Introduction
Nuclear protein 1 (NUPR1) is a multifunctional
protein that primarily acts as a transcriptional regulator. It has
also been shown to take part in cell cycle regulation (1), apoptosis (2), DNA damage response (3), and autophagy (4). NUPR1 is a highly sensitive
stress-inducible gene that responds to a number of biological and
chemical stressors including tumor necrosis factor (TNF) (5), transforming growth factor (TGF)-β
(6), serum starvation (7), amino acid deprivation (8), carbon tetrachloride (9), and hexavalent chromium [Cr(VI)]
(10). Moreover, NUPR1 is
overexpressed in lung, breast, colorectal, pancreatic, and many
other cancers, and plays a role in cell transformation,
tumorigenesis, metastasis, and chemotherapeutic resistance
(11). In lung cancer cell lines and
lung tumor tissues of different histopathological subtype, NUPR1
expression was found to be elevated (4,12).
Recently, NUPR1 was implicated in Cr(VI)-induced lung cell
transformation, which raises the question of whether NUPR1 may also
be involved in the carcinogenic process elicited by other
cancer-causing metals (10).
Nickel (Ni) is a naturally occurring element present
in rocks and sediment, and is released into the surrounding
environment through forest fires, volcanic emissions, and erosion
processes (13). Ni also occurs due
to anthropogenic activity largely attributable to stainless and
alloy steel, nonferrous alloy and superalloy, electroplating,
catalyst and chemical production and use (14). The vast majority of Ni is used to
produce stainless steel, followed by superalloys and nonferrous
alloys, which are predominately used in the aerospace industry
(14). Moreover, Ni is found in
dietary sources, and to a lesser degree, in drinking water
(15). Humans, therefore, may be both
occupationally and environmentally exposed to Ni. Exposure to Ni
occurs primarily via inhalation, and there are many health risks
associated with exposure to Ni, the majority of which impact the
respiratory system. Epidemiological evidence supporting a casual
role of Ni in respiratory cancers dates back to 1949, and since
then, has been well documented (13,15). Based
on epidemiological, mechanistic, and in vivo studies, Ni
compounds are carcinogenic to humans as classified by the
International Agency for Research on Cancer (IARC) (15). Exposure to Ni can also cause
deleterious health effects other than cancer such as asthma,
cardiovascular disease, dermatitis, and lung fibrosis (13). The mechanisms of Ni-mediated
carcinogenesis, however, have yet to be fully elucidated.
The role that NUPR1 plays in Ni-induced
carcinogenesis and the involvement of activator protein 1 (AP-1)
transcription factor in these processes were investigated in this
study. NUPR1 and AP-1 were both induced by Ni, and NUPR1 was
determined to be upregulated in Ni-transformed human bronchial
epithelial BEAS-2B cells. Furthermore, knockdown of AP-1 suppressed
NUPR1 induction by Ni. This suggests that AP-1 is a key
factor for NUPR1 induction by Ni and in the stress-response
directed by NUPR1. Since AP-1 is known to be induced by reactive
oxygen species (ROS), the possibility that ROS contributes to
NUPR1 induction following Ni exposure was investigated. ROS
were determined not to be a primary mechanism by which AP-1
regulates NUPR1 induction following Ni exposure.
Furthermore, NUPR1 transactivation was determined to be
enhanced by AP-1 component, JUN. To conclude, stable knockdown of
NUPR1 in Ni-transformed cells reduced cell proliferation and
anchorage-independent growth. In summary, NUPR1 induction via AP-1
in response to Ni represents a mechanism capable of conferring
carcinogenic potential to human bronchial epithelial cells exposed
to Ni.
Materials and methods
Chemicals and cell culture
Nickel chloride was purchased from Sigma
Aldrich/Merck KGaA (catalog no. N6136). Antioxidant treatments were
performed with (−)-epigallocatechin gallate (EGCG) (Sigma
Aldrich/Merck KGaA; catalog no. E4143), L-ascorbic acid (Asc;
Thermo Fisher Scientific, Inc.; catalog no. A-61), and
a-tocopherol/vitamin E (vit. E; Sigma Aldrich/Merck KGaA; catalog
no. T3251). Hydrogen peroxide was purchased from Sigma
Aldrich/Merck KGaA (catalog no. 216763). Immortalized human
bronchial epithelial cells (BEAS-2B) were obtained from ATCC
(ATCC® CRL-9609), adapted to serum growth immediately
after purchase, carefully maintained at below confluent density,
and were authenticated by short tandem repeats (STR) analysis.
Experiments were performed within approximately six passages (e.g.
two weeks) from the time of thawing, and replicates were performed
simultaneously. Ni-transformed BEAS-2B cells were previously
generated and characterized (16). In
brief, BEAS-2B cells were exposed to soluble Ni for 30 days, after
which transformed clonal populations were selected based upon
anchorage-independent growth of single cells in soft agar
accompanied by a corresponding footprint of cancer-related gene
expression changes (16). The 293
cell line was obtained from ATCC (ATCC® CRL-1573) and
authenticated by STR analysis. BEAS-2B, Ni-transformed BEAS-2B, and
293 cells were maintained in Dulbecco's modified Eagle's medium
(DMEM) supplemented with 10% heat-inactivated fetal bovine serum
(FBS; Atlanta Biologicals), and 1% penicillin-streptomycin (Gibco;
Thermo Fisher Scientific, Inc.; catalog no. 15140-122). Cells were
incubated at 37°C in a humidified atmosphere of 5% carbon
dioxide.
Cell transfection
Stable transfections in BEAS-2B cells were performed
using Lipofectamine LTX reagent with PLUS reagent (Thermo Fisher
Scientific, Inc.; catalog no. 15338030). Control shRNA (Santa Cruz
Biotechnology, Inc.; catalog no. sc-108060) and NUPR1 shRNA
(Santa Cruz Biotechnology, Inc.; catalog no. sc-40792-SH) were used
for stable knockdown. Transient transfections in BEAS-2B cells were
performed using Lipofectamine RNAiMAX transfection reagent (Thermo
Fisher Scientific, Inc.; catalog no. 13778030). Control siRNA
(Invitrogen; Thermo Fisher Scientific, Inc.; catalog no. 12935112),
siRNA against JUN (Invitrogen; Thermo Fisher Scientific,
Inc.), and siRNA against FOS (Invitrogen; Thermo Fisher
Scientific, Inc.) were used for transient knockdowns.
Cell lysate preparation and western
blot analysis
Cells were washed with ice-cold PBS 2X, lysed using
RIPA buffer (150 mM NaCl, 1.0% NP-40, 0.5% sodium deoxycholate,
0.1% SDS, 50 mM Tris, pH 8.0, 2 mM EDTA, 1 mM PMSF, and protease
inhibitors; Roche Diagnostics; catalog no. 11 836 170 001) or
boiling buffer (1% SDS, 4 mM Na3VO4, and 10
mM Tris-HCl (pH 7.4). Cells lysed with RIPA buffer were collected
and incubated at 4°C for 30 min with constant agitation. Lysates
were then centrifuged for 20 min at 4°C, 20,000 × g, transferred,
and frozen at −80°C until use. Cells lysed with boiling buffer were
collected, denatured at 100°C for 5–10 min, sonicated for 10 min,
centrifuged at 20,000 × g for 15 min, transferred, and stored at
−80°C until use. Protein was quantified using Pierce™ BCA protein
assay kit (Thermo Fisher Scientific, Inc.; catalog no. 23225)
according to the manufacturer's instructions. Approximately 30–70
µg of total protein was separated on 10–18% SDS-PAGE gels by
electrophoresis, and proteins were transferred to a 0.2 µM
nitrocellulose membrane (Bio-Rad) for 2 h at 100 V or overnight at
20 V. Membranes were blocked for non-specific binding sites in
Tris-buffered saline containing 0.1% Tween-20 and 5% non-fat dry
milk at room temperature. The membranes were immunoblotted with
primary antibodies overnight at 4°C (JUN, 1:400, Santa Cruz
Biotechnology, Inc., catalog no. sc-1694; FOS, 1:500, Cell
Signaling Technology, Inc., catalog no. 2250; β-tubulin, 1:20,000,
Proteintech, catalog no. 66240-1-Ig; β-actin, 1:15,000, Proteintech
catalog no. 66009-1-Ig; and NUPR1, 1:200, Sigma Aldrich/Merck KGaA,
catalog no. SAB1104559). Membranes were then incubated with HRP- or
AP-conjugated secondary antibodies (HRP, goat anti-rabbit IgG,
1:5,000, Cell Signaling Technology, Inc., catalog no. 7074; HRP,
goat anti-mouse IgG, 1:5,000, Santa Cruz Biotechnology, Inc.,
catalog no. sc-2005; AP, goat anti-rabbit, 1:5,000, Promega Corp.,
catalog no. S3731). Protein detection was performed using
chemiluminescence (Thermo Fisher Scientific, Inc., catalog no.
32106) or chemifluorescence (Cytiva, catalog no. RPN5785) and
developed using autoradiography or imaged using a Typhoon imager
(GE Healthcare, model no. FLA 7000).
RNA extraction and real-time
quantitative PCR
Cells were collected in Tri reagent (Molecular
Research Center, catalog no. RT 111), and either stored at −80°C or
processed immediately for RNA isolation. The quantity and purity of
RNA extracted from each sample were determined by UV absorbance
spectroscopy on a NanoDrop 2000 spectrophotometer system. Reverse
transcription was performed using either LunaScript RT SuperMix
(New England BioLabs, catalog no. E3010) or ProtoScript First
Strand cDNA Synthesis (New England BioLabs, catalog no. E6300) with
500 ng of RNA in a final volume of 10 µl. Following denaturation of
the first-strand cDNA product for 5 min, quantitative real-time PCR
analysis was performed using Power SYBR Green PCR Master Mix
(Thermo Fisher Scientific, Inc., catalog no. 4367659) on an Applied
Biosystems QuantStudio 6 Flex system (Thermo Fisher Scientific,
Inc.). Relative gene expression levels were normalized to an
endogenous control and calculated using the DDCq method (17). The following primers were used:
NUPR1: 5′-CTGGCCCATTCCTACCTCG-3′ (forward) and
5′-TCTCTTGGTGCGACCTTTC-3′ (reverse); JUN:
5′-GAGCTGGAGCGCCTGATAAT-3′ (forward) and 5′-CCCTCCTGCTCATCTGTCAC-3′
(reverse); FOS: 5′-GAATCCGAAGGGAAAGGAATAAG-3′ (forward) and
5′-TCCGCTTGGAGTGTATCAGTCA-3′ (reverse); ATF2:
5′-CATGGCCCACCAGCTAGAAA-3′ (forward) and
5′-GTATTGCCTGGCAGAATTCACA-3′ (reverse); FOSL1:
5′-CCTTGTGAACGAATCAGCCC-3′ (forward) and 5′-GTCGGTCAGTTCCTTCCTCC-3′
(reverse); TUBULIN: 5′-GCAAGGTATCCTAAG-3′ (forward) and
5′-CTCGTCCTGGTTGGGAAACA-3′ (reverse); and ACTIN:
5′-TGACGTGGACATCCGCAAAG-3′ (forward) and 5′-CTGGAAGGTGGACAGCGAGG-3′
(reverse), and GAPDH: 5′-TCAAGAAGGTGGTGAAGCAGG-3′ (forward)
and 5′-AGCGTCAAAGGTGGAGGAGTG-3′ (reverse).
NUPR1 promoter cloning and
NUPR1-luciferase vector construction
Two degenerate primers, 915NUPR1FD and 2394NUPR1RD
(Table SI) were used initially to
amplify BEAS-2B genomic DNA. A 1,480-base pair DNA fragment was
restricted with BamHI (present in the primer sequences), and
cloned into the BamHI site of pUC19 (pNUPR264). The presence
of NUPR1 promoter fragment nucleotide 925 to nucleotide 2384
of NCBI GenBank (https://www.ncbi.nlm.nih.gov/genbank/) submission
AF069074 was confirmed by sequencing. In order to include more
NUPR1 promoter DNA sequences in this clone, the BamHI
site was destroyed using Bal 31 on both ends to generate two
separate clones (pNUPR287 for 3′ and pNUPR288 for 5′ deletion)
(18). A 1.6-kb PCR product was
amplified from BEAS-2B genomic DNA with primers 72NUPR1FD and
1724NUPR1NR, restricted with BamHI (present in 72NUPR1FD)
and EcoNI (present in the 3′ end of the PCR product) and
inserted into corresponding sites of pNUPR287. This new recombinant
(pNUPR289) included an additional 842 base pairs of promoter
sequences at the 5′ end of the initial clone (pNUPR264). Similarly,
1,125 base pairs of DNA were amplified with primers 2099NUPR1NF and
3223NUPR1NRD, restricted with EcoRV (present in the 5′ end
of the PCR product) and BamHI (present in the primer
3223NUPR1NRD) and inserted into corresponding sites of pNUPR288.
This new recombinant (pNUPR290) contained an additional 699 base
pairs of promoter sequence at the 3′end.
For NUPR1-luciferase vector construction, a
2,054 base pair KpnI-EcoRV 5′ promoter fragment from
pNUPR289 and 1,074-base pair EcoRV-HindIII 3′
fragment from pNUPR290 was inserted into a pGL4.17 vector (Promega
Corp.) into their KpnI-HindIII sites to generate the
full length NUPR1 promoter construct, pNUPR308 (Fig. S1A). The recombinant construct was
sequenced in both directions and is identical to the published
sequence, except for an additional 19 base pairs
(CAAGTATCCTGTCTTCACT) after nucleotide 957 of GenBank submission of
AF069074. To further investigate promoter activity in the cloned
sequence using promoter bashing technique, the 5′ end of this
sequence was deleted sequentially using Bal 31 as previously
described (18). Six recombinants
(NUPR309-314) with increasing amount of deletions at the 5′ end of
the NUPR1 promoter were selected and sequenced to determine
the extent of deletion (Table
SII).
Luciferase gene reporter assay
Cells were stably transfected with pGL4-basic
reporter vector containing the full length NUPR1 promoter or
the NUPR1 promoter with deletions varying in length and
position located upstream of the luciferase gene (Fig. S1A). Approximately 13,500 cells were
seeded overnight into each well of a 48-well plate. After seeding,
the medium was refreshed, and the cells were unexposed or exposed
to Ni for 24 h. Luciferase reporter system (Promega Corp., catalog.
no. E1500) was used to detect the luminescence intensity, and cell
lysate preparation and luminescence measurements were conducted
according to the manufacturer's protocol. All measurements were
adjusted for total protein, normalized to pGL4 control, and
performed in triplicate.
Site-directed mutagenesis assay
ALGGEN PROMO software V 3.0.2, a virtual laboratory
for the identification of putative transcription factor binding
sites in DNA sequences, was used to determine the transcription
factor binding sites within the NUPR1 promoter region
(19). Cells were stably transfected
with a pGL4 vector containing the full length NUPR1 promoter
(pNUPR308) or NUPR1 full length promoter with JUN
transcription factor binding site deleted (−2339 to −2333) in the
NUPR1 promoter positioned upstream of the luciferase gene
(Fig. S1A). Deletions were made
using the Q5® site-directed mutagenesis assay kit (New
England BioLabs), and the resulting plasmid was sequenced (Genewiz,
http://www.genewiz.com/) to confirm the deletion
of the JUN binding site in question (Fig. S1B). The luciferase gene reporter
assay was conducted as described herein. All luciferase
measurements were adjusted for total protein, normalized to pGL4
vector control, and performed in triplicate.
Anchorage-independent growth
assay
Anchorage-independent growth was determined by the
ability of cells to grow in soft agar. A bottom layer of 0.5%
2-hydroxyethylagarose (Sigma Aldrich/Merck KGaA, catalog no.
A4018), and top layer containing 5,000 cells in 0.35%
2-hydroxyethylagarose was place in a 6-well plate. After two weeks,
the wells were stained with 500 µl INT/BCIP (Roche Diagnostics,
catalog no. 11 681 460 001), and prepared according to the
manufacturer's instructions. Images of each stained well were
acquired using a Bio-Rad Molecular Imager Gel-Doc XR+
system and Image Lab software (Bio-Rad Laboratories, Inc.). Colony
numbers were calculated using ImageJ (NIH, V 1.52q). When seeding
cells in soft agar, 200 cells were simultaneously seeded into a
100-mm dish in order to determine the plating efficiency in
monolayer culture, which is defined as the ratio of the number of
colonies (those formed in a cell culture dish) vs. the number of
cells seeded. After a 12-day incubation, the plates were fixed and
stained overnight with 5% Giemsa in a 5:6 methanol:glycerol
solution. After destaining, all cell colony numbers were counted
using ImageJ (NIH) and plating efficiencies were determined. All
soft-agar assays were adjusted for plating efficiency and performed
in triplicate.
Colony formation assay
The cells were rinsed with PBS, briefly trypsinized,
and neutralized with complete medium. Following neutralization, the
cell suspension was then passed through a 40-µm cell strainer
(Celltreat, catalog no. 229482) to eliminate cell clumps. Two
hundred cells were then reseeded into each of three 100-mm dishes,
and grown for 3 weeks. Surviving colonies were stained with Giemsa
and counted using ImageJ (NIH, V 1.52q). All colony formation
assays were conducted in triplicate.
Graphical depictions and statistical
analyses
ImageJ (NIH) was used to quantify western blots, and
to determine cell colony numbers. Western blot images were
converted into 8-bit JPEG images, and the intensity of protein
bands were normalized to the loading control. For colony formation
assays, images were captured using a Bio-Rad Molecular Imager
Gel-Doc XR+ system and Image Lab software (V 2.0.1.,
Bio-Rad Laboratories, Inc.). Images were converted into 8-bit JPEG
images, and the background was reduced using a median filter to
eliminate non-colonies. Graphical depictions and statistical
analyses were performed using GraphPad Prism 9 (GraphPad Software,
Inc.). Differences between groups were compared using either a
Student's t-test or ANOVA followed by a Tukey post hoc multiple
comparisons test. Differences were considered statistically
significant at P<0.05. *P<0.05 and **P<0.01, as shown in
the figures.
Results
Nickel is an inducer of NUPR1
Cr(VI), an IARC group I human lung carcinogen,
induces NUPR1 mRNA and protein levels in human bronchial
epithelial (BEAS-2B) cells (10). Ni
is also a group I human lung carcinogen; however, the mechanisms of
carcinogenesis between Cr(VI) and Ni differ in many respects.
Cr(VI) is a classical carcinogen in the sense that it strongly
binds DNA and proteins, creating protein and/or DNA adducts and
subsequently, mutations; however, it also produces a high degree of
oxidative stress through intracellular reduction to Cr(III) and
acts carcinogenically through epigenetic mechanisms (20,21). Ni,
on the other hand, is often thought of as a nonclassical
carcinogen, and acts carcinogenically by mimicking hypoxia, which
is common in tumors, as well as by dysregulating the epigenetic
program of cells (13). Both Ni and
Cr have been shown to induce AP-1, and multiple AP-1 sites reside
in the promoter region of NUPR1 [reviewed in ref. (21)] (22–24).
Therefore, the possibility of whether Ni is capable of inducing
NUPR1 expression in BEAS-2B cells was investigated.
BEAS-2B cells were acutely exposed to 0, 0.25, 0.5,
0.75, and 1.0 mM Ni for 12 h, and the mRNA levels of NUPR1
were measured by real-time PCR and protein expression was
determined by western blot analysis. The levels of NUPR1
mRNA were dose-dependently increased following exposure of BEAS-2B
cells to Ni (Fig. 1A) as were the
protein levels of NUPR1 (Fig. 1B).
Dose-dependent induction of NUPR1 protein and mRNA was also evident
in BEAS-2B cells after 6 h of Ni exposure (data not shown). BEAS-2B
cells were then exposed to 0, 0.05, and 0.1 mM Ni to explore
whether extended exposure to Ni can induce NUPR1 expression. As
shown in Fig. 1C, extended exposure
to 0.1 mM Ni was capable of inducing NUPR1 protein levels by
3.1-fold after 7 days. NUPR1 mRNA levels were unchanged,
however (data not shown). In order to determine if Ni induces
NUPR1 in a time-dependent manner, BEAS-2B cells were exposed
to 1.0 mM Ni for various time points up to 12 h. After 3 h there
was only a 2.9-fold increase in NUPR1 mRNA expression
(Fig. 1D). Surprisingly, however,
NUPR1 mRNA was highly induced at 9 h (17.3-fold), and
decreased through 12 h, after having peaked at 9 h. There was no
significant time-dependent difference in NUPR1 mRNA
expression in unexposed BEAS-2B cells over 12 h (data not shown).
Collectively, these results show that NUPR1 is induced at both the
protein and mRNA levels following dose- and time-dependent exposure
to Ni. Furthermore, this suggests that NUPR1 is quickly and
transiently induced by Ni, which is characteristic of early
stress-response genes (25).
 | Figure 1.NUPR1 mRNA and protein expression is
induced by nickel (Ni). (A) Acute Ni exposure induced NUPR1
mRNA expression in a dose-dependent manner. Total RNA was extracted
from BEAS-2B cells after 12 h of exposure to 0, 0.25, 0.5, 0.75,
and 1.0 mM Ni. (B) Acute Ni exposure induced NUPR1 protein
expression in a dose-dependent manner. BEAS-2B cells were exposed
to 0, 0.5, 0.75, and 1.0 mM Ni for 12 h. (C) Extended Ni exposure
induced NUPR1 protein expression. BEAS-2B cells were exposed to 0,
0.05, and 0.1 mM Ni for 7 days. (D) Acute Ni exposure induced
NUPR1 mRNA in a time-dependent manner. Total RNA was
extracted from BEAS-2B cells following exposure to 1.0 mM Ni after
0, 3, 6, 9, and 12 h. NUPR1 mRNA expression was assessed using
RT-qPCR. Gene expression levels were normalized to actin, and are
presented as fold change relative to the control group. NUPR1
protein expression was analyzed by western blot analysis using
antibodies against NUPR1. Band intensities were quantified using
ImageJ software, and are presented as fold change relative to the
control group after normalizing for β-actin. All data shown are the
mean ± SD from qPCRs performed in triplicate. *P<0.05 and
**P<0.01. NUPR1, nuclear protein 1. |
Nickel induces transcription factor
AP-1
BEAS-2B cells were exposed to 0, 0.125, 0.25, 0.5,
and 0.75 mM Ni for 24 h, and the protein expression of AP-1
subunits, JUN, FOS, and ATF2, was investigated. Both JUN and ATF2
protein expression were found to be dose-dependently increased
following Ni exposure (Fig. 2A).
However, unlike JUN and ATF2, FOS protein expression was undetected
after 24 h of exposure to Ni at increasing doses. Instead, FOS
protein expression was found to be quickly and transiently induced
by 1.0 mM Ni after only 1 h, and its expression proceeded to
increase until approximately 6 h when its expression then started
to decrease (Fig. 2B). This was not
determined to be the case for both JUN and ATF2, as their protein
expression was unaltered over the course of 9 h in response to 1.0
mM Ni (Fig. 2B).
 | Figure 2.AP-1 is induced by nickel (Ni). (A)
Acute Ni exposure induced JUN and ATF2 protein expression. BEAS-2B
cells were exposed to 0, 0.125, 0.25, 0.5, and 0.75 mM Ni for 24 h.
(B) Acute Ni exposure induced FOS protein expression in a
time-dependent manner. BEAS-2B cells were exposed to 1.0 mM Ni for
12 h. JUN, ATF2, and FOS protein expression were analyzed by
western blot analysis using antibodies against JUN, ATF2, and FOS.
Band intensities were quantified using ImageJ software, and are
presented as fold change relative to the control group after
normalizing for b-actin or b-tubulin. (C) Acute Ni exposure induced
FOSL1 mRNA expression. BEAS-2B cells were exposed to 0 and
0.5 mM Ni for 24 h. (D) NUPR1 is overexpressed in
Ni-transformed BEAS-2B cells. Total RNA was extracted from BEAS-2B
cells following exposure to Ni. NUPR1 and FOSL1 mRNA
levels were assessed using RT-qPCR. Gene expression levels were
normalized to actin, and are presented as fold change relative to
the control group. All data shown are the mean ± SD from qPCRs
performed in triplicate. **P<0.01. AP-1, activator protein 1;
NUPR1, nuclear protein 1. |
Transcription factor AP-1 is a heterodimer complex
consisting of subunits from numerous protein subfamilies [reviewed
in ref. (26)]. In lieu of this, the
expression of additional AP-1 subunits was investigated following
exposure to Ni. BEAS-2B cells were exposed to a single dose of 0.5
mM Ni for 24 h. FOSL1, a FOS-related member of the AP-1
complex, was found to be transcriptionally induced by Ni (Fig. 2C). However, no other AP-1 subunits
investigated (i.e. JUNB, JUND, FOSL2, FOSB, and JDP2)
were found to be highly elevated (data not shown). Collectively,
these results showed that following Ni exposure, AP-1 subunits, JUN
and ATF2, were dose-dependently induced and remained induced
through 24 h in response to Ni exposure and likely contribute to
NUPR1 induction over this time period. FOS, however, was quickly
and transiently induced in response to Ni exposure, which indicates
that FOS may be an early-response gene responsible for increased
NUPR1 expression during the first 12 h of exposure.
NUPR1 is induced in nickel-transformed
BEAS-2B cells
NUPR1 was previously demonstrated to be important in
Cr(VI)-induced cell transformation (10). Therefore, NUPR1 expression was
measured in Ni-transformed cells. Real-time PCR was used to measure
the mRNA expression of NUPR1 in Ni-transformed clones, and
we observed increased expressions of NUPR1 mRNA in 4 out of
5 Ni-transformed clones, with extremely high levels in Ni-2 clones
(Fig. 2D). Overall, the ability for
Ni to induce NUPR1 in BEAS-2B cells and elevated expression of
NUPR1 in BEAS-2B cells transformed by Ni suggests that NUPR1 likely
plays a role in Ni-mediated carcinogenesis.
Knockdown of AP-1 suppresses NUPR1
induction by nickel
Since both AP-1 and NUPR1 were determined to be
induced by Ni (Figs. 1 and 2) and AP-1 binding sites are present in the
NUPR1 promoter, transcriptional induction of NUPR1 by AP-1
was investigated. Both JUN and FOS were transiently
knocked down in BEAS-2B and 293 cells, and were exposed to 0.5 mM
Ni for 24 h. JUN mRNA expression was reduced by at least 70%
in both BEAS-2B and 293 cells (Fig.
3A). In both BEAS-2B and 293 cells silencing of JUN
attenuated NUPR1 induction by Ni from 8.8 to 5.1-fold and 10
to 0.4-fold, respectively (Fig. 3B).
FOS silencing was found to be highly lethal in both BEAS-2B
and 293 cells, and knockdown efficiencies of only approximately 20%
were achievable (Fig. 3C). Despite
the low knockdown efficiency, FOS knockdown in both BEAS-2B
and 293 cells also reduced NUPR1 induction from 14.1 to
2.8-fold and from 7.0 to 1.9-fold, respectively (Fig. 3D). These data support the notion that
AP-1 is important in NUPR1 mRNA induction in response to Ni
exposure in both BEAS-2B and 293 cells.
Reactive oxygen species induce NUPR1
and antioxidants do not prevent NUPR1 induction by nickel
ROS upregulates AP-1, specifically JUN and FOS at
the transcriptional level [reviewed in ref. (26)]. In addition, Ni exposure produces
time- and dose-dependent increases in ROS (26). Albeit, ROS generation by Ni is
relatively low compared to that generated by other metals such as
cobalt or chromate, and is generally not considered a major
mechanism of Ni-mediated carcinogenesis (20,26).
Therefore, due to the connection between ROS and AP-1 induction and
the capacity for Ni to generate some degree of ROS, the possibility
that ROS induces NUPR1 was investigated. BEAS-2B cells were
treated with 0.1 mM of hydrogen peroxide
(H2O2) for 6 h, and NUPR1 mRNA
expression was measured. As shown in Fig.
4A, H2O2 was capable of inducing
NUPR1 in BEAS-2B cells; however, compared to 8-fold increase
on NUPR1 mRNA by 6 h Ni exposure, NUPR1 induction by
H2O2 (2.5-fold) was considerably lower. In
order to further investigate a possible connection between ROS
production by Ni and NUPR1 induction, BEAS-2B cells were
co-treated with antioxidant and ROS scavenger, epigallocatechin
gallate (EGCG; 0.025 mM), and Ni (1.0 mM) for 6 h. Co-treatment
with EGCG was not able to significantly prevent NUPR1
induction by Ni (Fig. 4B). In
addition, BEAS-2B cells were pre-treated for 2 h with 0.1 mM (Asc)
or 0.1 mM vitamin E (vit. E) and then exposed to 1.0 mM Ni for 6 h.
Pre-treatment with either antioxidant was also unable to prevent
NUPR1 induction by Ni (Fig. 4C and
D). On the contrary, pre-treatment with ascorbate followed by
Ni exposure actually significantly increased NUPR1
expression compared to Ni alone (Fig.
4C). This may be due to the fact that tissue culture media
lacks the amount of ascorbate found in vivo, and usually
only contains approximately 0.05 mM ascorbate, which is the amount
in 10% fetal bovine serum. High levels of ascorbate can be toxic to
tissue culture cells because ascorbate can also be a pro-oxidant.
Collectively, these results suggest that NUPR1 induction by
Ni primarily occurs through a mechanism other than ROS
generation.
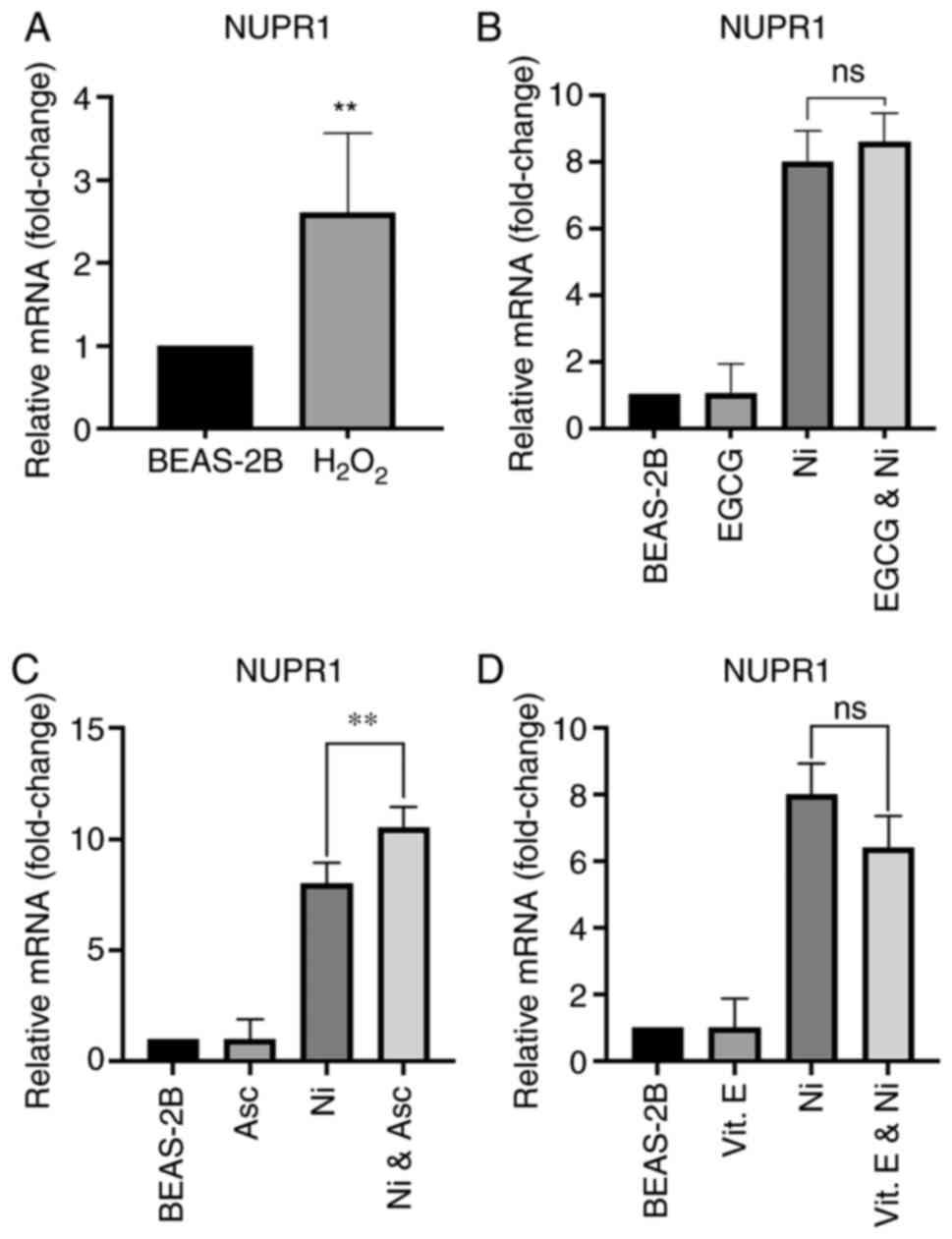 | Figure 4.ROS induces NUPR1 and antioxidants do
not prevent NUPR1 induction by nickel (Ni). (A) Acute exposure to
H2O2 induced NUPR1 expression. Total
RNA was extracted from unexposed BEAS-2B cells and BEAS-2B cells
exposed to 0.1 mM H2O2 for 6 h. (B) EGCG does
not prevent NUPR1 induction by Ni. Total RNA was extracted from
unexposed BEAS-2B cells, BEAS-2B cells exposed to 1.0 mM Ni for 6
h, BEAS-2B cells exposed to 0.025 mM EGCG for 6 h, and BEAS-2B
cells co-treated with 0.025 mM EGCG and 1.0 mM Ni for 6 h. (C)
Ascorbate (Asc) does not prevent NUPR1 induction by Ni. Total RNA
was extracted from unexposed BEAS-2B cells, BEAS-2B cells exposed
to 1.0 mM Ni for 6 h, BEAS-2B cells exposed to 100 µM ascorbate for
6 h, and BEAS-2B cells pre-treated with 100 µM ascorbate for 2 h
and then exposed to 1.0 mM Ni for 6 h. (D) Vitamin E (Vit. E) was
unable to significantly reduce NUPR1 induction by Ni. Total RNA was
extracted from unexposed BEAS-2B cells, BEAS-2B cells exposed to
1.0 mM Ni for 6 h, BEAS-2B cells exposed to 100 µM vit. E for 6 h,
and BEAS-2B cells pre-treated with 100 µM vit. E for 2 h and then
exposed to 1.0 mM Ni for 6 h. NUPR1 mRNA levels were measured by
RT-qPCR. Gene expression levels were normalized to actin, and are
presented as fold change relative to the control group. All data
shown are the mean ± SD from qPCRs performed in triplicate.
**P<0.01; ns, not significant. ROS, reactive oxygen species;
NUPR1, nuclear protein 1; EGCG, (−)-epigallocatechin gallate. |
Transcriptional regulation of
NUPR1
The NUPR1 promoter contains binding sites for
numerous transcription factors involved in cellular stress
including xenobiotic response elements, antioxidant response
elements, and binding sites for x-box-binding protein 1, which
classically functions in the unfolded protein response. In order to
fully elucidate the mechanism for NUPR1 transcriptional
activation and induction following exposure to Ni, deleted segments
of varying length of the NUPR1 promoter were made and
BEAS-2B cells were stably transfected with luciferase gene reporter
constructs containing either the full-length NUPR1 promoter
(construct pNUPR308), or a series of luciferase gene reporter
constructs containing the NUPR1 promoter with deleted
segments, which ranged in size from 299–2074 base pairs in length
(Fig. S1A). Stable transfectants
were treated with 1.0 mM Ni for 24 h to determine the regions
responsible for NUPR1 transactivation. Fig. 5 shows NUPR1 promoter activity
of the full-length NUPR1 promoter (pNUPR308) with and
without Ni exposure. Luciferase activity in Ni-treated
transfectants with the full-length promoter (pNUPR308) was
significantly higher than unexposed full-length promoter
transfectants (128-fold vs. 58-fold, respectively), which
demonstrates that the NUPR1 promoter is highly active
following exposure to Ni and is consistent with increased
NUPR1 expression.
Luciferase activity in stable transfectants
containing the NUPR1 promoter with various deletions
following exposure to 1.0 mM Ni for 24 h was subsequently
evaluated. A striking reduction in luciferase activity for both
unexposed and Ni-exposed stable transfectants containing the
NUPR1 promoter with the upstream most 299 base pairs deleted
(pNUPR309; NUPR1 promoter −2135 to +713) was found (Fig. 5). From these results, it was inferred
that this region must be important for high levels of NUPR1
transactivation. However, to determine which transcription
factor(s) is critical to NUPR1 transactivation following Ni
exposure, the NUPR1 promoter requires extensive examination
by deleting specific transcription factor binding sites.
AP-1 subunits, JUN and FOS, were determined to be
induced following Ni exposure in BEAS-2B cells. Additionally,
knockdown of JUN and FOS significantly suppressed
NUPR1 induction by Ni. Both JUN and FOS (i.e. AP-1) have
broad transcriptional repertoires, and are involved in many aspects
of tumor biology including tumor cell proliferation, apoptosis and
survival of tumor cells, and invasive growth and angiogenesis
(reviewed in (25). Moreover, a
putative JUN binding site is located in this upstream most region
of the NUPR1 promoter (−2339 to −2333). Consequently, the
role that JUN plays in the transcriptional upregulation of
NUPR1 was investigated. An NUPR1 promoter construct
without the JUN binding site corresponding to position (−2339 to
−2333) was generated (pNUPR_JUN; Fig.
S1B). BEAS-2B cells were then stably transfected with the
construct, and exposed to 1.0 mM Ni for 24 h or were unexposed.
Fig. 5 shows that upon deleting the
JUN binding site at (−2339 to −2333), a significant reduction in
promoter activity in both the Ni exposed and unexposed cells was
detected. This decrease in promoter activity mimics that which was
generated using the NUPR1 promoter construct deletion
corresponding to pNUPR309; NUPR1 promoter (−2135 to +713).
Therefore, we conclude that the JUN binding site at position (−2339
to −2333) in the NUPR1 promoter is important for
NUPR1 transactivation and that JUN acts by enhancing
NUPR1 transactivation.
NUPR1 knockdown suppresses
proliferation and colony formation in Ni-transformed BEAS-2B
cells
It has been reported on several occasions that NUPR1
expression is associated with tumorigenesis in vivo, and in
non-small cell lung cancer cells, NUPR1 knockdown has been
reported to reduce cell proliferation and colony formation ability
(4,12). Therefore, the role that NUPR1 plays in
cell proliferation in Ni-transformed cells was investigated.
Ni-transformed cells were originally generated by treating BEAS-2B
cells with Ni for 30 days and selecting single colonies that
developed the ability to grow anchorage-independently in soft agar
over a 4 week period (16).
Ni-transformed cells were stably transfected with either control
shRNA (ctrl) or NUPR1 shRNA (NUPR1 KD). Knockdown
efficiencies in two Ni-transformed cell lines were determined to be
approximately 50% (Fig. 6A). A colony
formation assay was performed with NUPR1 knockdown
Ni-transformed BEAS-2B cells to determine if NUPR1 knockdown
suppresses colony formation and cell proliferation. As shown in
Fig. 6B, colony formation was
significantly suppressed in both Ni-transformed cell lines upon
NUPR1 knockdown. Representative images are shown in Fig. S2A. Therefore, NUPR1 likely played an
important role in cell proliferation in both Ni-transformed BEAS-2B
cells.
In normal epithelial cells the occurrence of
anoikis, or programmed cell death by disruption of the interactions
with the extracellular matrix in suspension culture, renders cells
anchorage-dependent (27–29). In most squamous epithelial cells and
their transformed counterparts, growth in single-cell suspension is
prevented, as cells undergo differentiation and ultimately anoikis
(29). Anchorage-independent growth
is considered a hallmark of cancer cell growth, particularly with
respect to metastatic potential, and therefore, was assessed as an
indicator of cell transformation and chemical carcinogenesis
(30,31). NUPR1 overexpression was previously
shown to induce cell transformation and knockdown of NUPR1
prevented Cr(VI)-induced cell transformation (10). Since Ni-transformed cells showed
reduced cell proliferation when NUPR1 was stably knocked
down, we sought to determine if Ni-transformed-NUPR1
knockdown cells would forfeit their ability to grow
anchorage-independently. Results from the anchorage-independent
growth assay showed reduced colony formation in soft agar in NUPR1
knockdown Ni-transformed cells compared to Ni-transformed shRNA
control cells (Fig. 6C).
Representative images of soft agar colonies are also depicted in
Fig. S2B. From this it can be
concluded that NUPR1 likely plays an important role in
anchorage-independent growth in Ni-transformed BEAS-2B cells.
Discussion
Nuclear protein 1 (NUPR1) is a highly-sensitive
stress response protein that can be induced by a myriad of chemical
and biological stressors, and is upregulated in many cancers
including those originating in the lungs, breast, pancreas, and
colon, among others (11). However,
the mechanisms and stressors capable of inducing NUPR1 remain
largely understood. NUPR1 upregulation is significant because NUPR1
can permit cells to cope with high degrees of cellular stress and
damage, and inadvertently confer a growth advantage under such
conditions. In tumor cells, NUPR1 upregulation may facilitate an
adaptive response to unfavorable conditions, paving the way for
cancer progression. As described herein, Ni is a well-known human
carcinogen that induces NUPR1, yet the mechanisms of Ni-mediated
carcinogenesis have not yet been fully described. This is the first
study showing that Ni can induce NUPR1 and that explores the
mechanism of NUPR1 induction by Ni.
NUPR1 mRNA and protein expression levels were
highly induced in BEAS-2B cells following acute exposure to Ni,
which indicates that Ni is a potent inducer of NUPR1. Notably, no
significant time-dependent difference in NUPR1 mRNA
expression in unexposed BEAS-2B cells over 12 h was detected (data
not shown). This contrasts a previous report whereby NUPR1
mRNA expression was shown to be induced following cell culture
medium change in murine embryonic fibroblast (NIH 3T3) cells
(32). Induction of NUPR1 by routine
medium change was determined to be due to the presence of
thermolabile factors in conditioned medium that block activation of
stress-sensitive protein kinases and NUPR1 expression (32). Due to the fact that NUPR1 is
overexpressed in lung cancers (11),
NUPR1 expression was evaluated in Ni-transformed BEAS-2B cells.
NUPR1 was determined to be upregulated in 4 out of 5 Ni-transformed
clones. Since NUPR1 was induced following Ni exposure in BEAS-2B
cells and was upregulated in Ni-transformed BEAS-2B cells, it is
likely that NUPR1 plays a role in Ni-mediated carcinogenesis.
Multiple AP-1 binding sites are located in the
NUPR1 promoter, and Ni was previously shown to induce AP-1
(22–24). In addition, carcinogenic metal Cr(VI)
was previously shown to open chromatin around AP-1 sites in the
NUPR1 promoter (33). ChIP-seq
data (ENCODE) also show the binding of AP-1 around the promoter
region of NUPR1. Therefore, the possibility that Ni induces
AP-1, namely JUN, FOS, and ATF2, was investigated. Protein and mRNA
expression analysis confirmed that AP-1 subunits (i.e. JUN, FOS,
and ATF2) were also induced following Ni exposure. FOS was quickly
and transiently induced after only 1 h, and JUN and ATF2 proteins
were found to be induced after 12 h of Ni exposure. These
observations suggest that FOS is one of the AP-1 subunits involved
in the initial response to Ni, and that additional AP-1 subunits
respond over time to Ni exposure. Therefore, it is suspected that
AP-1 subunits (i.e. JUN, FOS, and ATF2) may be responsible for
NUPR1 transactivation.
To confirm that AP-1 transcriptionally regulates
NUPR1 in response to Ni, both JUN and FOS were silenced in
BEAS-2B cells and cells were then exposed to Ni. NUPR1
expression was significantly suppressed after silencing either JUN
or FOS in BEAS-2B cells and exposed to Ni for 24 h. The involvement
of JUN in NUPR1 transcription was further demonstrated by
cloning the NUPR1 promoter, and creating a series of deletions in
the promoter region. By subsequently deleting a JUN binding site in
the NUPR1 promoter region corresponding to position (−2339 to
−2333), it was determined that JUN enhanced NUPR1
transactivation.
Reactive oxygen species (ROS) are capable of
inducing AP-1, particularly JUN and FOS at the transcriptional
level, and although Ni only generates a relatively small amount of
oxidative stress compared to other carcinogenic metals, such as
Cr(VI) and cobalt, this relationship was investigated (34). BEAS-2B cells were treated with
H2O2 (100 µM for 6 h), and NUPR1 mRNA
levels were increased, albeit to a much lesser magnitude than
observed following Ni exposure. BEAS-2B cells were then either
co-treated or pre-treated with ROS scavenger EGCG, ascorbate, or
vitamin E, and exposed to Ni. NUPR1 induction by Ni was
unable to be prevented by any of the antioxidants tested.
NUPR1 expression remained highly elevated in BEAS-2B cells
co- or pre-treated with antioxidants and EGCG. Therefore,
NUPR1 was capable of being induced by ROS, but oxidative
stress is likely not responsible for NUPR1 induction by
Ni.
Silencing of NUPR1 in human lung cancer
cells was previously shown to reduce cell proliferation (12). It has also been shown that NUPR1 acts
as an oncogene, in part, by enhancing cell proliferation in cells
overexpressing NUPR1 (35).
Therefore, the colony formation ability of NUPR1 knockdown
Ni-transformed BEAS-2B cells was assessed, and reduced colony
formation was observed. This suggests that NUPR1 may aid in cell
proliferation ability in Ni-transformed BEAS-2B cells, thereby
conferring carcinogenic potential. In Cr(VI)-treated BEAS-2B cells,
NUPR1 knockdown was previously shown to reduce
anchorage-independent growth, which is considered a hallmark of
carcinogenesis (10). In
Ni-transformed BEAS-2B cells, NUPR1 knockdown also was able to
significantly reduce anchorage-independent growth. Overall, the
data presented demonstrate that AP-1 is partly responsible for
NUPR1 transactivation following Ni exposure, and that NUPR1
plays an important role in Ni-mediated carcinogenesis. However, the
precise mechanism that lies upstream of AP-1 which governs
NUPR1 induction following Ni exposure remains unclear.
AP-1 is regulated by the activation of mitogen
activated protein kinases (MAPK), which are classically thought of
as first responders to environmental signals (36). Activation of the MAPK pathway
comprises phosphorylation and activation of key proteins in MAPK
signaling: ERK1/2, JNK1/2/3, and p38 MAPK (36). Ni has, on several occasions, been
shown to influence MAPK signaling, which may therefore be
responsible for NUPR1 induction (24,37–39).
Likewise, Ni has also been shown to potentiate NF-κB signaling,
which cross talks with AP-1 and is therefore another potential
candidate that may mediate NUPR1 induction by Ni (22,24,38,40).
Furthermore, both HIF1-α and AP-1 are induced by hypoxia signaling,
which is mimicked by Ni, and both HIF1-α and AP-1 have been shown
to cooperate in the transcription of various genes associated with
cancer development and progression (13,41,42).
Therefore, it is plausible that Ni induces the AP-1/NUPR1 signaling
axis by activating of one of these upstream mediators. Further
research is needed in order to unveil the specific mechanism
responsible for induction of the AP-1 and subsequently, NUPR1, by
Ni. In conclusion, Ni induces NUPR1 through AP-1 (i.e. JUN
and FOS), and the activation of this pathway has the potential to
contribute to carcinogenesis brought on by Ni and the progression
of cancers associated with Ni exposure. Given that NUPR1 is also
induced by Cr(VI), NUPR1 induction may be a shared phenomenon among
carcinogenic metals that contributes to their carcinogenic
potential. This possibility requires additional research, but if
discovered, would be a significant finding because it would help
bridge the knowledge gap in how exposure to various metals causes
cancer.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This research was funded by the National Institutes
of Health (ES000260, ES022935, ES023174, and ES026138 to MC;
ES029359, ES026138, and ES030583 to CJ).
Availability of data and materials
The datasets used and/or analyzed during the
current study are available from the corresponding author on
reasonable request.
Authors' contributions
AM designed and performed experiments, analyzed the
data, and wrote the manuscript. NR cloned the NUPR1 promoter. MC
and CJ conceptualized the study. MC, CJ, and HS contributed to the
interpretation of the results, and assisted in troubleshooting. All
authors discussed the results, and contributed to the writing and
revision of the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors state that they have no competing
interests.
References
|
1
|
Malicet C, Hoffmeister A, Moreno S, Closa
D, Dagorn JC, Vasseur S and Iovanna JL: Interaction of the stress
protein p8 with Jab1 is required for Jab1-dependent p27
nuclear-to-cytoplasm translocation. Biochem Biophys Res Commun.
339:284–289. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Malicet C, Giroux V, Vasseur S, Dagorn JC,
Neira JL and Iovanna JL: Regulation of apoptosis by the
p8/prothymosin alpha complex. Proc Natl Acad Sci USA.
103:2671–2676. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Aguado-Llera D, Hamidi T, Doménech R,
Pantoja-Uceda D, Gironella M, Santoro J, Velázquez-Campoy A, Neira
JL and Iovanna JL: Deciphering the binding between Nupr1 and MSL1
and their DNA-repairing activity. PLoS One. 8:e781012013.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mu Y, Yan X, Li D, Zhao D, Wang L, Wang X,
Gao D, Yang J, Zhang H, Li Y, et al: NUPR1 maintains autolysosomal
efflux by activating SNAP25 transcription in cancer cells.
Autophagy. 14:654–670. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Goruppi S, Patten RD, Force T and Kyriakis
JM: Helix-loop-helix protein p8, a transcriptional regulator
required for cardiomyocyte hypertrophy and cardiac fibroblast
matrix metalloprotease induction. Mol Cell Biol. 27:993–1006. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
García-Montero AC, Vasseur S, Giono LE,
Canepa E, Moreno S, Dagorn JC and Iovanna JL: Transforming growth
factor beta-1 enhances Smad transcriptional activity through
activation of p8 gene expression. Biochem J. 357:249–23. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mallo GV, Fiedler F, Calvo EL, Ortiz EM,
Vasseur S, Keim V, Morisset J and Iovanna JL: Cloning and
expression of the rat p8 cDNA, a new gene activated in pancreas
during the acute phase of pancreatitis, pancreatic development, and
regeneration, and which promotes cellular growth. J Biol Chem.
272:32360–32369. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Averous J, Lambert-Langlais S, Cherasse Y,
Carraro V, Parry L, B'chir W, Jousse C, Maurin AC, Bruhat A and
Fafournoux P: Amino acid deprivation regulates the stress-inducible
gene p8 via the GCN2/ATF4 pathway. Biochem Biophys Res Commun.
413:24–29. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Taïeb D, Malicet C, Garcia S, Rocchi P,
Arnaud C, Dagorn JC, Iovanna JL and Vasseur S: Inactivation of
stress protein p8 increases murine carbon tetrachloride
hepatotoxicity via preserved CYP2E1 activity. Hepatology.
42:176–182. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen D, Kluz T, Fang L, Zhang X, Sun H,
Jin C and Costa M: Hexavalent chromium (Cr(VI)) down-regulates
acetylation of histone H4 at lysine 16 through induction of
stressor protein Nupr1. PLoS One. 11:e01573172016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Murphy A and Costa M: Nuclear protein 1
imparts oncogenic potential and chemotherapeutic resistance in
cancer. Cancer Lett. 494:132–141. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Guo X, Wang W, Hu J, Feng K, Pan Y, Zhang
L and Feng Y: Lentivirus-mediated RNAi knockdown of NUPR1 inhibits
human nonsmall cell lung cancer growth in vitro and in vivo. Anat
Rec (Hoboken). 295:2114–2121. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chen QY, DesMarais T and Costa M: Metals
and mechanisms of carcinogenesis. Annu Rev Pharmacol Toxicol.
59:537–554. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
McRae ME: Mineral commodity summaries
2020: Nickel, in Mineral Commodity Summaries. Reston, VA: pp.
2042020
|
|
15
|
IARC Working Group on the Evaluation of
Carcinogenic Risks to Humans, . Arsenic, metals, fibres, and dusts.
IARC Monogr Eval Carcinog Risks Hum. 100:11–465. 2012.PubMed/NCBI
|
|
16
|
Clancy HA, Sun H, Passantino L, Kluz T,
Muñoz A, Zavadil J and Costa M: Gene expression changes in human
lung cells exposed to arsenic, chromium, nickel or vanadium
indicate the first steps in cancer. Metallomics. 4:784–793. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Slatko B, Heinrich P, Nixon BT and Voytas
D: Constructing nested deletions for use in DNA sequencing. Curr
Protoc Mol Biol Chapter. 7:Unit7.2. 2001. View Article : Google Scholar
|
|
19
|
Messeguer X, Escudero R, Farré D, Núñez O,
Martínez J and Albà MM: PROMO: Detection of known transcription
regulatory elements using species-tailored searches.
Bioinformatics. 18:333–334. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen QY, Murphy A, Sun H and Costa M:
Molecular and epigenetic mechanisms of Cr(VI)-induced
carcinogenesis. Toxicol Appl Pharmacol. 377:1146362019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Costa M and Murphy A: Chapter 11-overview
of chromium(III) toxicology. The Nutritional Biochemistry of
Chromium (III). Vincent JB: 2nd edition. Elsevier; pp. 341–359.
2019, View Article : Google Scholar
|
|
22
|
Cruz MT, Gonçalo M, Figueiredo A, Carvalho
AP, Duarte CB and Lopes MC: Contact sensitizer nickel sulfate
activates the transcription factors NF-kB and AP-1 and increases
the expression of nitric oxide synthase in a skin dendritic cell
line. Exp Dermatol. 13:18–26. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Andrew AS, Klei LR and Barchowsky A:
AP-1-dependent induction of plasminogen activator inhibitor-1 by
nickel does not require reactive oxygen. Am J Physiol Lung Cell Mol
Physiol. 281:L616–L623. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ding J, Huang Y, Ning B, Gong W, Li J,
Wang H, Chen CY and Huang C: TNF-alpha induction by nickel
compounds is specific through ERKs/AP-1-dependent pathway in human
bronchial epithelial cells. Curr Cancer Drug Targets. 9:81–90.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Eferl R and Wagner EF: AP-1: A
double-edged sword in tumorigenesis. Nat Rev Cancer. 3:859–868.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Salnikow K, Su W, Blagosklonny MV and
Costa M: Carcinogenic metals induce hypoxia-inducible
factor-stimulated transcription by reactive oxygen
species-independent mechanism. Cancer Res. 60:3375–3378.
2000.PubMed/NCBI
|
|
27
|
Maeno K, Masuda A, Yanagisawa K, Konishi
H, Osada H, Saito T, Ueda R and Takahashi T: Altered regulation of
c-jun and its involvement in anchorage-independent growth of human
lung cancers. Oncogene. 25:271–277. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Frisch SM and Francis H: Disruption of
epithelial cell-matrix interactions induces apoptosis. J Cell Biol.
124:619–626. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kantak SS and Kramer RH: E-cadherin
regulates anchorage-independent growth and survival in oral
squamous cell carcinoma cells. J Biol Chem. 273:16953–16961. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Mori S, Chang JT, Andrechek ER, Matsumura
N, Baba T, Yao G, Kim JW, Gatza M, Murphy S and Nevins JR:
Anchorage-independent cell growth signature identifies tumors with
metastatic potential. Oncogene. 28:2796–2805. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Guadamillas MC, Cerezo A and Del Pozo MA:
Overcoming anoikis-pathways to anchorage-independent growth in
cancer. J Cell Sci. 124:3189–1397. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Garcia-Montero A, Vasseur S, Mallo GV,
Soubeyran P, Dagorn JC and Iovanna JL: Expression of the
stress-induced p8 mRNA is transiently activated after culture
medium change. Eur J Cell Biol. 80:720–725. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ovesen JL, Fan Y, Zhang X, Chen J,
Medvedovic M, Xia Y and Puga A: Formaldehyde-assisted isolation of
regulatory elements (FAIRE) analysis uncovers broad changes in
chromatin structure resulting from hexavalent chromium exposure.
PLoS One. 9:e978492014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Marinho HS, Real C, Cyrne L, Soares H and
Antunes F: Hydrogen peroxide sensing, signaling and regulation of
transcription factors. Redox Biol. 2:535–562. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang L, Jiang F, Xia X and Zhang B: LncRNA
FAL1 promotes carcinogenesis by regulation of miR-637/NUPR1 pathway
in colorectal cancer. Int J Biochem Cell Biol. 106:46–56. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Atsaves V, Leventaki V, Rassidakis GZ and
Claret FX: AP-1 transcription factors as regulators of immune
responses in cancer. Cancers (Basel). 11:10372019. View Article : Google Scholar
|
|
37
|
Gu Y, Wang Y, Zhou Q, Bowman L, Mao G, Zou
B, Xu J, Liu Y, Liu K, Zhao J and Ding M: Inhibition of nickel
nanoparticles-induced toxicity by epigallocatechin-3-gallate in JB6
cells may be through down-regulation of the MAPK signaling
pathways. PLoS One. 11:e01509542016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Magaye R, Zhou Q, Bowman L, Zou B, Mao G,
Xu J, Castranova V, Zhao J and Ding M: Metallic nickel
nanoparticles may exhibit higher carcinogenic potential than fine
particles in JB6 cells. PLoS One. 9:e924182014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhang D, Li J, Wu K, Ouyang W, Ding J, Liu
ZG, Costa M and Huang C: JNK1, but not JNK2, is required for COX-2
induction by nickel compounds. Carcinogenesis. 28:883–891. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Fujioka S, Niu J, Schmidt C, Sclabas GM,
Peng B, Uwagawa T, Li Z, Evans DB, Abbruzzese JL and Chiao PJ:
NF-kappaB and AP-1 connection: Mechanism of NF-kappaB-dependent
regulation of AP-1 activity. Mol Cell Biol. 24:7806–7819. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Michiels C, Minet E, Michel G, Mottet D,
Piret JP and Raes M: HIF-1 and AP-1 cooperate to increase gene
expression in hypoxia: Role of MAP kinases. IUBMB Life. 52:49–53.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Damert A, Ikeda E and Risau W:
Activator-protein-1 binding potentiates the
hypoxia-induciblefactor-1-mediated hypoxia-induced transcriptional
activation of vascular-endothelial growth factor expression in C6
glioma cells. Biochem J. 327:419–423. 1997. View Article : Google Scholar : PubMed/NCBI
|




















