Introduction
Thyroid cancer, the most frequent type of cancer of
the endocrine system, originates from parafollicular thyroid cells
or follicular thyroid cells (1).
There are several different histologic subtypes of thyroid cancer,
such as anaplastic thyroid carcinoma, papillary thyroid carcinoma
(PTC), and follicular thyroid carcinoma (2). PTC, which stems from thyroid follicular
cells, accounts for the majority of thyroid cancer cases (3). The overall five-year survival rate in
patients at early stages of PTC is as high as 97%, whereas, in
those in advanced stages, the overall five-year survival rate is
estimated to be approximately 59% (4). Therefore, identifying the molecular
mechanisms underlying the progression of PTC is of great importance
in order to optimize the prognosis in patients with PTC.
Long non-coding RNAs (lncRNAs) are a subtype of
non-coding transcripts resulting from the lack of open reading
frames (ORFs). Their length is >200 nt and are encoded by the
mammalian genome (5). It has been
reported that the dysregulation of lncRNAs is associated with
carcinogenesis. For example, lncRNA HOTAIR mediated the metastasis
of breast cancer cells via targeting genome-wide retargeting of
polycomb repressive complex 2 (PRC2) (6). Additionally, the dysregulation of lncRNA
KIAA0125 was involved in the development of gallbladder carcinoma
via regulating β-catenin and vimentin (7). Several lncRNAs have been identified to
serve an important role in the development of thyroid cancer. For
instance, H19, which is overexpressed in thyroid cancer tissues and
cells, induced cell growth, migration, and invasion by serving as a
competitive endogenous RNA (ceRNA) for miR-17-5p and regulating the
miR-17-5p/YES1 axis (8). Furthermore,
the expression of actin filament-associated protein 1-antisense RNA
1 (AFAP1-AS1) was revealed to be increased in thyroid cancer
tissues, and its downregulation attenuated cell proliferation and
invasion, and induced cell apoptosis (9). In 2018, a study demonstrated that the
lncRNA double homeobox A pseudogene 8 (DUXAP8) was differentially
expressed in human thyroid cancer using genome-wide analysis
(10). lncRNA DUXAP8 has been
identified to promote the growth of numerous cancers, including
non-small cell lung cancer (11),
pancreatic carcinoma (12), and renal
cell carcinoma (13). However, the
role of lncRNA DUXAP8 in PTC has not been investigated.
Mechanistically, some studies have demonstrated that
lncRNAs serve as microRNA (miRNA) sponges through ceRNA activity,
thus promoting the expression of the downstream mRNAs (14,15); and
some studies have indicated that lncRNAs and miRNAs interact
(16,17). miRNAs are a group of small,
non-coding, and single-chain molecules, which regulate the
translation or degradation of their target mRNAs (18). Notably, miRNAs play important roles in
regulating several biological behaviors, including cell
proliferation, apoptosis, and differentiation (19). However, the function of the lncRNA
DUXAP8-miRNA network in the pathogenesis of PTC remains
elusive.
Materials and methods
Sample collection
Tumor tissues and the adjacent normal tissues (>5
cm away from the tumor) from 60 patients (31 males and 29 females;
aged 25–66) with PTC were collected at the First Hospital of Jilin
University, between January 2016 and December 2018. The
inclusion/exclusion criteria were as follows: No chemotherapy or
radiotherapy received before the surgery. The clinical stage of
each patient after surgery was evaluated based on the 7th edition
of the American Joint Committee on Cancer (AJCC)
tumor-node-metastasis staging system (20). The adjacent normal tissues served as
matched controls for the PTC tissues. Written informed consent was
provided by all patients prior to enrollment in this study. All
experimental procedures were approved by the Ethics Committee of
the First Hospital of Jilin University.
Bioinformatics analysis
The dysregulated lncRNAs were analyzed in The Cancer
Genome Atlas (TCGA)-thyroid carcinoma (THCA) data. TCGA-THCA data
were retrieved from the Broad Institute Genome Data Analysis Center
(GDAC) Firehose as previously reported (21). The interaction between miR-20b-5p and
DUXAP8 was predicted by the miRDB (http://mirdb.org/) bioinformatics analysis tool. The
association between DUXAP8 and disease stage as well as with the
prognosis of patients with PTC was evaluated with the Gene
Expression Profiling Interactive Analysis (GEPIA) database
(22). GEPIA downloaded level 3 gene
expression data (RNA sequencing data after alignment and
standardization by RSEM method) and patient information from TCGA
database. All patients from TCGA-THCA (with survival information
and gene expression data) were used to investigate the association
between DUXAP8 expression and overall survival by Kaplan-Meier Plot
analysis with log-rank testing on GEPIA database, which was
employed to divide the patients into a high-DUXAP8 expression group
and a low-DUXAP8 expression group according to the median
expression of DUXAP8. The correlation between DUXAP8 and son of
sevenless 1 (SOS1), c-Myc, or cyclin D1 (CCND1) was analyzed in the
TCGA-THCA data.
Cell culture
The human PTC cell lines, CGTH W-3 and TPC1, and the
human thyroid follicular epithelial cell line, Nthy-ori 3-1, were
obtained from the American Type Culture Collection (ATCC). Cells
were maintained in Roswell Park Memorial Institute (RPMI)-1640
medium (Invitrogen; Thermo Fisher Scientific, Inc.), supplemented
with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific,
Inc.), 100 U/ml penicillin, and 100 µg/ml streptomycin (Invitrogen;
Thermo Fisher Scientific, Inc.) in a humidified incubator at 37°C
with 5% CO2.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was isolated from PTC tissues and cells
using TRIzol (Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. Total RNA was reverse transcribed into
cDNA with the TaqMan microRNA Reverse Transcription kit (Applied
Biosystems; Thermo Fisher Scientific, Inc.). Subsequently, RT-qPCR
was carried out using Bio-Rad cFX96 Real-Time PCR System (Bio-Rad
Laboratories, Inc.) and SYBR Premix Ex Taq kit (Takara Bio, Inc.)
according to the manufacturer's instructions. The thermocycling
conditions were as follows: 95°C for 5 min, followed by 45 cycles
at 95°C for 10 sec, 55°C for 30 sec, and a melting curve every
0.2°C from 55 to 95°C for 2 min. The expression levels of DUXAP8
and SOS1 were normalized to GAPDH, and those of miR-20b-5p to U6
expression with the 2−ΔΔCq method (23). The primers were as follows: DUXAP8F
forward, 5′-AGGATGGAGTCTCGCTGTATTGC-3′ and reverse,
5′-GGAGGTTTGTTTTCTTCTTTTTT-3′; SOS1 forward,
5′-ACCACAGATGTTTGCAGTGTATTTG-3′ and reverse,
5′-GCAGATTCTGGTCGTCTTCGTG-3′; GAPDH forward,
5′-GCACCGTCAAGGCTGAGAAC-3′ and reverse, 5′-TGGTGAAGACGCCAGTGGA-3′;
miR-20b-5p forward, 5′-CAAAGTGCTCATAGTGCAGGT-3′ and reverse,
Uni-miR qPCR (Takara Biotechnology Co., Ltd.); U6 forward,
5′-CTCGCTTCGGCAGCACA-3′ and reverse,
5′-AACGCTTCACGAATTTGCGT-3′.
Western blot analysis
Total proteins were extracted from PTC tissues and
cells using RIPA lysis buffer (Roche Diagnostics) containing
protease inhibitors, following the manufacturer's instructions. The
supernatant was harvested by centrifugation at 13,282 × g for 10
min at 4°C. The protein concentration was measured with a BCA
Protein Assay kit (Beyotime Institute of Biotechnology). The
protein extracts (20 µg) were separated by SDS-PAGE (8%) and were
then blotted onto a polyvinylidene fluoride (PVDF) membrane.
Subsequently, the proteins on the PVDF membrane were blocked with
5% non-fat milk for 2 h at 25°C prior to incubation with primary
antibodies against SOS1 (1:1,000; product no. 12409), extracellular
signal-regulated kinase 1/2 (ERK1/2; 1:1,000; product no. 9101) and
GAPDH (1:1,000; cat. no. 5174; all from Cell Signaling Technology,
Inc.) overnight at 4°C. The membrane was then incubated with a
horseradish peroxidase-conjugated secondary antibody (1:2,000; cat.
no. 7074; Cell Signaling Technology, Inc.) for 2 h at 25°C. The
blots were visualized using an ECL reagent (Beyotime Institute of
Biotechnology) and analyzed with ImageJ software 1.48u (National
Institutes of Health). The target-protein expression was normalized
to that of GAPDH.
Downregulation of DUXAP8
The small interfering RNA (siRNA) technology was
applied for DUXAP8 downregulation in PTC cells. The DUXAP8 siRNAs
(si- DUXAP8; 5′-AAGAUAAAGGUGGUUUCCACAAGAATT-3′) and control siRNA
(siRNA-NC; 5′-AGCUUGAUACGACAAAGCUTT-3′) were purchased from
Shanghai GenePharma Co., Ltd. PTC cells were transfected with
DUXAP8 siRNAs or siRNA-NC (50 nM) using Lipofectamine 2000 (Thermo
Fisher Scientific, Inc.) following the manufacturer's protocol, and
were then incubated for 48 h according to the manufacturer's
instructions.
Overexpression of SOS1
The pcDNA3.1 and pcDNA3.1-SOS1 plasmids (2 µg) were
purchased from GenePharma Co., Ltd. PTC cells were transfected with
pcDNA3.1 or pcDNA3.1-SOS1 using Lipofectamine 2000 (Thermo Fisher
Scientific, Inc.) following manufacturer's protocol, and incubated
for 48 h following the manufacturer's protocol.
Cell proliferation assay
A Cell Counting Kit-8 (CCK-8) assay was carried out
to measure cell proliferation, following the manufacturer's
protocol. Briefly, cells (density, 1×104 cells/well)
were seeded into 96-well plates, and were supplemented with 10 µl
of CCK-8 reagent (Dojindo Molecular Technologies, Inc.) for 2 h at
37°C. The optical density at 450 nm was measured in each well at 0,
24, 48 and 72 h using a microplate reader (Thermo Fisher
Scientific, Inc.).
Cell apoptosis assay
Briefly, PTC cells (density, 1×106
cells/ml) were first treated with 100 µl of 1X binding buffer. The
mixture was supplemented with 2 µl of Annexin V-FITC and maintained
for 15 min at room temperature. Subsequently, 400 µl of PBS and 1
µl propidium iodide (PI) were added and cells were incubated for 5
min at room temperature. Following treatment with Annexin
V-FITC/PI, cells were subjected to flow cytometric analysis on a
FACSCalibur flow cytometer (BD Biosciences) with the CellQuest
software (version 6.0; BD Biosciences). Finally, the data were
analyzed using the FlowJo software (version 10.2; FlowJo, LLC).
RNA immunoprecipitation (RIP)
assay
To verify the association between DUXAP8 and
miR-20b-5p a RIP assay was carried out using the EZMagna RIP
RNA-binding protein immunoprecipitation kit (EMD Millipore)
following the manufacturer's instructions. Briefly, PTC cells were
lysed with RNA lysis buffer supplemented with protease and RNase
inhibitors. The cell lysate was then treated with magnetic beads
conjugated with human Ago2 antibody (cat. no. 10686-1-AP;
ProteinTech Group, Inc.) or a negative control IgG (product code
ab133470; Abcam). Following incubation at 4°C overnight, the
co-precipitated RNAs were subjected to RT-qPCR analysis as
previously described.
Dual luciferase reporter assay
The wild-type or mutant DUXAP8 3′untranslated region
(3′UTR; Shanghai GeneChem Co., Ltd.) was sub-cloned into the pGL3
vector (Promega Corporation). Subsequently, the plasmids were
transfected into PTC cells for 24 h at 37°C using Lipofectamine
2000 (Thermo Fisher Scientific, Inc.). Finally, the luciferase
activity was measured using the dual-luciferase reporter assay
system (Promega Corporation), and was normalized to that of
Renilla.
Statistical analysis
Data are expressed as the mean ± standard deviation
(SD). The GraphPad Prism 6 (GraphPad Software, Inc.) software was
applied for all statistical analyses. All the experiments were
repeated 3 times. The comparisons between two groups were carried
out using unpaired Student's t-test, while one-way analysis of
variance (ANOVA) followed by Bonferroni's correction as a post hoc
test was performed for the comparisons among multiple groups. The
association between DUXAP8 and the clinicopathological
characteristics of PTC patients was evaluated by Chi-square test.
The association between the expression of DUXAP8 and that of SOS1,
c-Myc and cyclin D1 (CCND1) in the TCGA-THCA dataset or PTC tumor
tissues was analyzed by Spearman's correlation test. P<0.05 was
considered to indicate a statistically significant difference.
Results
DUXAP8 is upregulated in PTC
tissues
The results from the TCGA-THCA revealed that DUXAP8
was one of the most significantly upregulated lncRNAs in patients
with PTC (Fig. 1A). In addition, it
was confirmed that DUXAP8 was increased in our collected PTC
samples compared with the adjacent normal tissues (Fig. 1B). Furthermore, the data from the
GEPIA online software revealed that the DUXAP8 expression levels
were higher in the high-grade (IV) PTC samples compared with those
in low-grade (I) samples (Fig. 1C).
In addition, the GEPIA database revealed that high DUXAP8
expression was associated with poor overall survival in patients
with PTC (Fig. 1D).
As revealed in Table
I, the expression of DUXAP8 was associated with tumor size and
TNM stage, but not with sex, age, extrathyroidal extension or lymph
node metastasis in patients with PTC.
 | Table I.Association between DUXAP8 expression
level and clinicopathological characteristics of patients with
PTC. |
Table I.
Association between DUXAP8 expression
level and clinicopathological characteristics of patients with
PTC.
|
|
| DUXAP8
expression |
|
|---|
|
|
|
|
|
|---|
|
Characteristics | Cases | Low | High | P-value |
|---|
| Sex |
|
|
| 0.606 |
|
Male | 31 | 14 | 17 |
|
|
Female | 29 | 16 | 13 |
|
| Age (years) |
|
|
| 0.204 |
|
≤45 | 31 | 13 | 18 |
|
|
>45 | 29 | 17 | 12 |
|
| Extrathyroidal
extension |
|
|
| 0.438 |
|
Yes | 28 | 12 | 16 |
|
| No | 32 | 18 | 14 |
|
| Lymph node
metastasis |
|
|
| 0.601 |
| N0 | 25 | 11 | 14 |
|
| N1 | 35 | 19 | 16 |
|
| Tumor size
(cm) |
|
|
| 0.019 |
|
<2 | 32 | 21 | 11 |
|
|
2-4 | 28 | 9 | 19 |
|
| TNM stage |
|
|
| 0.017 |
|
I–II | 24 | 17 | 7 |
|
|
III–IV | 26 | 13 | 23 |
|
DUXAP8 knockdown inhibits cell
proliferation and induces cell apoptosis
DUXAP8 was also upregulated in the PTC cell lines,
CGTH W-3 and TPC1, compared with the human thyroid follicular
epithelial cells, Nthy-ori 3-1 (Fig.
2A). Subsequently, the expression of DUXAP8 was silenced in PTC
cell lines using the si-RNA technology in order to investigate the
effect of DUXAP8 in PTC. The results revealed that compared with
the si-normal control (si-NC) group, transfection of CGTH W-3 and
TPC1 PTC cell lines with si-DUXAP8 significantly reduced DUXAP8
expression levels (Fig. 2B).
Furthermore, DUXAP8 silencing significantly attenuated CGTH W-3 and
TPC1 cell proliferation compared with the si-NC group (Fig. 2C and D). However, compared with the
si-NC group, transfection with si-DUXAP8 significantly induced CGTH
W-3 and TPC1 cell apoptosis (Fig. 2E and
F).
DUXAP8 knockdown inactivates the
MAPK/ERK pathway
It has been reported that the constitutive
activation of the MAPK/ERK pathway contributes to the malignant
progression of PTC (22). However,
the effects of DUXAP8 on the PTC-associated signaling pathways and
molecules have not been studied. Therefore, the results of the
present study demonstrated that DUXAP8 downregulation reduced the
phosphorylation levels of mitogen-activated protein kinase kinase
1/2 (p-MEK1/2) and ERK1/2 (p-ERK1/2) in the CGTH W-3 and TPC1 PTC
cell lines (Fig. 3A-C). Additionally,
the mRNA and protein expression levels of CCND1 and c-Myc, two
downstream target genes of the MAPK/ERK pathway (24), were also downregulated following
silencing of DUXAP8 in PTC cell lines (Fig. 3A and D-G).
 | Figure 3.DUXAP8 knockdown inactivates the
MAPK/ERK pathway. (A-E) Western blotting was applied for the
detection of p-MEK1/2, p-ERK1/2, CCND1 and c-Myc level in PTC
cells. (F and G) RT-qPCR was applied for the detection of mRNA
expression of (F) CCND1 and (G) c-Myc. *P<0.05 and **P<0.01,
siRNA-DUXAP8 compared with siRNA-NC. DUXAP8, double homeobox A
pseudogene 8; p-, phosphorylated; MEK1/2, mitogen-activated protein
kinase kinase 1/2; ERK1/2, extracellular signal-regulated kinase
1/2; cyclin D1, CCND1; RT-qPCR, reverse transcription-quantitative
polymerase chain reaction; t-, total; siRNA, small interfering RNA;
NC, negative control. |
DUXAP8 negatively regulates
miR-20b-5p
Subsequently, the present study investigated whether
specific miRNAs could be sponged by DUXAP8. Screening in the miRDB
online database revealed numerous miRNAs complementary to DUXAP8.
Among them, miR-20b-5p attracted our attention. It has been
reported that miR-20b-5p is downregulated in PTC, and inhibits the
progression of the disease (25)
(Fig. 4A). Therefore, the association
between DUXAP8 and miR-20b-5p was further verified. Transfection of
the CGTH W-3 and TPC1 PTC cell lines with miR-20b-5p mimic
significantly increased miR-20b-5p expression levels (Fig. 4B), indicating that the transfection
procedure was efficiently performed. miR-20b-5p overexpression
reduced the luciferase activity of DUXAP8 (Fig. 4C and D) and downregulated the
expression of DUXAP8 in PTC cells (Fig.
4E), which was consistent with the reciprocal suppression
between miRNA and lncRNA as recently reported (26). Furthermore, DUXAP8 knockdown
significantly increased the expression of miR-20b-5p in CGTH W-3
and TPC1 cells (Fig. 4F). The RIP
assay results in the PTC cell lines revealed that DUXAP8 and
miR-20b-5p were enriched in the Ago2-pulled down protein complex
(Fig. 4G and H). The abovementioned
findings suggested that DUXAP8 could directly target miR-20b-5p in
PTC cells.
 | Figure 4.DUXAP8 negatively regulates
miR-20b-5p. (A) The online database miRDB (http://mirdb.org/) was used to predict the
complementary sites between DUXAP8 and miR-20b-5p. (B) RT-qPCR was
applied for the detection of miR-20b-5p in PTC cells. (C and D)
Dual luciferase reporter assay was used to detect the association
between miR-20b-5p and DUXAP8 in PTC cells. (E and F) RT-qPCR was
applied for the detection of DUXAP8 and miR-20b-5p in PTC cells. (G
and H) RIP assay was used to further confirm the interaction
between miR-20b-5p and DUXAP8 in PTC cells. **P<0.01, miR-20b-5p
mimic compared with miR-NC mimic; siRNA-DUXAP8 compared with
siRNA-NC; and Ago 2 compared with IgG. ***P<0.001, miR-20b-5p
mimic compared with miR-NC mimic; siRNA-DUXAP8 compared with
siRNA-NC. DUXAP8, double homeobox A pseudogene 8; miR, microRNA;
RT-qPCR, reverse transcription-quantitative polymerase chain
reaction; PTC, papillary thyroid carcinoma; RIP, RNA
immunoprecipitation; siRNA, small interfering RNA; NC, negative
control; WT, wild-type; MUT, mutant. |
DUXAP8 is positively associated with
miR-20b-5p-related target genes
It has been reported that miR-20b-5p regulates the
MAPK/ERK signaling and PTC cell proliferation via targeting SOS1
(25). In the TCGA-THCA data,
Spearman's correlation analysis demonstrated that the expression of
DUXAP8 was positively associated with the SOS1 mRNA levels
(Fig. 5A). In addition, in the
TCGA-THCA dataset, the expression of DUXAP8 was strongly associated
with the downstream target genes of the MAPK/ERK signaling,
including c-Myc and CCND1 (Fig. 5B and
C). Moreover, in our collected tissues, SOS1 was increased in
PTC tumor tissues compared with the adjacent normal tissues
(Fig. 5D), and was positively
correlated with DUXAP8 level in the PTC tumor tissues (Fig. 5E).
 | Figure 5.DUXAP8 is positively correlated with
miR-20b-5p-related target genes. TCGA-THCA dataset was used to
exhibit the association between DUXAP8 level and mRNA level of (A)
SOS1, (B) c-Myc, and (C) CCND1. (D) RT-qPCR was applied for the
examination of SOS1 mRNA level in PTC tumor tissues and the normal
tissues. (E) Spearman's correlation analysis was used to analyze
the correlation between SOS1 mRNA level and DUXAP8 level in the PTC
tumor tissues. (F) The binding sites among DUXAP8, miR-20b-5p and
SOS1 are presented. (G) RT-qPCR was applied for the detection of
mRNA expression of SOS1. **P<0.01, siRNA-DUXAP8 compared with
siRNA-NC. ***P<0.001, PTC tumor tissues compared to healthy
tissues. DUXAP8, double homeobox A pseudogene 8; miR, microRNA;
TCGA, The Cancer Genome Atlas; THCA, thyroid carcinoma; SOS1, son
of sevenless 1; cyclin D1, CCND1; RT-qPCR, reverse
transcription-quantitative polymerase chain reaction; PTC,
papillary thyroid carcinoma; siRNA, small interfering RNA; NC,
negative control; WT, wild-type; MUT, mutant. |
As revealed in Fig. 5F
SOS1 could share the same binding sites with DUXAP8 on miR-20b-5p.
Subsequently, the effect of DUXAP8 on the expression of SOS1 was
evaluated. As anticipated, DUXAP8 knockdown downregulated the mRNA
expression levels of SOS1 (Fig. 5G)
in PTC cells.
DUXAP8 knockdown inhibits cell
proliferation and induces cell apoptosis via regulating SOS1
To explore the effect of DUXAP8 and SOS1 on the
progression of PTC, SOS1 was overexpressed in PTC cell lines.
Therefore, transfection of CGTH W-3 and TPC1 cells with
pcDNA3.1-SOS1, increased the mRNA and protein levels of SOS1
(Fig. 6A-C). In addition, si-DUXAP8
inhibited PTC cell proliferation (Fig. 6D
and E) and induced apoptosis (Fig. 6F
and G). These effects were reversed following transfection of
CGTH W-3 and TPC1 cells with the SOS1 overexpression plasmid
pcDNA3.1-SOS1.
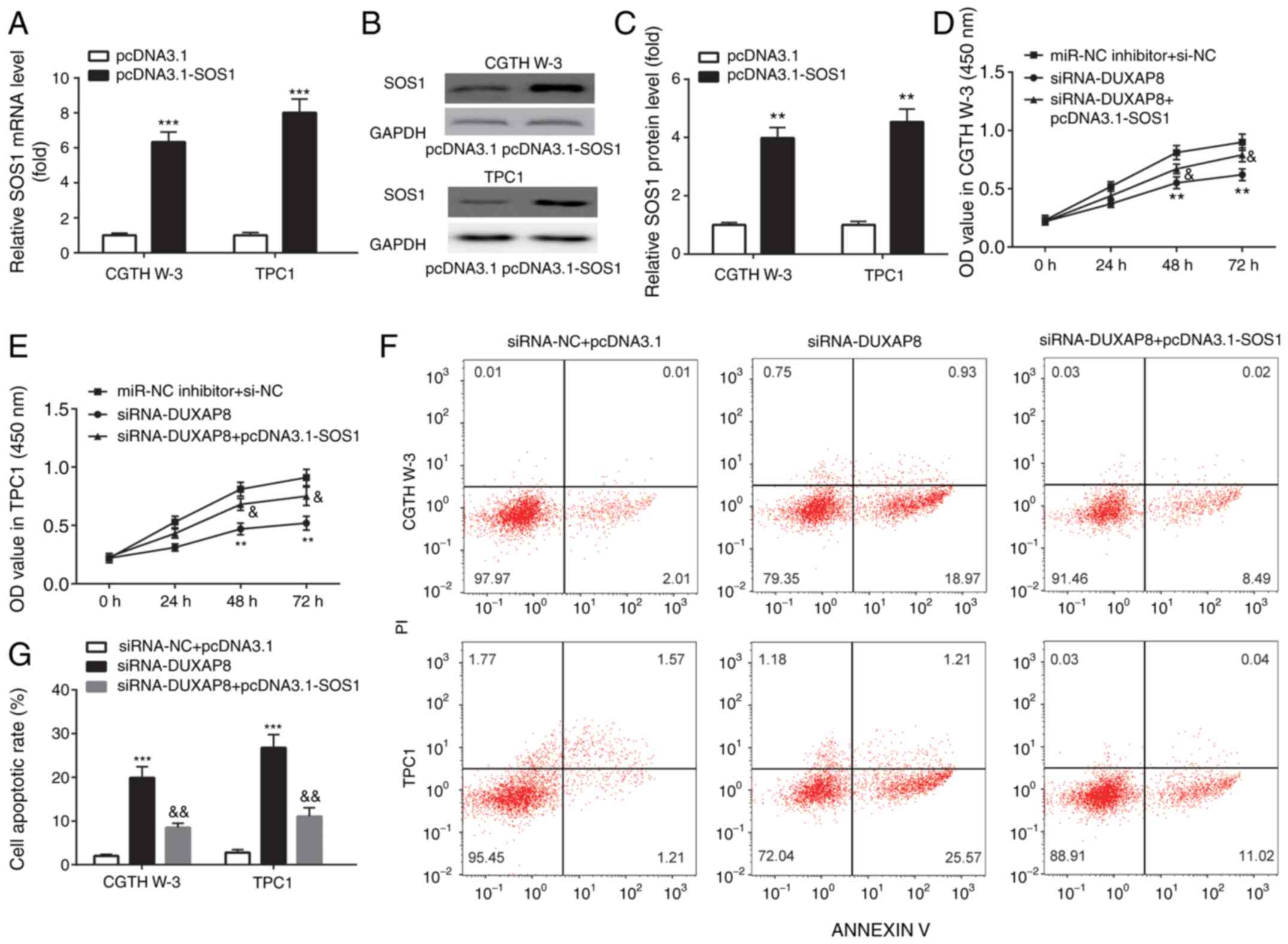 | Figure 6.DUXAP8 knockdown inhibits cell
proliferation and induces cell apoptosis by regulation of SOS1.
(A-C) RT-qPCR and western blotting were applied for the examination
of mRNA and protein expression of SOS1 in PTC cell lines. (D and E)
CCK-8 assays were used to detect cell proliferation, and (F and G)
flow cytometric assays were used to detect cell apoptosis in PTC
cell lines. **P<0.01, pcDNA3.1-SOS1 compared with pcDNA3.1;
siRNA-DUXAP8 compared with miR-NC inhibitor+si-NC. ***P<0.001,
pcDNA3.1-SOS1 compared with pcDNA3.1; siRNA-DUXAP8 compared with
siRNA-NC+pcDNA3.1. &P<0.05 and
&&P<0.01, siRNA-DUXAP8+pcDNA3.1-SOS1 compared
with siRNA-DUXAP8. DUXAP8, double homeobox A pseudogene 8; SOS1,
son of sevenless 1; RT-qPCR, reverse transcription-quantitative
polymerase chain reaction; PTC, papillary thyroid carcinoma; CCK-8,
Cell Counting Kit-8; miR, microRNA; siRNA, small interfering RNA;
NC, negative control; PI, propidium iodide. |
Discussion
The incidence of PTC is growing globally (27). Several non-coding RNAs, including
lncRNAs and miRNAs, have been identified in recent years (28). It has been reported that lncRNA DUXAP8
promotes the growth of non-small cell lung cancer (11), pancreatic carcinoma (12), and renal cell carcinoma (13). However, the effect of lncRNA DUXAP8 on
PTC remains elusive. In 2018, a genome-wide analysis revealed that
lncRNA DUXAP8 was upregulated in human thyroid cancer (10). Consistently, the present study
demonstrated that the expression of DUXAP8 was increased in the PTC
TCGA-THCA data. This finding was further confirmed in tumor samples
from patients with PTC. Furthermore, transfection of PTC cell lines
with si-DUXAP8 inhibited cell proliferation and induced cell
apoptosis. This finding was also consistent with previous studies
on the role of DUXAP8 in other types of cancer (11–13). The
abovementioned results supported the promotional effects of DUXAP8
on the progression of PTC.
The MAPK/ERK signaling regulates cell viability and
apoptosis via phosphorylating numerous substrates (29), and serves a crucial role during the
tumorigenesis of multiple cancers, including PTC (30). ERK2 regulates several biological cell
processes via regulating the expression of downstream substrates of
the MAPK/ERK signaling, such as c-Myc and CCND1 (27). Consistently, in the present study,
transfection of PTC cells with si-DUXAP8 decreased the levels of
p-MEK1/2 and p-ERK1/2 and downregulated CCND1 and c-Myc expression,
resulting in the inactivation of the MAPK/ERK pathway.
As acknowledged, miRNAs regulate various biological
processes, including cell proliferation and apoptosis (19). However, the effect of the lncRNA
DUXAP8-miRNA network in the pathogenesis of PTC has not been
identified. Therefore, miRNAs, acting as tumor suppressors during
the progression of PTC, attracted the attention of the present
study. Bioinformatics analysis predicted that DUXAP8 was
complementary to miR-20b-5p. Regarding the role of miR-20b-5p in
PTC, a study demonstrated, using next-generation deep sequencing
and microarray analysis, that the expression of miR-20b-5p was
decreased in PTC compared with normal thyroid tissues (31). In addition, miR-20b acted as a tumor
suppressor in PTC via regulating the MAPK/ERK signaling (25). Furthermore, in 2018, a miRNA profiling
analysis revealed that the expression of miR-20b-5p was
downregulated and was associated with the aggressive behavior of
PTC (32). As acknowledged, a
reciprocal suppression between miRNA and lncRNA has been revealed
(26), for example, TUSC7 and miR-23b
in gastric cancer (33), as well as
lncRNA LOC728196 and miR-513c in glioma (34), and TUG1 and miR-145 in bladder cancer
(35). Herein, a reciprocal
suppression between DUXAP8 and miR-20b-5p was revealed, i.e.,
DUXAP8 and miR-20b-5p suppressed the expression of each other,
miR-20b-5p decreased the luciferase activity of DUXAP8, and RIP
assays revealed that miR-20b-5p could directly target DUXAP8.
Therefore, the present study is the first to the best of our
knowledge, to identify the effect of the DUXAP8/miR-20b-5p network
in PTC.
SOS1, a downstream target for miR-20b-5p in PTC
(25), also exerts an oncogenic
function in multiple types of cancer (36), such as ovarian cancer (37), glioblastoma (38) and colorectal cancer (39). However, the association between DUXAP8
and SOS1 in PTC has not been identified. Herein, DUXAP8 knockdown
downregulated the expression of SOS1. Additionally, the Spearman's
correlation analysis revealed that DUXAP8 was positively associated
with SOS1, c-Myc and CCND1 in the TCGA-THCA data. In addition,
DUXAP8 was positively correlated with SOS1 in our collected PTC
tumor tissues. Finally, SOS1 overexpression rescued the DUXAP8
downregulation-mediated inhibition of cell proliferation and
promotion of cell apoptosis in PTC cells. Therefore, these results
were the first to reveal the positive association between DUXAP8
and SOS1 expression in PTC.
Collectively, the present study, to the best of our
knowledge, was the first to identify DUXAP8 as a novel upregulated
lncRNA in PTC and provided new insights in understanding the role
of the DUXAP8/miR-20b-5p/SOS1 network in the progression of
PTC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The data and materials were available from the
corresponding author on special request.
Authors' contributions
Both authors, RP and SY performed the
experimentation and data analysis. SY designed the study and wrote
the manuscript. Both authors read and approved the manuscript and
agree to be accountable for all aspects of the research in ensuring
that the accuracy or integrity of any part of the work are
appropriately investigated and resolved.
Ethics approval and consent to
participate
All experimental procedures were approved by the
Ethics Committee of the First Hospital of Jilin University. Written
informed consent was provided by all patients prior to enrollment
in this study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Carling T and Udelsman R: Thyroid cancer.
Annu Rev Med. 65:125–137. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Fagin JA and Wells SA Jr: Biologic and
clinical perspectives on thyroid cancer. N Engl J Med.
375:1054–1067. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sipos JA and Mazzaferri EL: Thyroid cancer
epidemiology and prognostic variables. Clin Oncol (R Coll Radiol).
22:395–404. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kunavisarut T: Diagnostic biomarkers of
differentiated thyroid cancer. Endocrine. 44:616–622. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Li J, Tian H, Yang J and Gong Z: Long
noncoding RNAs regulate cell growth, proliferation, and apoptosis.
DNA Cell Biol. 35:459–470. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lv W, Wang L, Lu J, Mu J, Liu Y and Dong
P: Long noncoding RNA KIAA0125 potentiates cell migration and
invasion in gallbladder cancer. BioMed Res Int. 2015:1084582015.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liu L, Yang J, Zhu X, Li D, Lv Z and Zhang
X: Long noncoding RNA H19 competitively binds miR-17-5p to regulate
YES1 expression in thyroid cancer. FEBS J. 283:2326–2339. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Dai W, Tian Y, Jiang B and Chen W:
Down-regulation of long non- coding RNA AFAP1-AS1 inhibits tumor
growth, promotes apoptosis and decreases metastasis in thyroid
cancer. Biomed Pharmacother. 99:191–197. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lu W, Xu Y, Xu J, Wang Z and Ye G:
Identification of differential expressed lncRNAs in human thyroid
cancer by a genome-wide analyses. Cancer Med. 7:3935–3944. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sun M, Nie FQ, Zang C, Wang Y, Hou J, Wei
C, Li W, He X and Lu KH: The Pseudogene DUXAP8 promotes
non-small-cell lung cancer cell proliferation and invasion by
epigenetically silencing EGR1 and RHOB. Mol Ther. 25:739–751. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lian Y, Yang J, Lian Y, Xiao C, Hu X and
Xu H: DUXAP8, a pseudogene derived lncRNA, promotes growth of
pancreatic carcinoma cells by epigenetically silencing CDKN1A and
KLF2. Cancer Commun (Lond). 38:642018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chen J, Lou W, Ding B and Wang X:
Overexpressed pseudogenes, DUXAP8 and DUXAP9, promote growth of
renal cell carcinoma and serve as unfavorable prognostic
biomarkers. Aging (Albany NY). 11:5666–5688. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Thomson DW and Dinger ME: Endogenous
microRNA sponges: Evidence and controversy. Nat Rev Genet.
17:272–283. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tay Y, Rinn J and Pandolfi PP: The
multilayered complexity of ceRNA crosstalk and competition. Nature.
505:344–352. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Han P, Li J, Zhang B, Lv J, Li Y, Gu X, Yu
Z, Jia Y, Bai X, Li L, et al: The lncRNA CRNDE promotes colorectal
cancer cell proliferation and chemoresistance via
miR-181a-5p-mediated regulation of Wnt/β-catenin signaling. Mol
Cancer. 16:92017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ba Z, Gu L, Hao S, Wang X, Cheng Z and Nie
G: Downregulation of lncRNA CASC2 facilitates osteosarcoma growth
and invasion through miR-181a. Cell Prolif. 51:e124092018.
View Article : Google Scholar
|
|
18
|
Jazdzewski K, Liyanarachchi S, Swierniak
M, Pachucki J, Ringel Md, Jarzab B and de la chapelle A:
Polymorphic mature microRNAs from passenger strand of pre-miR-146a
contribute to thyroid cancer. Proc Natl Acad Sci USA.
106:1502–1505. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li M, Teruya-Feldstein J and Weinberg RA:
Tumour invasion and metastasis initiated by microRNA-10b in breast
cancer. Nature. 449:682–688. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Edge SB, Byrd DR, Compton CC, Fritz AG and
Greene FL: AJCC Cancer Staging Manual. 7th edition. Springer; New
York, NY: 2010
|
|
21
|
Rau A, Flister M, Rui H and Auer PL:
Exploring drivers of gene expression in the Cancer Genome Atlas.
Bioinformatics. 35:62–68. 2019.PubMed/NCBI
|
|
22
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Research.
45:W98–W102. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Deschenes-Simard X, Kottakis F, Meloche S
and Ferbeyre G: ERKs in cancer: Friends or foes? Cancer Res.
74:412–419. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hong S, Yu S, Li J, Yin Y, Liu Y, Zhang Q,
Guan H, Li Y and Xiao H: MiR-20b displays tumor-suppressor
functions in papillary thyroid carcinoma by regulating the MAPK/ERK
signaling pathway. Thyroid. 26:1733–1743. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Huang Y: The novel regulatory role of
lncRNA-miRNA-mRNA axis in cardiovascular diseases. J Cell Mol Med.
22:5768–5775. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Harrow J, Frankish A, Gonzalez JM,
Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa
A, Searle S, et al: GENCODE: The reference human genome annotation
for The ENCODE Project. Genome Res. 22:1760–1774. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Cargnello M and Roux PP: Activation and
function of the MAPKs and their substrates, the MAPK-activated
protein kinases. Microbiol Mol Biol Rev. 75:50–83. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Peyret V, Nazar M, Martín M, Quintar AA,
Fernandez EA, Geysels RC, Fuziwara CS, Montesinos MM, Maldonado CA,
Santisteban P, et al: Functional toll-like receptor 4
overexpression in papillary thyroid cancer by MAPK/ERK-induced ETS1
transcriptional activity. Mol Cancer Res. 16:833–845. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Swierniak M, Wojcicka A, Czetwertynska M,
Stachlewska E, Maciag M, Wiechno W, Gornicka B, Bogdanska M,
Koperski L, de la Chapelle A and Jazdzewski K: In-depth
characterization of the microRNA transcriptome in normal thyroid
and papillary thyroid carcinoma. J Clin Endocrinol Metab.
98:E1401–E1409. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Jahanbani I, Al-Abdallah A, Ali RH,
Al-Brahim N and Mojiminiyi O: Discriminatory miRNAs for the
management of papillary thyroid carcinoma and noninvasive
follicular thyroid neoplasms with papillary-like nuclear features.
Thyroid. 28:319–327. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Qi P, Xu MD, Shen XH, Ni SJ, Huang D, Tan
C, Weng WW, Sheng WQ, Zhou XY and Du X: Reciprocal repression
between TUSC7 and miR-23b in gastric cancer. Int J Cancer.
137:1269–1278. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang O, Huang Y, Wu H, Zheng B, Lin J and
Jin P: lncRNA LOC728196/miR-513c axis facilitates glioma
carcinogenesis by targeting TCF7. Gene. 679:119–125. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Tan J, Qiu K, Li M and Liang Y:
Double-negative feedback loop between long non-coding RNA TUG1 and
miR-145 promotes epithelial to mesenchymal transition and
radioresistance in human bladder cancer cells. FEBS Lett.
589:3175–3181. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
De S, Dermawan JK and Stark GR: EGF
receptor uses SOS1 to drive constitutive activation of B in cancer
cells. Proc Natl Acad Sci USA. 111:11721–11726. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chen H, Wu X, Pan ZK and Huang S:
Integrity of SOS1/EPS8/ABI1 tri-complex determines ovarian cancer
metastasis. Cancer Res. 70:9979–9990. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wang M, Wu Q, Fang M, Huang W and Zhu H:
miR-152-3p sensitizes glioblastoma cells towards cisplatin via
regulation of SOS1. Onco Targets Ther. 12:9513–9525. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Jiang H, Dong L, Gong F, Gu Y, Zhang H,
Fan D and Sun Z: Inflammatory genes are novel prognostic biomarkers
for colorectal cancer. Int J Mol Med. 42:368–380. 2018.PubMed/NCBI
|




















