Introduction
Renal cell carcinoma (RCC) is the most lethal
urologic tumor, accounting for 2–3% of global adult malignancies in
2017, and the annual morbidity due to RCC is constantly rising
(1,2).
RCC is two times more common in men than in women, and ~50% of
patients already have metastases at initial diagnosis (3–6). Clear
cell RCC (ccRCC) originates from the proximal tubule of the kidney
and is the most common subtype of RCC, accounting for 75–80% of all
cases (7–9). However, ccRCC is not sensitive to
radiotherapy or chemotherapy, and the sensitivity of ccRCC to
immunotherapy and targeted therapy remains to be studied (10–14). Thus,
partial nephrectomy remains the best approach to treat localized
tumors (15,16). An enhanced understanding of the
molecular pathogenesis of RCC is of great importance for
formulating tumor prevention strategies and guiding clinical
treatment (17–19).
Sirtuin 1 (SIRT1) is a NAD+-dependent deacetylase
belonging to the mammalian homolog of yeast Sir2 protein that
serves an important role in regulating several bioactive substances
(20–23). The activity of SIRT deacetylase
depends on the concentration of NAD+ in the cytoplasm, so that when
the concentration of NAD+ increases, SIRT1 deacetylase activity
also increases (24,25). AMP-activated protein kinase (AMPK) is
a key kinase involved in the regulation of cellular energy
metabolism (26,27). AMPK responds to the ratio of AMP/ATP
in the cell and accordingly adjusts the activity of the enzyme to
maintain an energy balance (28).
SIRT1 is considered an upstream activator of AMPK, and AMPK is
considered a downstream molecule of SIRT1, capable of regulating
energy metabolism in apoptosis (29).
Additionally, AMPK can enhance SIRT1 activity by increasing
intracellular NAD+ levels, leading to the deacetylation and
activation of the SIRT1 target protein, regulation of mitochondrial
function and maintenance of energy balance (30,31).
Conversely, current evidence supports the opposite effect of SIRT1
as a tumor protein or tumor suppressor in different types of tumor
(32–35).
Thus, the present study aimed to investigate the
role of SIRT1/AMPK signaling in RCC progression and the potential
mechanisms of its involvement. It was hypothesized that SIRT1 may
act as a tumor suppressor in RCC by increasing phosphorylated AMPK
expression.
Materials and methods
Study design and patients
The present study evaluated 100 patients (mean age,
57.6 years; age range, 43–81 years) who underwent radical
nephrectomy at The Second Affiliated Hospital of Anhui Medical
University (Hefei, China) between July 2016 and October 2019.
Patients who received chemotherapy or radiotherapy before surgery
were excluded from analyses. In total, 100 cases of primary RCC
tissues and matched adjacent normal tissues (1 cm from the tumor
margin) were obtained. After surgical resection, the tumor and
adjacent normal tissues were collected and stored at −80°C until
use. The study was approved by the Institutional Research Ethics
Committee of The Second Affiliated Hospital of Anhui Medical
University, and written informed consent was obtained from all
patients.
Tissue microarray (TMA)
immunohistochemical analysis
Immunohistochemical staining was performed on tissue
chips containing tumor and adjacent tissues from patients with RCC.
Surgically resected tumor specimens were fixed with 10% neutral
formalin and embedded in paraffin. The tissue block was then sliced
into 3-µm-thick sections. The slices were heated at 60°C for 60–120
min, then dewaxed in xylene, rehydrated with a series of graded
ethanol and boiled in a pressure cooker in 0.01 M citrate buffer
(pH 6.0) for 2 min. Hydrogen peroxide (0.3%) was used to block
endogenous peroxide activity, and the sections were incubated with
10% normal goat serum (Gibco; Thermo Fisher Scientific, Inc.) in
TBS with 0.5% Tween-20 for 1 h at room temperature to block
non-specific binding. The primary antibody (anti-SIRT1; cat. no.
ab110304; 1:30; Abcam) was incubated overnight at 4°C. The
biotin-labeled secondary antibody (cat. no. PV-6000; 1:500; OriGene
Technologies, Inc.) was incubated at room temperature for 1 h.
For quantitative scoring based on
immunohistochemistry results (positive light microscope;
magnification, ×400; Zen blue 3.1 software; Zeiss AG), the scoring
criteria were based on staining intensity (0, no staining; 1, weak
staining; 2, medium staining; and 3, strong staining) and the
percentage of positively stained cells (1, ≤25%; 2, 26–50%; 3,
51–75%; and 4, >75%). The final score was calculated by adding
these two scores. Total score cut-offs of ≤4 and >4 were used to
divide patients into low and high SIRT1 expression groups,
respectively.
Cell line and cell culture
The human RCC 786-O and ACHN cell lines were
obtained from The Cell Bank of Type Culture Collection of The
Chinese Academy of Sciences. The cells were cultured in RPMI-1640
medium supplemented with 10% FBS and 5% penicillin and streptomycin
(all Gibco; Thermo Fisher Scientific, Inc.) at 37°C in a 5%
CO2 incubator, and the medium was renewed every two
days.
SIRT1 overexpression and RNA
interference
SIRT1 overexpression plasmids (3 and 5 mg; OE-SIRT1)
and empty vector (OE-NC) (both from Shanghai GenePharma Co., Ltd.)
were transfected into 786-O and ACHN cells for 20 min at room
temperature using Lipofectamine® 2000 transfection
reagent (Beyotime Institute of Biotechnology) according to the
manufacturer's instructions.
Small interfering RNA (siRNA) targeting SIRT1
(si-SIRT#1, 5′-GGAGAUGAUCAAGAGGCAATT-3′ and si-SIRT#2,
5′-GGAAAUAUAUCCUGGACAATT-3′), AMPK (si-AMPK#1,
5′-UGACCUCAACUACAUGGUUTT-′3 and si-AMPK#2,
5′-GCGGGAAUCCAAAGGAUAATT-′3) and scrambled negative control
(si-NC#1, 5′-UUCUCCGAACGUGUCACGUTT-′3 and si-NC#2,
5′-UUCUCCGAACGUGUCACGUTT-′3) were also designed by Shanghai
GenePharma Co., Ltd. 786-O and ACHN cells were cultured in a 6-well
plate to 60% confluence, and were transfected with 50 nmol/l of
designated siRNA using Lipofectamine 2000 transfection reagent at
37°C for 6 h, according to the manufacturer's instructions. Western
blotting and reverse transcription-quantitative (RT-q)PCR were used
to detect gene knockout efficiency 48 h after transfection.
MTT assay
To assess cell proliferation, cells were plated into
96-well culture plates overnight at a density of 5×103
cells/well, and then overexpression, knockdown, and empty carrier
models were constructed and then cultured for 24, 48 or 72 h. The
control and transfected groups received the same amount of DMSO.
Subsequently, 10 µl MTT solution (2.5 mg/ml) was added to each
well, and the cells were incubated at 37°C for 4 h. DMSO (150 µl)
was added to dissolve the obtained crystals. The optical density
(OD) value was measured at 570 nm using an enzyme labeling
instrument (Thermo Fisher Scientific, Inc.). The cell proliferation
inhibition rate was calculated using the following formula: (1-OD
experimental group/OD control group) ×100%.
Transwell assay
Cell migration and invasion assays were performed
using Transwell chambers (Corning, Inc.). A total of
2×104 786-O and ACHN cells were resuspended in
serum-free medium and added to the upper chambers, while medium
containing 10% FBS was added to the bottom chambers. Transwell
chambers coated with Matrigel for 5 h at 37°C were used for cell
invasion. The cells were placed in an incubator at 37°C for 24 h.
Subsequently, the Transwell chambers were washed twice with PBS,
fixed with 4% paraformaldehyde at room temperature for 25 min,
stained with 0.4% crystal violet staining solution for 5 min and
rinsed with distilled water. Finally, the number of migrating or
invading cells was counted using a light inverted microscope
(magnification, ×400; Olympus Corporation).
Apoptosis assay
Apoptosis analysis was performed using the Annexin
V-FITC Apoptosis Detection kit (BD Biosciences). Briefly, cells
were seeded in 6-well plates at a cell density of 105
cells/well. Following overnight culture, cells were transfected as
aforementioned for 24 h. Cells from each group were collected and
centrifuged at 1,000 × g for 5 min at room temperature. After
washing twice with PBS, the supernatant was discarded, and the
cells were gently resuspended in 500 µl binding buffer. After
adding 5 µl Annexin V-FITC, 10 µl propidium iodide was further
added and mixed. After incubation at room temperature in the dark
for 15 min, the cells were immediately subjected to detection by
flow cytometry (NAVIOS; Beckman Coulter, Inc.) and analyzed using
FlowJo VX 10.6.2 (FlowJo LLC).
Western blotting
The mashed tissues and cells were lysed in RIPA
lysis buffer (150 mM NaCl, 0.5% sodium deoxycholate, 0.1% SDS, 1%
Igepal, 50 mM Tris-HCl pH 8.0 and 2 mM EDTA) in an ice bath for 30
min and centrifuged at 18,000 × g rpm for 30 min at 4°C. The
supernatant was stored at −80°C. A BCA kit was used to determine
the protein concentration. The same amount of protein (8 µg) was
separated via 10% SDS-PAGE and transferred to a PVDF membrane.
After the PVDF membrane was blocked in 5% BSA (cat. no. 9048-46-8;
Beijing Solarbio Science & Technology Co., Ltd.) at room
temperature for 1 h, it was incubated with anti-SIRT1 (cat. no.
19A7AB4; 1:2,000; Abcam), anti-AMPK (cat. no. D63G4; 1:1,000; Cell
Signaling Technology, Inc.), anti-phospho-AMPKa (cat. no. D4D6D;
1:1,000; Cell Signaling Technology, Inc.) and anti-β-actin
antibodies (cat. no. T0022; 1:3,000; Affinity Biosciences)
overnight at 4°C. Subsequently, it was incubated with
HRP-conjugated goat anti-rabbit IgG (cat. no. S0001) and goat
anti-mouse IgG (cat. no. S0002) secondary antibodies (both
1:10,000; Affinity Biosciences) at room temperature for 1 h. The
color of the PVDF membrane was developed using an enhanced
chemiluminescence kit (Thermo Fisher Scientific, Inc.), and the
gray value was analyzed using ImageJ software (version 1.8.0;
National Institutes of Health).
Cell immunofluorescence
A total of 10,000 cells were seeded on a glass slide
in a 6-well plate. The next day, the cells were fixed with 4%
paraformaldehyde at room temperature for 20 min. Subsequently, they
were permeabilized with 0.1% TBS-Triton (cat. no. T8200; Beijing
Solarbio Science & Technology Co., Ltd.) and blocked with 1%
normal goat serum (cat. no. SL038; Beijing Solarbio Science &
Technology Co., Ltd.) at room temperature for 1 h. The primary
antibodies (anti-SIRT1 and anti-phospho-AMPKa; 1:100) were
incubated overnight at 4°C. The Cy3-labeled goat anti-mouse IgG
(cat. no. A0521) and the FITC-labeled goat anti-rabbit IgG (cat.
no. A0562) secondary antibodies (both 1:200; Beyotime Institute of
Biotechnology) were incubated for 1 h at room temperature. Finally,
DAPI staining was performed for 2 min at room temperature and
observed under a fluorescence microscope (magnification, ×400).
RT-qPCR
Total RNA was extracted using TRIzol®
reagent (Invitrogen; Thermo Fisher Scientific, Inc.), and then
reverse transcribed into cDNA using a reverse transcription kit
(Takara Bio, Inc.), according to the manufacturer's instructions.
Subsequently, the SYBR-Green Real-time Polymerase Chain Reaction
kit (Takara Bio, Inc.) was used to perform qPCR using the CFX96
system (Bio-Rad Laboratories, Inc.). The thermocycling conditions
were as follows: Denaturation at 95°C for 5 min, followed by 35
cycles of denaturation at 95°C for 30 sec, annealing at 55°C for 30
sec and extension at 72°C for 1 min. The 2−ΔΔCq method
(36) was used to calculate the
changes in expression levels. All RT-qPCR data was normalized by
comparison to an endogenous β-actin control sample. The following
primers were used: SIRT1 forward, 5′-ATCTGACTTTGCTCCCCTTAACC-3′ and
reverse, 5′-GGGCCCTGGTTGCAAGA-3′; AMPK forward,
5′-CCCACCATCACTCCATCTCT-3′ and reverse, 5′-AGCCTGCTTGGCACACTTAT-3′;
β-actin forward, 5′-TCCTTCCTGGGCATGGAGT-3′ and reverse,
5′-ACTGTGTTGGCGTACAGGTC-3′.
Data sets
The mRNA expression levels of SIRT1 were analyzed in
different stages (stage I–V) of renal cell carcinoma using Gene
Expression Profiling Interactive Analysis, based on thousands of
samples data from The Cancer Genome Atlas (TCGA) database
(http://gepia.cancer-pku.cn/index.html).
Statistical analysis
All experiments were performed independently three
times. The experimental data are expressed as the mean ± standard
deviation. χ2 test and Fisher's exact test were used to
assess the association between SIRT1 expression and
clinicopathological characteristics of the patients with RCC.
Overall survival (OS) was evaluated using the Kaplan-Meier method
with the log-rank test. Paired Student's t-test was used to compare
two paired groups (clinical tumor samples and adjacent normal
tissues) and one-way ANOVA with Bonferroni post-hoc test was used
to analyze the differences between >2 groups. All statistical
analyses were performed using the SPSS 23.0 software package (IBM
Corp.). P<0.05 was considered to indicate a statistically
significant difference.
Results
High SIRT1 expression is associated
with a favorable prognosis in patients with RCC
All patients were pathologically confirmed to
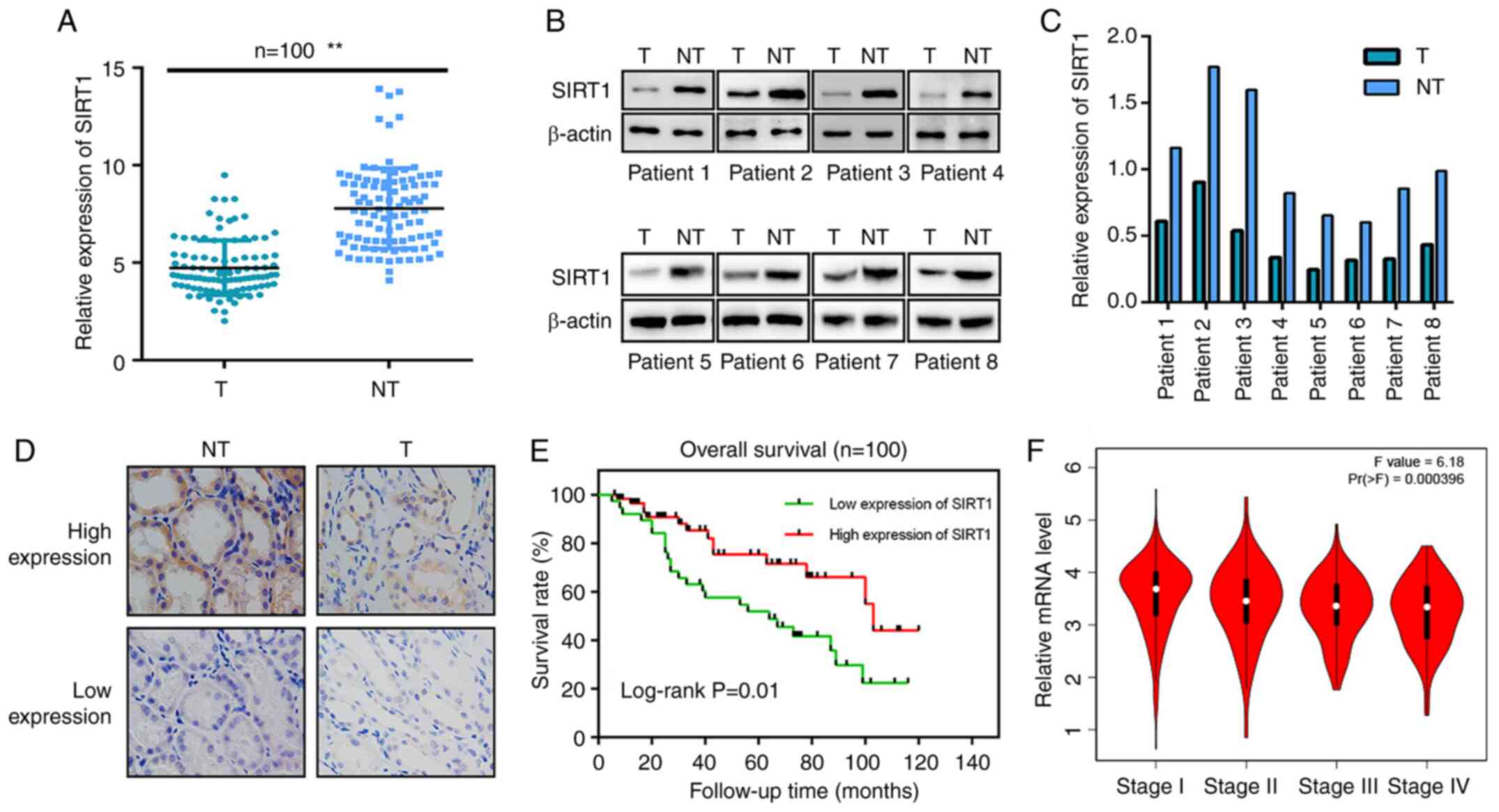
undergo radical resection for renal cancer. TMA immunohistochemical
staining results revealed lower SIRT1 expression in clinical tumor
samples than in adjacent normal tissues (Fig. 1A and D). A western blot assay using
samples from 8 patients revealed similar results, indicating that
SIRT1 expression in human renal carcinoma tissues was lower than
that in normal kidney tissues (Fig. 1B
and C). The clinical characteristics of the patients are shown
in Table I. All patients had complete
follow-up data (mean follow-up time, 47.5 months). The high and low
SIRT1 expression groups comprised 46 and 54 patients, respectively.
The Kaplan-Meier OS curves indicated significantly longer OS rates
in the high SIRT1 expression group. In terms of tumor stage, the
present statistical analysis revealed that the difference between
the high- and low-expression groups was statistically significant
(Table I). Low SIRT1 expression in
RCC cells was positively associated with advanced clinical stage
(Fig. 1F), higher World Health
Organization/International Society of Urologic Pathology (WHO/ISUP)
nuclear grade (37) and larger tumor
size (Table I).
 | Table I.Association between SIRT1 expression
and clinical characteristics of patients with RCC (n=100). |
Table I.
Association between SIRT1 expression
and clinical characteristics of patients with RCC (n=100).
| Clinical
characteristic | N | High
expression | Low expression | P-value |
|---|
| Age, years |
|
|
| 0.808 |
|
≤50 | 41 | 18 | 23 |
|
|
>50 | 59 | 28 | 31 |
|
| Sex |
|
|
| 0.752 |
|
Male | 43 | 19 | 24 |
|
|
Female | 57 | 27 | 30 |
|
| Affected
kidney |
|
|
| 0.929 |
|
Left | 45 | 21 | 24 |
|
|
Right | 55 | 25 | 30 |
|
| Tumor
classification |
|
|
| 0.629 |
|
ccRCC | 86 | 38 | 48 |
|
|
pRCC | 10 | 6 | 4 |
|
|
cRCC | 4 | 2 | 2 |
|
| WHO/ISUP grade |
|
|
| 0.001 |
| G1 | 28 | 23 | 5 |
|
|
G2-G3 | 65 | 21 | 44 |
|
| G4 | 7 | 2 | 5 |
|
| Clinical stage |
|
|
| 0.026 |
| I | 78 | 41 | 37 |
|
| II | 17 | 4 | 13 |
|
|
III–IV | 5 | 1 | 4 |
|
| Tumor size, cm |
|
|
| 0.044 |
|
<4 | 46 | 30 | 16 |
|
|
4-7 | 31 | 10 | 21 |
|
|
>7 | 23 | 6 | 17 |
|
SIRT1 inhibits the proliferation,
migration and invasion of RCC cells in vitro
Of the two siRNA knockdown models, si-SIRT1#1 and
si-SIRT1#2, it was observed that si-SIRT1#1 had a higher knockdown
efficiency in both 786-O and ACHN RCC cell lines (Fig. 2A). In addition, the overexpression
model demonstrated that, compared with the OE-NC, SIRT1
overexpression was positively associated with the amount of
transfected plasmid (3 and 5 mg; Fig.
2B). SIRT1 overexpression and knockdown models revealed that
following SIRT1 overexpression, the proliferation, migration and
invasion of RCC cells was inhibited, while SIRT1-knockdown enhanced
the proliferation, migration and invasion of cancer cells (Fig. 2C-E). The current transfected RCC model
exhibited differences in apoptosis, with increased early and late
apoptotic cells following SIRT1 overexpression and decreased early
and late apoptotic cells following SIRT1-knockdown; although no
significant difference was observed following SIRT1 overexpression
in 786-O cells, significant differences were observed for the other
conditions (Fig. 2F).
AMPK expression in RCC cells is
positively associated with SIRT1
Using western blotting and immunofluorescence
staining, it was revealed that following SIRT1 overexpression, the
levels of phosphorylated AMPK were significantly increased, while
when SIRT1 was knocked down, the levels of phosphorylated AMPK were
significantly decreased (Fig. 3A and
B).
SIRT1 inhibits the proliferation,
migration and invasion of RCC cells by targeting AMPK
In order to verify whether AMPK is a downstream
target of SIRT1, phenotypic response tests were performed. First,
the transfection efficiency of overexpressing SIRT1 and inhibiting
AMPK expression was verified by RT-qPCR and western blotting
(Fig. 4A-C). Si-AMPK#1 and si-AMPK#2
were used to interfere with AMPK expression, and since si-AMPK#1
exhibited higher interference efficiency, it was used in subsequent
experiments. Phenotypic rescue assays revealed that, in terms of
phosphorylated AMPK protein expression, AMPK inhibition reversed
the effect induced by SIRT1 overexpression (Fig. 4D). Additionally, MTT, Transwell and
apoptosis assays demonstrated that after AMPK inhibition, the
effects of SIRT1 overexpression on proliferation, invasion,
migration and apoptosis of RCC cells were reversed (Fig. 4E-G), indicating that AMPK may be a
downstream target of SIRT1.
 | Figure 4.SIRT1 regulates the proliferation,
migration and invasion of renal cell carcinoma cells by targeting
AMPK. (A) Reverse transcription-quantitative PCR was performed to
verify the transfection efficiency of AMPK siRNA (si-AMPK#1 and
si-AMPK#2). Western blotting was used to assess (B) the levels of
p-AMPK following AMPK inhibition and (C) SIRT1 expression following
SIRT1 overexpression. (D) Differences in protein expression levels
following SIRT1 overexpression and AMPK inhibition were detected
via western blotting in 786-O cells. (E) MTT, (F) Transwell
(magnification, ×400) and (G) apoptosis assays were performed to
detect differences in the proliferation, migration, invasion and
apoptosis of cells after AMPK inhibition in 786-O cells with SIRT1
overexpression. *P<0.05 and **P<0.01 vs. respective NC or
control. p, phosphorylated; AMPK, AMP-activated protein kinase; OE,
overexpression; NC, negative control; si, small interfering RNA;
SIRT1, sirtuin 1. |
Discussion
Previous studies (38,39) have
emphasized SIRT1 as a potential tumor suppressor protein to inhibit
RCC tumorigenesis, but the role of SIRT1/AMPK signaling in RCC
progression is yet to be clarified (40). The present study used western blotting
and immunohistochemistry to detect the expression levels of SIRT1
in clinical samples, revealing lower SIRT1 expression in cancerous
renal tissues than in normal renal tissues. Low SIRT1 expression in
RCC tissues was associated with advanced clinical stage, higher
WHO/ISUP nuclear grade and larger tumor size, which are negative
prognostic features. The current results indicated that SIRT1 may
serve a biological role in the development of RCC. Furthermore, a
lower OS rate was associated with low SIRT1 expression than with
high SIRT1 expression. Therefore, low SIRT1 expression indicated a
poor prognosis in patients with RCC, consistent with the different
expression levels of SIRT1 at different tumor stages according to
TCGA database. However, previous studies have revealed that in
esophageal squamous cell carcinoma and osteosarcoma, high SIRT1
expression is associated with a poor prognosis, which suggests that
SIRT1 may serve different roles in different types of tumor
(41–43).
SIRT1 is a type III histone deacetylase involved in
regulating various physiological processes, including gene
transcription, energy metabolism, cell cycle and apoptosis
(44–47). Through TCGA database, it was revealed
that among the numerous downstream signals of SIRT1, AMPK
expression was closely associated with SIRT1 expression in kidney
cancer (48). SIRT1 activation leads
to deacetylation of lysine residues on liver kinase B1 (LKB1),
which in turn enhances LKB1 kinase activity and leads to AMPK
phosphorylation (49). Concurrently,
the AMP/ATP ratio increases during insufficient cellular energy
levels, subsequently activating AMPK (50). Activated AMPK increases the expression
and activity of nicotinamide phosphoribosyl transferase, leading to
an increase in NAD+ concentration, and further SIRT1 activation
(51). Furthermore, the present
results suggested that phosphorylated AMPK may have an anticancer
role as a downstream target of SIRT1.
The present study used a plasmid transfection in
vitro system to overexpress and knockdown SIRT1 in RCC cells.
Results of Transwell and MTT assays revealed that the
proliferation, migration and invasion of RCC cells were inhibited
after SIRT1 overexpression, while knockdown of SIRT1 expression
mediated the opposite effects. Apoptosis experiments indicated that
in terms of early apoptosis, SIRT1 overexpression significantly
promoted apoptosis in both cell lines. However, in terms of late
apoptosis, the effect of SIRT1 overexpression in 786-O cells was
not statistically significant. A previous study has revealed that
SIRT1 may play a role by affecting the energy metabolism of tumor
cells (52). AMPK is an enzyme
closely associated with cell energy metabolism, and as a downstream
molecule of SIRT1, it has a strong association with it in kidney
cancer (53). Western blotting and
immunofluorescence were used in the present study to confirm that
when regulating SIRT1 expression, the levels of phosphorylated AMPK
also changed in the same way. Therefore, the current results
suggested that AMPK may act as a key downstream protein of SIRT1 to
inhibit RCC cells.
Previous studies have revealed that AMPK activation
directly phosphorylates its target proteins or transcriptionally
controls its target genes, such as PGC1α and p300 (54,55). The
present aforementioned results indicated that SIRT1 was positively
associated with the levels of phosphorylated AMPK. The next step
was to clarify whether AMPK is a downstream molecule of SIRT1 and
the specific mechanism by which SIRT1 regulates AMPK. Therefore, a
phenotypic rescue assay was designed to confirm the interference
efficiency of AMPK siRNA, and revealed that AMPK inhibition
reversed the increase in phosphorylated AMPK caused by
overexpression of SIRT1. Using MTT and Transwell assays, it was
confirmed that AMPK inhibition reversed the increase in cell
proliferation, migration and invasion after overexpression of
SIRT1, suggesting that SIRT1 may regulate cell proliferation and
migration through AMPK, and that AMPK may be a downstream target of
SIRT1. Therefore, the current results suggested that the SIRT1/AMPK
signaling pathway may serve as a potential therapeutic target in
the treatment of kidney cancer. However, the sample size of the
present study was limited and should be expanded for a deeper
understanding of the mechanisms of SIRT1 in future studies.
In conclusion, SIRT1 expression was downregulated in
RCC, and SIRT1 may serve an antitumor role by activating AMPK.
Additionally, high SIRT1 expression predicted a favorable prognosis
in patients with RCC. The current findings may help to develop new
potential treatment strategies for RCC.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant no. 81572507).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request. The datasets generated and/or analyzed during the current
study are available in The Cancer Genome Atlas repository
(http://gepia.cancer-pku.cn/index.html).
Authors' contributions
XW conceived and designed the study, wrote the
original draft and acquired the data. YL acquired the data and was
involved in drafting the manuscript. ZT statistically analyzed the
data and revised it critically for important intellectual content.
LB contributed to conceiving the study and gave final approval of
the version to be published. YZ made substantial contributions to
study design and acquisition of data, and was involved in drafting
the manuscript. HZ contributed to analyzing and interpreting the
data, and critically revised the manuscript for important
intellectual content. ZC made substantial contributions to
conception and design of the study, and statistically analyzed the
data. LP analyzed the data and was involved in drafting the
manuscript. DY contributed to conception and design of the study,
and revised it critically for important intellectual content. XW
and LB are responsible for confirming the authenticity of the data.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The study was approved by the Institutional Research
Ethics Committee of The Second Affiliated Hospital of Anhui Medical
University (Hefei, China), and written informed consent was
obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
AMPK
|
AMP-activated protein kinase
|
|
OD
|
optical density
|
|
OS
|
overall survival
|
|
RCC
|
renal cell carcinoma
|
|
TCGA
|
The Cancer Genome Atlas
|
|
WHO/ISUP
|
World Health
Organization/International Society of Urologic Pathology
|
|
SIRT1
|
sirtuin 1
|
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
Statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rini BI, Campbell SC and Escudier B: Renal
cell carcinoma. Lancet. 373:1119–1132. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gumz ML, Zou H, Kreinest PA, Childs AC,
Belmonte LS, LeGrand SN, Wu KJ, Luxon BA, Sinha M, Parker AS, et
al: Secreted frizzled-related protein 1 loss contributes to tumor
phenotype of clear cell renal cell carcinoma. Clin Cancer Res.
13:4740–4749. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bao Y, Wang Z, Liu B, Lu X, Xiong Y, Shi
J, Li P, Chen J, Zhang Z, Chen M, et al: A feed-forward loop
between nuclear translocation of CXCR4 and HIF-1α promotes renal
cell carcinoma metastasis. Oncogene. 38:881–895. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Haber T, Jöckel E, Roos FC, Junker K,
Prawitt D, Hampel C, Thüroff JW and Brenner W; German Renal Cell
Tumor Network, : Bone metastasis in renal cell carcinoma is
preprogrammed in the primary tumor and caused by AKT and Integrin
α5 Signaling. J Urol. 194:539–546. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang Y, Thayele Purayil H, Black JB,
Fetto F, Lynch LD, Masannat JN and Daaka Y: Prostaglandin E2
receptor 4 mediates renal cell carcinoma intravasation and
metastasis. Cancer Lett. 391:50–58. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
DeSantis CE, Lin CC, Mariotto AB, Siegel
RL, Stein KD, Kramer JL, Alteri R, Robbins AS and Jemal A: Cancer
treatment and survivorship statistics, 2014. CA Cancer J Clin.
64:252–271. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Obeng RC, Arnold RS, Ogan K, Master VA,
Pattaras JG, Petros JA and Osunkoya AO: Molecular characteristics
and markers of advanced clear cell renal cell carcinoma: Pitfalls
due to intratumoral heterogeneity and identification of genetic
alterations associated with metastasis. Int J Urol. 27:790–797.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Xue YJ, Chen SN, Chen WG, Wu GQ, Liao YF,
Xu JB, Tang H, Yang SH, He SY, Luo YF, et al: Cripto-1 expression
in patients with clear cell renal cell carcinoma is associated with
poor disease outcome. J Exp Clin Cancer Res. 38:3782019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kasuya G, Tsuji H, Nomiya T, Makishima H,
Haruyama Y, Kobashi G, Ebner DK, Hayashi K, Omatsu T, Kishimoto R,
et al: Updated long-term outcomes after carbon-ion radiotherapy for
primary renal cell carcinoma. Cancer Sci. 109:2873–2880. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang Q, Shi J, Yuan F, Wang H, Fu W, Pan
J, Huang Y, Yu J, Yang J and Chen Z: Higher expression of XPF is a
critical factor in intrinsic chemotherapy resistance of human renal
cell carcinoma. Int J Cancer. 139:2827–2837. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Diamond E, Molina AM, Carbonaro M, Akhtar
NH, Giannakakou P, Tagawa ST and Nanus DM: Cytotoxic chemotherapy
in the treatment of advanced renal cell carcinoma in the era of
targeted therapy. Crit Rev Oncol Hematol. 96:518–526. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Rini BI, Battle D, Figlin RA, George DJ,
Hammers H, Hutson T, Jonasch E, Joseph RW, McDermott DF, Motzer RJ,
et al: The society for immunotherapy of cancer consensus statement
on immunotherapy for the treatment of advanced renal cell carcinoma
(RCC). J Immunother Cancer. 7:3542019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bhindi B, Habermann EB, Mason RJ, Costello
BA, Pagliaro LC, Thompson RH, Leibovich BC and Boorjian SA:
Comparative survival following initial cytoreductive nephrectomy
versus initial targeted therapy for metastatic renal cell
carcinoma. J Urol. 200:528–534. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gershman B, Thompson RH, Boorjian SA,
Lohse CM, Costello BA, Cheville JC and Leibovich BC: Radical versus
partial nephrectomy for cT1 renal cell carcinoma. Eur Urol.
74:825–832. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tabayoyong W, Abouassaly R, Kiechle JE,
Cherullo EE, Meropol NJ, Shah ND, Dong S, Thompson RH, Smaldone MC,
Zhu H, et al: Variation in surgical margin status by surgical
approach among patients undergoing partial nephrectomy for small
renal masses. J Urol. 194:1548–1553. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Weiss J, Notohamiprodjo M, Bedke J,
Nikolaou K and Kaufmann S: Imaging response assessment of
immunotherapy in patients with renal cell and urothelial carcinoma.
Curr Opin Urol. 28:35–41. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Caliò A, Harada S, Brunelli M, Pedron S,
Segala D, Portillo SC, Magi-Galluzzi C, Netto GJ, Mackinnon AC and
Martignoni G: TFEB rearranged renal cell carcinoma. A
clinicopathologic and molecular study of 13 cases. Tumors harboring
MALAT1-TFEB, ACTB-TFEB, and the novel NEAT1-TFEB translocations
constantly express PDL1. Mod Pathol. 34:842–850. 2021. View Article : Google Scholar
|
|
19
|
de Velasco G, Culhane AC, Fay AP, Hakimi
AA, Voss MH, Tannir NM, Tamboli P, Appleman LJ, Bellmunt J, Kimryn
Rathmell W, et al: Molecular subtypes improve prognostic value of
International Metastatic Renal Cell Carcinoma Database Consortium
Prognostic Model. Oncologist. 22:286–292. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Vaziri H, Dessain SK, Ng Eaton E, Imai SI,
Frye RA, Pandita TK, Guarente L and Weinberg RA: hSIR2(SIRT1)
functions as an NAD-dependent p53 deacetylase. Cell. 107:149–159.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Stamatovic SM, Martinez-Revollar G, Hu A,
Choi J, Keep RF and Andjelkovic AV: Decline in Sirtuin-1 expression
and activity plays a critical role in blood-brain barrier
permeability in aging. Neurobiol Dis. 126:105–116. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Islam S, Uehara O, Matsuoka H, Kuramitsu
Y, Adhikari BR, Hiraki D, Toraya S, Jayawardena A, Saito I,
Muthumala M, et al: DNA hypermethylation of sirtuin 1 (SIRT1)
caused by betel quid chewing-a possible predictive biomarker for
malignant transformation. Clin Epigenetics. 12:122020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ming M, Zhao B, Shea C, Shea CR, Shah P,
Qiang L, White SR, Sims DM and He YY: Loss of sirtuin 1 (SIRT1)
disrupts skin barrier integrity and sensitizes mice to epicutaneous
allergen challenge. J Allergy Clin Immunol. 135:936–945.e4. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kalous KS, Wynia-Smith SL, Olp MD and
Smith BC: Mechanism of Sirt1 NAD+-dependent protein deacetylase
inhibition by Cysteine S-Nitrosation. J Biol Chem. 291:25398–25410.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chandrasekaran K, Anjaneyulu M, Choi J,
Kumar P, Salimian M, Ho CY and Russell JW: Role of mitochondria in
diabetic peripheral neuropathy: Influencing the NAD-dependent
SIRT1-PGC-1α-TFAM pathway. Int Rev Neurobiol. 145:177–209. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Okamoto S, Asgar NF, Yokota S, Saito K and
Minokoshi Y: Role of the α2 subunit of AMP-activated protein kinase
and its nuclear localization in mitochondria and energy
metabolism-related gene expressions in C2C12 cells. Metabolism.
90:52–68. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kanazawa I, Takeno A, Tanaka KI, Notsu M
and Sugimoto T: Osteoblast AMP-activated protein kinase regulates
glucose metabolism and bone mass in adult mice. Biochem Biophys Res
Commun. 503:1955–1961. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Jiang S, Wang Y, Luo L, Shi F, Zou J, Lin
H, Ying Y, Luo Y, Zhan Z, Liu P, et al: AMP-activated protein
kinase regulates cancer cell growth and metabolism via nuclear and
mitochondria events. J Cell Mol Med. 23:3951–3961. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hou X, Xu S, Maitland-Toolan KA, Sato K,
Jiang B, Ido Y, Lan F, Walsh K, Wierzbicki M, Verbeuren TJ, et al:
SIRT1 regulates hepatocyte lipid metabolism through activating
AMP-activated protein kinase. J Biol Chem. 283:20015–20026. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bae UJ, Park J, Park IW, Chae BM, Oh MR,
Jung SJ, Ryu GS, Chae SW and Park BH:
Epigallocatechin-3-Gallate-Rich Green tea extract ameliorates fatty
liver and weight gain in mice fed a high fat diet by activating the
Sirtuin 1 and AMP activating protein kinase pathway. Am J Chin Med.
46:617–632. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Shentu TP, He M, Sun X, Zhang J, Zhang F,
Gongol B, Marin TL, Zhang J, Wen L, Wang Y, et al: AMP-Activated
Protein Kinase and Sirtuin 1 Coregulation of cortactin contributes
to endothelial function. Arterioscler Thromb Vasc Biol.
36:2358–2368. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chou X, Ding F, Zhang X, Ding X, Gao H and
Wu Q: Sirtuin-1 ameliorates cadmium-induced endoplasmic reticulum
stress and pyroptosis through XBP-1s deacetylation in human renal
tubular epithelial cells. Arch Toxicol. 93:965–986. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Guo H, Ding H, Tang X, Liang M, Li S,
Zhang J and Cao J: Quercetin induces pro-apoptotic autophagy via
SIRT1/AMPK signaling pathway in human lung cancer cell lines A549
and H1299 in vitro. Thorac Cancer. May 11–2021.(Epub ahead of
print). doi: 10.1111/1759-7714.13925. View Article : Google Scholar
|
|
34
|
Subbaramaiah K, Iyengar NM, Morrow M,
Elemento O, Zhou XK and Dannenberg AJ: Prostaglandin E2
down-regulates sirtuin 1 (SIRT1), leading to elevated levels of
aromatase, providing insights into the obesity-breast cancer
connection. J Biol Chem. 294:361–371. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
George J, Nihal M, Singh CK and Ahmad N:
4′-Bromo-resveratrol, a dual Sirtuin-1 and Sirtuin-3 inhibitor,
inhibits melanoma cell growth through mitochondrial metabolic
reprogramming. Mol Carcinog. 58:1876–1885. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Warren AY and Harrison D: WHO/ISUP
classification, grading and pathological staging of renal cell
carcinoma: Standards and controversies. World J Urol. 36:1913–1926.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Miranda-Gonçalves V, Lameirinhas A,
Macedo-Silva C, Lobo J, C Dias P, Ferreira V, Henrique R and
Jerónimo C: Lactate increases renal cell carcinoma aggressiveness
through Sirtuin 1-dependent epithelial mesenchymal transition axis
regulation. Cells. 9:10532020. View Article : Google Scholar
|
|
39
|
Jeh SU, Park JJ, Lee JS, Kim DC, Do J, Lee
SW, Choi SM, Hyun JS, Seo DH, Lee C, et al: Differential expression
of the sirtuin family in renal cell carcinoma: Aspects of
carcinogenesis and prognostic significance. Urol Oncol.
35:675.e9–675.e15. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Chen Y, Zhu Y, Sheng Y, Xiao J, Xiao Y,
Cheng N, Chai Y, Wu X, Zhang S and Xiang T: SIRT1 downregulated FGB
expression to inhibit RCC tumorigenesis by destabilizing STAT3. Exp
Cell Res. 382:1114662019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Feng H, Guo P, Wang J, Xu J, Xie C and Gao
F: Expression of Leptin and Sirtuin-1 is associated with poor
prognosis in patients with osteosarcoma. Pathol Res Pract.
212:319–324. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yan L, Zhao Q, Liu L, Jin N, Wang S and
Zhan X: Expression of SIRT1 and survivin correlates with poor
prognosis in esophageal squamous cell carcinoma. Medicine
(Baltimore). 99:e216452020. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Chung SY, Jung YY, Park IA, Kim H, Chung
YR, Kim JY, Park SY, Im SA, Lee KH, Moon HG, et al: Oncogenic role
of SIRT1 associated with tumor invasion, lymph node metastasis, and
poor disease-free survival in triple negative breast cancer. Clin
Exp Metastasis. 33:179–185. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Dong YJ, Liu N, Xiao Z, Sun T, Wu SH, Sun
WX, Xu ZG and Yuan H: Renal protective effect of sirtuin 1. J
Diabetes Res. 2014:8437862014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zhang X, Chen J, Sun L and Xu Y: SIRT1
deacetylates KLF4 to activate Claudin-5 transcription in ovarian
cancer cells. J Cell Biochem. 119:2418–2426. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Atkins KM, Thomas LL, Barroso-González J,
Thomas L, Auclair S, Yin J, Kang H, Chung JH, Dikeakos JD and
Thomas G: The multifunctional sorting protein PACS-2 regulates
SIRT1-mediated deacetylation of p53 to modulate p21-dependent
cell-cycle arrest. Cell Rep. 8:1545–1557. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Han Y, Luo H, Wang H, Cai J and Zhang YJ:
SIRT1 induces resistance to apoptosis in human granulosa cells by
activating the ERK pathway and inhibiting NF-κB signaling with
anti-inflammatory functions. Apoptosis. 22:1260–1272. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45:W98–W102.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Fu H, Song W, Chen X, Guo T, Duan B, Wang
X, Tang Y, Huang L and Zhang C: MiRNA-200a induce cell apoptosis in
renal cell carcinoma by directly targeting SIRT1. Mol Cell Biochem.
437:143–152. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Kahn BB, Alquier T, Carling D and Hardie
DG: AMP-activated protein kinase: Ancient energy gauge provides
clues to modern understanding of metabolism. Cell Metab. 1:15–25.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Wang LF, Wang XN, Huang CC, Hu L, Xiao YF,
Guan XH, Qian YS, Deng KY and Xin HB: Inhibition of NAMPT
aggravates high fat diet-induced hepatic steatosis in mice through
regulating Sirt1/AMPKα/SREBP1 signaling pathway. Lipids Health Dis.
16:822017. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Dong HW, Zhang LF and Bao SL: AMPK
regulates energy metabolism through the SIRT1 signaling pathway to
improve myocardial hypertrophy. Eur Rev Med Pharmacol Sci.
22:2757–2766. 2018.PubMed/NCBI
|
|
53
|
Tian L, Cao W, Yue R, Yuan Y, Guo X, Qin
D, Xing J and Wang X: Pretreatment with Tilianin improves
mitochondrial energy metabolism and oxidative stress in rats with
myocardial ischemia/reperfusion injury via AMPK/SIRT1/PGC-1 alpha
signaling pathway. J Pharmacol Sci. 139:352–360. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Wang L, Quan N, Sun W, Chen X, Cates C,
Rousselle T, Zhou X, Zhao X and Li J: Cardiomyocyte-specific
deletion of Sirt1 gene sensitizes myocardium to ischaemia and
reperfusion injury. Cardiovasc Res. 114:805–821. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Cao Y, Bojjireddy N, Kim M, Li T, Zhai P,
Nagarajan N, Sadoshima J, Palmiter RD and Tian R: Activation of
γ2-AMPK suppresses ribosome biogenesis and protects against
myocardial ischemia/reperfusion injury. Circ Res. 121:1182–1191.
2017. View Article : Google Scholar : PubMed/NCBI
|