 promoter. Additionally, the GENT database revealed
that MEF2D mRNA expression was significantly upregulated in NSCLC
tissues compared with in normal tissues and was positively
correlated with NUSAP1 expression (Fig.
4G). In the present study, MEF2D mRNA expression in the 35
pairs of human NSCLC and adjacent normal tissues was determined via
RT-qPCR. As shown in Fig. 4H, MEF2D
mRNA expression was significantly higher in NSCLC tissues compared
with that in normal tissues (P<0.001). In addition, MEF2D mRNA
expression was positively correlated with NUSAP1 mRNA expression
(Fig. 4H).
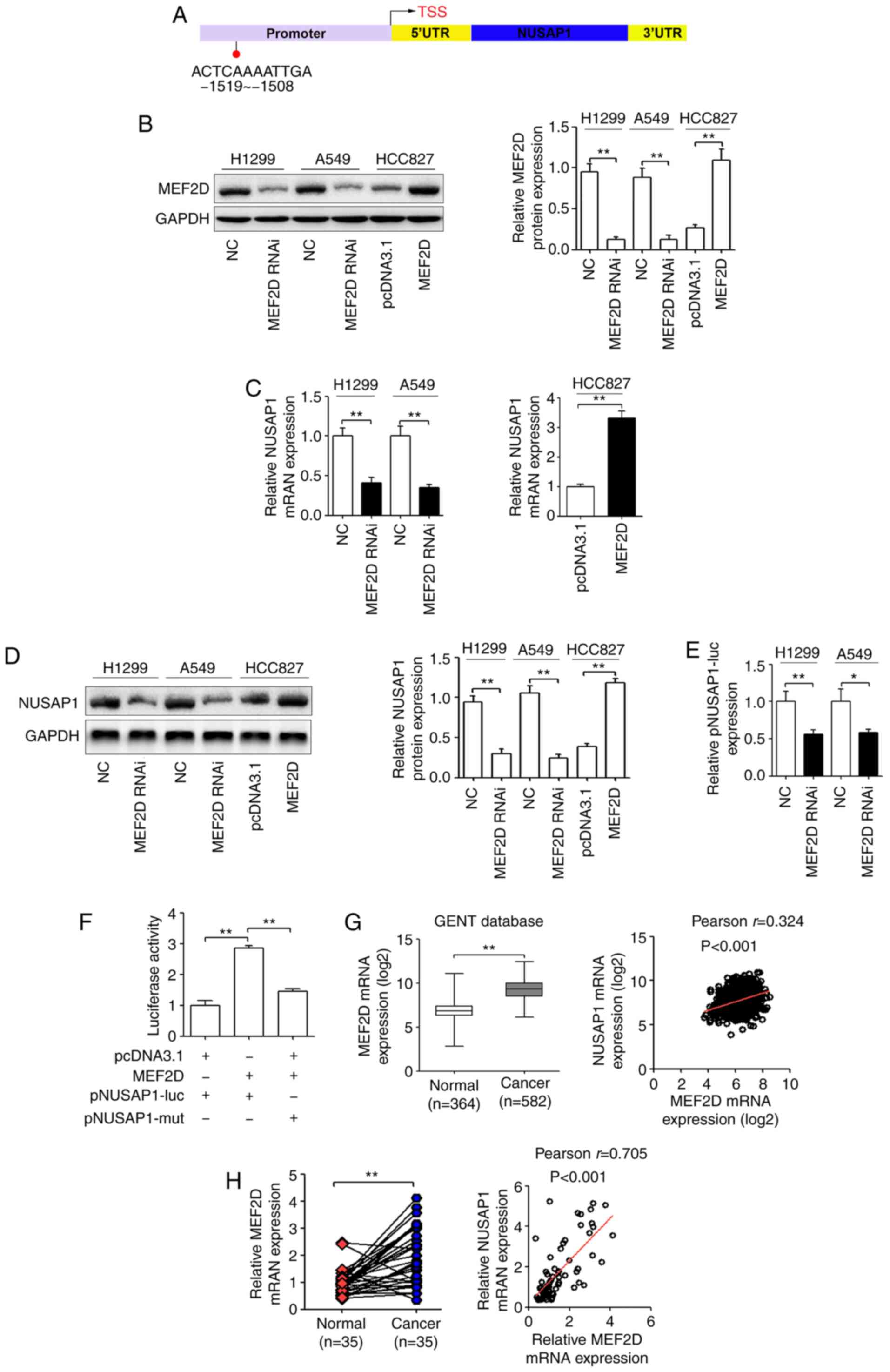 | Figure 4.NUSAP1 serves as the effector of
MEF2D in NSCLC cells. (A) Prediction of the putative MEF2D-binding
site in the promoter of NUSAP1 based on the JASPAR database. H1299
and A549 cells were transfected with a MEF2D shRNA plasmid (MEF2D
RNAi) or a universal NC, and HCC827 cells were transfected with a
MEF2D overexpression plasmid or a pcDNA3.1 control plasmid. (B)
Protein expression levels of MEF2D. (C) mRNA and (D) protein
expression levels of NUSAP1 determined via RT-qPCR and western
blotting, respectively. (E) Luciferase reporter assay demonstrated
that MEF2D-knockdown significantly inhibited the luciferase
activity of the wild-type NUSAP1 promoter reporter gene
(pNUSAP1-luc). (F) MEF2D overexpression induced the promoter
activity of NUSAP1 in HCC827 cells, while the mutated NUSAP1
promoter reversed the effect of MEF2D overexpression. (G) MEF2D
expression was upregulated in lung cancer tissues compared with in
normal lung tissues from the GENT database and was positively
correlated with NUSAP1 expression in lung cancer according to this
database. (H) RT-qPCR analysis of MEF2D mRNA expression in NSCLC
and matched adjacent normal lung tissues. Pearson's correlation
analysis was used to explore the association between MEF2D and
NUSAP1 expression. *P<0.05; **P<0.01. MEF2D, myocyte enhancer
factor 2D; NC, negative control; NSCLC, non-small cell lung cancer;
NUSAP1, nucleolar and spindle-associated protein 1; RT-qPCR,
reverse transcription-quantitative PCR; shRNA, short hairpin RNA;
RNAi, RNA interference; UTR, untranslated region; mut, mutated;
GENT, Gene Expression of Normal and Tumor tissues. |
NUSAP1 restoration reverses the
effects of MEF2D-knockdown on NSCLC cells
To confirm that MEF2D exerts its effects on NSCLC
cells via NUSAP1, NUSAP1 overexpression plasmids were constructed
and transfected into H12299 and A549 cells. As shown in Fig. 5A, NUSAP1 protein expression was
significantly increased in H12299 and A549 cells. Subsequently,
MEF2D RNAi together with NUSAP1 overexpression plasmids were
transfected into H12299 and A549 cells. As confirmed via western
blotting, NUSAP1 expression was restored in MEF2D RNAi-transfected
NSCLC cells using the NUSAP1 expression plasmid (Fig. 5B). In addition, it was observed that
the restoration of NUSAP1 expression rescued the proliferation,
colony formation and invasion of NSCLC cells suppressed by
MEF2D-knockdown (Fig. 5C-E).
Both MEF2D and NUSAP1 promote the
activation of the Wnt/β-catenin signaling pathway
In NSCLC, Wnt/β-catenin signaling is involved in
promoting tumor aggressiveness and resistance to chemotherapy and
radiation (25). In addition,
NUSAP1 induces cervical cancer cell metastasis by activating
Wnt/β-catenin signaling (13). In
gastric cancer, MEF2D contributes to cancer cell proliferation by
regulating the activation of the Wnt/β-catenin signaling pathway
(26). Thus, Wnt/β-catenin
signaling may contribute to MEF2D/NUSAP1-regulated NSCLC
progression. The results of the present study revealed that either
MEF2D- or NUSAP1-knockdown repressed the accumulation and nuclear
translocation of β-catenin and the translation of Wnt targets,
including cyclin D1, c-Myc and MMP7 (Fig. 6A and B).
 | Figure 6.Both MEF2D and NUSAP1 activate the
Wnt/β-catenin signaling pathway in NSCLC. (A) H1299 and A549 cells
were transfected with a NUSAP1 shRNA plasmid (NUSAP1 RNAi), a MEF2D
shRNA plasmid (MEF2D RNAi) or a universal NC plasmid. The
accumulation and nuclear translocation of β-catenin and the
translation of Wnt targets, including cyclin D1, c-Myc and MMP7,
were determined by western blotting. (B) Quantification of western
blotting results from triplicates. **P<0.01. MEF2D, myocyte
enhancer factor 2D; NC, negative control; NSCLC, non-small cell
lung cancer; NUSAP1, nucleolar and spindle-associated protein 1;
shRNA, short hairpin RNA; RNAi, RNA interference; MMP, matrix
metallopeptidase. |
MEF2D promotes the activation of the
Wnt/β-catenin signaling pathway via NUSAP1
To further determine the role of NUSAP1 in the
effects of MEF2D on the activation of the Wnt/β-catenin signaling
pathway, NUSAP1 expression was rescued using NUSAP1 overexpression
plasmid vectors in cells transfected with MEF2D RNAi. As shown in
Fig. 7A and B, NUSAP1
overexpression reversed the suppressive effects of MEF2D
downregulation on the activation of the Wnt/β-catenin signaling
pathway. Overall, the present findings indicated that MEF2D may
promote NSCLC progression via the NUSAP1/Wnt/β-catenin signaling
axis.
Discussion
NSCLC remains one of the most lethal types of cancer
worldwide, largely due to the majority of patients being diagnosed
at the locally advanced or metastatic stage (27). Thus, exploring new biomarkers and
elucidating the underlying mechanisms of NSCLC progression are
fundamental for the development of novel therapeutic approaches for
NSCLC. As reported by Li et al (13), NUSAP1 is upregulated in cervical
cancer tissues and cell lines, and is associated with metastasis
and poor clinical outcomes of patients. Additionally, a previous
study revealed that NUSAP1 is upregulated in lung cancer (15). Thus, NUSAP1 may be a novel
prognostic marker of NSCLC. In the present study, the results from
the GENT database, RT-qPCR analysis and IHC staining demonstrated
that the mRNA and protein expression levels of NUSAP1 were
significantly upregulated in NSCLC tissues compared with those in
normal tissues. High NUSAP1 expression was significantly associated
with large tumor size, advanced TNM stage and lymph node
metastasis. Furthermore, multivariate Cox regression analysis
revealed that NUSAP1 expression was an independent risk factor for
shorter overall survival. The present results indicated the
prognostic significance of NUSAP1 in NSCLC.
Recently, accumulating evidence has indicated that
NUSAP1 acts as an oncogene in different types of malignant tumor
(9,13,14,28,29).
For example, NUSAP1 inhibition in HCC Huh7 cells repressed their
proliferation, survival, migration and growth as xenograft tumors
in nude mice (14). Overexpression
of NUSAP1 can modulate the expression levels of family with
sequence similarity 101, member B, thereby enhancing the invasion
and migration of prostate cancer cells (9). Furthermore, NUSAP1-knockdown represses
cell proliferation, migration and invasion in human colorectal
cancer (28). In addition, NUSAP1
enhances the invasive ability of astrocytoma cells by activating
the Hedgehog signaling pathway (29). Xu et al (16) demonstrated that NUSAP1-knockdown
inhibits NSCLC cell proliferation, migration and invasion by
regulating BTG2/PI3K/Akt signaling. The results of the present
study revealed that NUSAP1 downregulation suppressed NSCLC cell
proliferation, colony formation and invasion. Moreover, NUSAP1
downregulation in NSCLC cells markedly decreased their
tumorigenicity in nude mice. Thus, NUSAP1 functioned as an oncogene
in NSCLC cells. However, the mechanism underlying the role of
NUSAP1 in the regulation of NSCLC progression remains unknown.
Therefore, further studies are required to explore the potential
mechanism of action of NUSAP1 in NSCLC progression.
Several studies have reported that transcription
factors, such as E2F1 and lin-9 DREAM MuvB core complex component
can transcriptionally regulate NUSAP1 by binding to the NUSAP1
promoter region (30,31). Thus, regulation by transcription
factors may be a key mechanism underlying the high expression
levels of NUSAP1 in NSCLC. To further explore the mechanism of
action of NUSAP1 in NSCLC, known transcription factors were
searched using the JASPAR database and GeneCard. Several putative
transcription factors, including MEF2D, ZEB1 and E2F1, were
selected. It was previously reported that MEF2D directly binds to
the region upstream from the transcription sites of reprimo
TP53-dependent G2 arrest mediator homolog, cyclin-dependent kinase
inhibitor 1A and growth arrest and DNA damage inducible α/β
(GADD45A/B), and regulates the activity of these promoters in HCC
(32). Additionally, MEF2D is
involved in the regulation of tumor cell proliferation, invasion
and migration in NSCLC (33,34).
ZEB1 expression is associated with tumor grade and metastasis in
lung cancer, and has been reported to induce invasion and
metastases in NSCLC (35). E2F1
expression is significantly increased in lung cancer and is
required for tumor growth (36). In
the present study, MEF2D-knockdown significantly inhibited the mRNA
expression levels of NUSAP1; however, neither ZEB1-nor
E2F1-knockdown repressed the mRNA expression levels of NUSAP1. In
addition, MEF2D-knockdown inhibited NUSAP1 expression at both the
mRNA and protein level, while MEF2D overexpression increased the
mRNA and protein expression levels of NUSAP1. Further investigation
indicated that NUSAP1 was a direct target of MEF2D in NSCLC. First,
the luciferase assay revealed that MEF2D inhibition significantly
decreased the promoter activity of NUSAP1. Second, MEF2D
overexpression induced the promoter activity of NUSAP1 in HCC827
cells, while the mutant NUSAP1 promoter abolished the promotional
effect of MEF2D overexpression on luciferase activity. Third, MEF2D
expression was significantly upregulated in NSCLC and was
positively correlated with NUSAP1 expression. Fourth,
MEF2D-knockdown exerted an effect similar to that of NUSAP1
depletion, including inhibition of NSCLC cell proliferation, colony
formation and invasion. Furthermore, NUSAP1 restoration reversed
the inhibition of proliferation, colony formation and invasion of
NSCLC cells caused by MEF2D-knockdown. The current results
indicated that MEF2D regulated the proliferation, colony formation
and invasion of NSCLC cells via NUSAP1.
It is well known that the Wnt/β-catenin signaling
pathway serves a pivotal role in regulating NSCLC cell
proliferation, motility and invasion (25). Additionally, NUSAP1 upregulation
promotes cervical cancer cell metastasis by activating
Wnt/β-catenin signaling (13). In
vascular smooth muscle cells, MEF2D induces cell proliferation via
regulating the Wnt/β-catenin signaling pathway (37). In gastric cancer, MEF2D-knockdown
inhibits cell proliferation by regulating the activation of
Wnt/β-catenin signaling (26).
Consistently with the aforementioned studies, the present study
revealed that either MEF2D- or NUSAP1-knockdown repressed the
accumulation and nuclear translocation of β-catenin and the
translation of Wnt targets, including cyclin D1 and c-Myc, which
are associated with cell proliferation (38,39),
and MMP7, which is associated with cell mobility (40). Hence, it was inferred that MEF2D may
activate the Wnt/β-catenin signaling pathway by regulating NUSAP1
expression in NSCLC cells. The current results demonstrated that
NUSAP1 overexpression reversed the effect of MEF2D-knockdown on the
accumulation and nuclear translocation of β-catenin and cyclin D1,
c-Myc and MMP7 protein expression. Overall, the present findings
suggested that the regulation of NUSAP1 by MEF2D may contribute to
the activation of the Wnt/β-catenin signaling pathway.
p53, a tumor suppressor, serves an important role in
repressing malignant progression and is mutated in >50% of human
primary tumors (41). In addition,
mutant p53 occurs in ~50% of NSCLC cases and promotes NSCLC cell
proliferation, invasion and EMT (42,43).
Notably, mutant p53 (R175H) regulates the transcriptional
expression of MEF2D in SKOV3 cells (44). In the present study, NUSAP1 was
involved in the regulation of proliferation and invasion. This
property may be shared by mutant p53. Thus, the association between
mutant p53 and the MEF2D/NUSAP1 pathway requires further
exploration. However, there are some limitations in the present
study. For example, the association between mutant p53 and MEF2D
expression was not investigated in NSCLC tissues. In addition,
whether mutant p53 may drive NSCLC progression by directly
regulating MEF2D requires further confirmation.
In conclusion, NUSAP1 expression was upregulated in
NSCLC tissues. Additionally, either NUSAP1 or MEF2D depletion
inhibited the proliferation, colony formation and invasion of NSCLC
cells. Mechanistically, MEF2D directly regulated the
transcriptional expression of NUSAP1 by binding to its promoter in
NSCLC cells. In addition, MEF2D may regulate NSCLC progression via
modulating the NUSAP1/Wnt/β-catenin signaling axis, which may
indicate new targets for NSCLC therapy in the future.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the Science and
Technology Project of Yuzhong, Chongqing (grant no. 20150128), the
Natural Science Foundation of Guangxi (grant no. 2019GXNSFAA245066)
and the Education Department Science Foundation of Guangxi Zhuang
Autonomous Region (grant no. 2017KY0511).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
BL, WH and SH designed the study. BL, PW, JX, XF, GY
and SL performed the experiments. BL, BC, ZZ, WL and HW analyzed
and interpreted the data. BL and WH wrote the manuscript. SH
supervised the study. All authors have read and approved the final
version of the manuscript and agree to be accountable for all
aspects of the research in ensuring that the accuracy and integrity
of any part of the work are appropriately investigated and
resolved.
Ethics approval and consent to
participate
The protocol of the present study was approved by
the Ethics Committee of Chongqing General Hospital (Chongqing,
China). All patients provided written informed consent. All animal
studies were approved by the Animal Care Committee of Youjiang
Medical University for Nationalities (Guangxi, China) and complied
with the recommendations of the Chinese Guidelines for the Care and
Use of Laboratory Animals.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 67:7–30. 2018. View Article : Google Scholar
|
|
2
|
Domvri K, Zarogoulidis P, Darwiche K,
Browning RF, Li Q, Turner JF, Kioumis I, Spyratos D, Porpodis K,
Papaiwannou A, et al: Molecular targeted drugs and biomarkers in
NSCLC, the evolving role of individualized therapy. J Cancer.
4:736–754. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kim KW, Myers CJ, Jung DK and Lu B:
NVP-BEZ-235 enhances radiosensitization via blockade of the
PI3K/mTOR pathway in cisplatin-resistant non-small cell lung
carcinoma. Genes Cancer. 5:293–302. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hussain S, Benavente SB, Nascimento E,
Dragoni I, Kurowski A, Gillich A, Humphreys P and Frye M: The
nucleolar RNA methyltransferase Misu (NSun2) is required for
mitotic spindle stability. J Cell Biol. 186:27–40. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Li C, Zhang Y, Yang Q, Ye F, Sun SY, Chen
ES and Liou YC: NuSAP modulates the dynamics of kinetochore
microtubules by attenuating MCAK depolymerisation activity. Sci
Rep. 6:187732016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ribbeck K, Groen AC, Santarella R,
Bohnsack MT, Raemaekers T, Köcher T, Gentzel M, Görlich D, Wilm M,
Carmeliet G, et al: NuSAP, a mitotic RanGTP target that stabilizes
and cross-links microtubules. Mol Biol Cell. 17:2646–2660. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kokkinakis DM, Liu X and Neuner RD:
Modulation of cell cycle and gene expression in pancreatic tumor
cell lines by methionine deprivation (methionine stress):
Implications to the therapy of pancreatic adenocarcinoma. Mol
Cancer Ther. 4:1338–1348. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ryu B, Kim DS, DeLuca AM and Alani RM:
Comprehensive expression profiling of tumor cell lines identifies
molecular signatures of melanoma progression. PLoS One. 2:e5942007.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gordon CA, Gong X, Ganesh D and Brooks JD:
NUSAP1 promotes invasion and metastasis of prostate cancer.
Oncotarget. 8:29935–29950. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen DT, Nasir A, Culhane A, Venkataramu
C, Fulp W, Rubio R, Wang T, Agrawal D, McCarthy SM, Gruidl M, et
al: Proliferative genes dominate malignancy-risk gene signature in
histologically-normal breast tissue. Breast Cancer Res Treat.
119:335–346. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu R, Guo CX and Zhou HH: Network-based
approach to identify prognostic biomarkers for estrogen
receptor-positive breast cancer treatment with tamoxifen. Cancer
Biol Ther. 16:317–324. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cuzick J, Swanson GP, Fisher G, Brothman
AR, Berney DM, Reid JE, Mesher D, Speights VO, Stankiewicz E,
Foster CS, et al: Prognostic value of an RNA expression signature
derived from cell cycle proliferation genes in patients with
prostate cancer: A retrospective study. Lancet Oncol. 12:245–255.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li H, Zhang W, Yan M, Qiu J, Chen J, Sun
X, Chen X, Song L and Zhang Y: Nucleolar and spindle associated
protein 1 promotes metastasis of cervical carcinoma cells by
activating Wnt/β-catenin signaling. J Exp Clin Cancer Res.
38:332019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Roy S, Hooiveld GJ, Seehawer M, Caruso S,
Heinzmann F, Schneider AT, Frank AK, Cardenas DV, Sonntag R, Luedde
M, et al: microRNA 193a-5p regulates levels of Nucleolar- and
spindle-associated Protein 1 to suppress hepatocarcinogenesis.
Gastroenterology. 155:1951–1966. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bidkhori G, Narimani Z, Hosseini Ashtiani
S, Moeini A, Nowzari-Dalini A and Masoudi-Nejad A: Reconstruction
of an integrated genome-scale co-expression network reveals key
modules involved in lung adenocarcinoma. PLoS One. 8:e675522013.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xu Z, Wang Y, Xiong J, Cui F, Wang L and
Peng H: NUSAP1 knockdown inhibits cell growth and metastasis of
non-small-cell lung cancer through regulating BTG2/PI3K/Akt
signaling. J Cell Physiol. 235:3886–3893. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kay FU, Kandathil A, Batra K, Saboo SS,
Abbara S and Rajiah P: Revisions to the tumor, node, metastasis
staging of lung cancer (8th edition): Rationale, radiologic
findings and clinical implications. World J Radiol. 9:269–279.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Pinheiro C, Longatto-Filho A, Scapulatempo
C, Ferreira L, Martins S, Pellerin L, Rodrigues M, Alves VA,
Schmitt F and Baltazar F: Increased expression of monocarboxylate
transporters 1, 2, and 4 in colorectal carcinomas. Virchows Arch.
452:139–146. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lin JX, Xie XS, Weng XF, Qiu SL, Xie JW,
Wang JB, Lu J, Chen QY, Cao LL, Lin M, et al: Overexpression of
IC53d promotes the proliferation of gastric cancer cells by
activating the AKT/GSK3β/cyclin D1 signaling pathway. Oncol Rep.
41:2739–2752. 2019.PubMed/NCBI
|
|
20
|
Wang CJ, Zhou ZG, Holmqvist A, Zhang H, Li
Y, Adell G and Sun XF: Survivin expression quantified by Image
Pro-Plus compared with visual assessment. Appl Immunohistochem Mol
Morphol. 17:530–535. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Knutsen A, Adell G and Sun XF: Survivin
expression is an independent prognostic factor in rectal cancer
patients with and without preoperative radiotherapy. Int J Radiat
Oncol Biol Phys. 60:149–155. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
National Research Council (US) Committee
for the Update of the Guide for the Care and Use of Laboratory
animals, . Guide for the Care and Use of Laboratory Animals. 8th
edition. National Academies Press; Washington, DC: 2011, https://www.ncbi.nlm.nih.gov/books/NBK54050/
|
|
24
|
Rhodes DR, Ateeq B, Cao Q, Tomlins SA,
Mehra R, Laxman B, Kalyana-Sundaram S, Lonigro RJ, Helgeson BE,
Bhojani MS, et al: AGTR1 overexpression defines a subset of breast
cancer and confers sensitivity to losartan, an AGTR1 antagonist.
Proc Natl Acad Sci USA. 106:10284–10289. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Stewart DJ: Wnt signaling pathway in
non-small cell lung cancer. J Natl Cancer Inst. 106:djt3562014.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Xu K and Zhao YC: MEF2D/Wnt/β-catenin
pathway regulates the proliferation of gastric cancer cells and is
regulated by microRNA-19. Tumour Biol. 37:9059–9069. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
I H and Cho JY: Lung cancer biomarkers.
Adv Clin Chem. 72:107–170. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Han G, Wei Z, Cui H, Zhang W, Wei X, Lu Z
and Bai X: NUSAP1 gene silencing inhibits cell proliferation,
migration and invasion through inhibiting DNMT1 gene expression in
human colorectal cancer. Exp Cell Res. 367:216–221. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wu X, Xu B, Yang C, Wang W, Zhong D, Zhao
Z, He L, Hu Y, Jiang L, Li J, et al: Nucleolar and spindle
associated protein 1 promotes the aggressiveness of astrocytoma by
activating the Hedgehog signaling pathway. J Exp Clin Cancer Res.
36:1272017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gulzar ZG, McKenney JK and Brooks JD:
Increased expression of NuSAP in recurrent prostate cancer is
mediated by E2F1. Oncogene. 32:70–77. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Reichert N, Wurster S, Ulrich T, Schmitt
K, Hauser S, Probst L, Götz R, Ceteci F, Moll R, Rapp U and Gaubatz
S: Lin9, a subunit of the mammalian DREAM complex, is essential for
embryonic development, for survival of adult mice, and for tumor
suppression. Mol Cell Biol. 30:2896–2908. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ma L, Liu J, Liu L, Duan G, Wang Q, Xu Y,
Xia F, Shan J, Shen J, Yang Z, et al: Overexpression of the
transcription factor MEF2D in hepatocellular carcinoma sustains
malignant character by suppressing G2-M transition genes. Cancer
Res. 74:1452–1462. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhu HX, Shi L, Zhang Y, Zhu YC, Bai CX,
Wang XD and Zhou JB: Myocyte enhancer factor 2D provides a
cross-talk between chronic inflammation and lung cancer. J Transl
Med. 15:652017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhang R, Zhang Y and Li H:
miR-1244/myocyte enhancer factor 2D regulatory loop contributes to
the growth of lung carcinoma. DNA Cell Biol. 34:692–700. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Su L, Luo Y, Yang Z, Yang J, Yao C, Cheng
F, Shan J, Chen J, Li F, Liu L, et al: MEF2D transduces
microenvironment stimuli to ZEB1 to promote epithelial-mesenchymal
transition and metastasis in colorectal cancer. Cancer Res.
76:5054–5067. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chen L, Wei T, Si X, Wang Q, Li Y, Leng Y,
Deng A, Chen J, Wang G, Zhu S and Kang J: Lysine acetyltransferase
GCN5 potentiates the growth of non-small cell lung cancer via
promotion of E2F1, Cyclin D1, and Cyclin E1 expression. J Biol
Chem. 288:14510–14521. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Li K, Pan J, Wang J, Liu F and Wang L:
MiR-665 regulates VSMCs proliferation via targeting FGF9 and MEF2D
and modulating activities of Wnt/β-catenin signaling. Am J Transl
Res. 9:4402–4414. 2017.PubMed/NCBI
|
|
38
|
Shang S, Hua F and Hu ZW: The regulation
of β-catenin activity and function in cancer: Therapeutic
opportunities. Oncotarget. 8:33972–33989. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhou P, Jiang W, Zhang YJ, Kahn SM,
Schieren I, Santella RM and Weinstein IB: Antisense to cyclin D1
inhibits growth and reverses the transformed phenotype of human
esophageal cancer cells. Oncogene. 11:571–580. 1995.PubMed/NCBI
|
|
40
|
Jiang Q, He M, Ma MT, Wu HZ, Yu ZJ, Guan
S, Jiang LY, Wang Y, Zheng DD, Jin F and Wei MJ: MicroRNA-148a
inhibits breast cancer migration and invasion by directly targeting
WNT-1. Oncol Rep. 35:1425–1432. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yue X, Zhao Y, Xu Y, Zheng M, Feng Z and
Hu W: Mutant p53 in cancer: Accumulation, gain-of-function, and
therapy. J Mol Biol. 429:1595–1606. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Sankala H, Vaughan C, Wang J, Deb S and
Graves PR: Upregulation of the mitochondrial transport protein,
Tim50, by mutant p53 contributes to cell growth and
chemoresistance. Arch Biochem Biophys. 512:52–60. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Muller PA, Trinidad AG, Timpson P, Morton
JP, Zanivan S, van den Berghe PV, Nixon C, Karim SA, Caswell PT,
Noll JE, et al: Mutant p53 enhances MET trafficking and signalling
to drive cell scattering and invasion. Oncogene. 32:1252–1265.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Buganim Y, Kalo E, Brosh R, Besserglick H,
Nachmany I, Rais Y, Stambolsky P, Tang X, Milyavsky M, Shats I, et
al: Mutant p53 protects cells from
12-O-tetradecanoylphorbol-13-acetate-induced death by
attenuating activating transcription factor 3 induction. Cancer
Res. 66:10750–10759. 2006. View Article : Google Scholar : PubMed/NCBI
|