Introduction
At present, lung cancer has the highest mortality
rate among all malignant tumors (1).
According to statistics, as of 2018, lung cancer accounted for
11.6% of all cancer cases and cancer-related mortality (2). It is well known that smoking, air
pollution and chronic lung disease are all common causes of lung
cancer (3,4). Although early screening for lung cancer
is widespread, most patients are diagnosed at the terminal stage
and have a poor prognosis. Despite current popular targeted therapy
and immunotherapy, it remains difficult to achieve satisfactory
results (5,6). Determining the stage of lung cancer is
required for treatment but it also important for evaluating
prognosis. Based on the 8th edition of the American Joint Committee
on Cancer, the size of the tumor, number of lymph nodes and sites
of distant metastasis are the three major factors for determining
cancer stage (7). Lung cancer can be
divided into non-small cell lung cancer (NSCLC) and SCLC, then
NSCLC can be further characterized into lung adenocarcinoma, lung
squamous cell cancer and large cell cancer (8). Moreover, NSCLC is the most common
pathological type. At present, providing a novel target for the
molecular diagnosis and treatment of NSCLC has become a hotspot in
basic research.
Thyroid hormone receptor-interacting protein 13
(TRIP13) is one of the most important members of the ATPase family
associated with various cellular activities (AAA+) ATPase family,
which is associated with cell cycle checkpoint and DNA repair
(9,10). TRIP13 participates in numerous
cellular physiological processes, including chromosome checkpoints,
DNA break repair and chromosome synapsis (11). The spindle assembly checkpoint (SAC),
which is closely associated with chromosome repair, ensures that
all chromosomes produce bipolar spindle structures before mitosis
occurs (12,13). The participation of TRIP13 affects the
formation of SAC structure. For example, Tao et al (14) reported that overexpression of TRIP13
promoted multiple myeloma cell proliferation by invalidating
checkpoints via the Akt pathway. TRIP13 is an oncogene considered
to be involved in the progression of multiple tumor types, such as
hepatocellular carcinoma (15,16),
colorectal cancer (17), bladder
cancer (18) and prostate cancer
(19), and is associated with a poor
prognosis. Zhang et al (20)
revealed that TRIP13 may promote the malignant progression of NSCLC
and may be a potential target. Furthermore, TRIP13 knockdown could
arrest lung cancer cells in the G2/M phase, and it could
regulate the expression levels of genes associated with cell cycle
checkpoints. However, the mechanism of TRIP13 influencing the
invasion and metastasis of NSLCL has not been fully elucidated.
Epithelial-mesenchymal transformation (EMT) serves an important
role in tumor metastasis by enhancing the motility of cancer cells
(21). At present, the mechanism of
TRIP13 influencing EMT process remains unknown and should be
further investigated.
The present study aimed to examine the association
of TRIP13 with NSCLC. Reverse transcription-quantitative (RT-qPCR)
and western blot analyses were conducted to assess the role of
TRIP13 in regulating apoptosis, proliferation, invasion and
migration. Furthermore, knockdown or overexpression of TRIP13 may
affect the EMT during the progression of tumors. Studying the
molecular biological mechanism underlying the occurrence and
development of NSCLC and identifying effective biomarkers are
extremely important.
Materials and methods
Clinical information and bioinformatic
analyses
The clinical information of patients and mRNA
expression levels of TRIP13 were obtained for NSCLC tissues (n=515)
and adjacent normal tissues (n=59) from The Cancer Genome Atlas
(TCGA) database (https://cancer.gov/). These tissues
were obtained from 238 men and 276 women. The age of patients
ranged from 40–80 years. The relationships between TRIP13 and
different stages of NSCLC, as well as numbers of axillary lymph
node metastases are presented. Kaplan-Meier analysis was performed
to evaluate the prognostic value of TRIP13 at high level (n=124)
compared with low level (n=378) according to data from TCGA
database. The threshold of optimal gene expression was set as the
cut-off. The log-rank test was used to further evaluate the
P-value.
Cell culture and cell
transfection
Human NSCLC cell lines, A549, H1299 and H661, were
purchased from the Chinese Academy of Sciences. Cells were cultured
in DMEM (Gibco; Thermo Fisher Scientific, Inc.) supplemented with
10% FBS (Cell Sciences) in a humidified atmosphere with 5%
CO2 at 37°C. Small interfering (si)RNA negative control
(si-NC) and TRIP13 siRNA molecules (si1 and si2), pcDNA-3.1 vector
(empty vector) and TRIP13-overexpressing plasmid were purchased
from Suzhou GenePharma Co., Ltd. The cells were maintained in
6-well plates until they reached 60–70% confluence. According to
the manufacturer's instructions, the siRNAs (2.5 µg) and
overexpression plasmid (2.5 µg) were transfected into A549 and
H1299 cells using Lipofectamine® 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.) at room temperature and the medium
with serum was changed after 6 h. RNA was extracted 48 h after
transfection to verify the knockdown and overexpression effects.
All experiments were repeated three times. The siRNA sequences
were: si1, 5′-GACCAGAAAUGUGCAGUCU-3′; si2,
5′-GCAAAUCACUGGGUUCUAC-3′; and si-NC,
5′-UUCUCCGAACGUGUCACGUTT−3′.
RT-qPCR
Total RNA was extracted from NSCLC cell lines and
tumor tissues using TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.). Then, the RNA was reverse transcribed
into cDNA using PrimeScript RT Master mix (Takara Bio, Inc.). The
RT conditions were 37°C for 15 min, 85°C for 5 sec and 4°C
preservation. The expression level of TRIP13 was determined via
RT-qPCR using SYBR Green Master mix (Vazyme Biotech Co., Ltd.) and
a Bio-Rad Real-Time PCR detection system (Bio-Rad Laboratories,
Inc.). The complete thermocycling conditions were: Initial
denaturation at 95C° for 2 min, followed by 40 cycles of
denaturation at 95°C for 10 sec, annealing and elongation at 60°C
for 30 sec, and the final extension at 72°C for 7 min. β-actin was
the internal control. The expression level of the relevant genes
was calculated using the formula 2−∆∆Cq (22). All experiments were repeated three
times. The primer sequences were as follows: TRIP13 forward,
5′-GCGTGGTCAATGCTGTCTTG-3′ and reverse, 5′-CACGTCGATCTTCTCGGTGA-3′
and β-actin forward, 5′-CCAACCGCGAGAAGATGA−3′ and reverse,
5′-CCAGAGGCGTACAGGGATAG-3′.
Immunohistochemistry (IHC)
Paraffin-embedded NSCLC tissues (n=10) were fixed
with 4% paraformaldehyde for 24 h at room temperature, then cut
into 5-µm paraffin sections and attached to slides. After dewaxing
with xylene for 30 min at room temperature, the slices were boiled
for 10 min in citric acid buffer solution (pH 6.0) for antigen
repair, blocked with 10% non-immune goat serum (LMAI Bio) for 30
min at room temperature and incubated with an antibody against
TRIP13 (1:1,000; cat. no. ab204331; Abcam) overnight at room
temperature. Then, the slices were incubated with HRP-conjugated
secondary antibody (1:2,000; cat. no. ab205718; Abcam) for 30 min
at room temperature. After diaminobenzidine staining, the degree of
staining was observed under a fluorescent microscope. There was a
total of 10 patients, consisting of 5 men and 5 women. The age of
patients ranged 44–70 years. The following samples were collected:
Five tissues in right upper lobe, five tissues in right lower lobe,
one tissue in left upper lobe and one tissue in left lower lobe.
Moreover, the paired adjacent normal tissues were also obtained
from these patients. All tissues were obtained between March 2020
and November 2020. All samples came from patients at The Third
People's Hospital of Nantong Affiliated to Nantong University.
Informed consent was obtained from each patient. This study was
approved by the Ethics Committee of The Third People's Hospital of
Nantong Affiliated to Nantong University. All experiments were
repeated three times.
Cell apoptosis assay
The apoptotic rate was measured via flow cytometry
and included the percent of early and late apoptotic cells. After
transfection, the dead cells were collected and stained with 5 µl
Annexin V PE and 5 µl 7-AAD (BD Biosciences) in the dark at room
temperature for 15 min. In total, ~400 µl diluted binding buffer
was added to each sample. Then, the apoptotic rate was measured
using a FACSCalibur flow cytometer (BD Biosciences), and the
results were analyzed using FlowJo software 10.7 (FlowJo LLC). All
experiments were repeated three times.
Cell cycle assay
Human A549 and H1299 cells were transfected with the
siRNAs or overexpression plasmid for 48 h. The cells were
resuspended in 70% ethanol and incubated in a −20°C freezer for
>24 h. The samples were stained with 20 µg/ml PI and 20 µg/ml
RNase A (Sigma-Aldrich; Merck KGaA) for 15 min at room temperature.
Flow cytometry (BD Biosciences) was used to analyze cell cycle
progression, and the data were processed using FlowJo software 10.7
(FlowJo LLC). All experiments were repeated three times.
Cell proliferation assay
The proliferation of the NSCLC cells was measured
using a Cell Counting Kit-8 (CCK-8; Dojindo Molecular Technologies,
Inc.) assay, according to the manufacturer's instructions. A total
of 4×103 cells per well were seeded in 96-well plates
and cultured for 0, 24, 48 and 72 h. After incubation with 10 µl
CCK-8 solution for 2 h at 37°C, the absorbance values were measured
at 450 nm. All experiments were repeated three times.
Transwell assay
Transwell assays were performed to detect cell
invasion and migration. For the invasion assay, a total of 40 µl
Matrigel (BD Biosciences) was diluted with serum-free medium and
placed in the upper chamber (pore size, 3 µm; MilliporeSigma) for
30 min at 37°C, and 2×105 cells were plated in the upper
chamber with 200 µl serum-free medium. DMEM with 20% FBS was added
to the lower chamber. For the migration assay, Matrigel was not
required in the upper chamber. After incubation for 48 h at 37°C,
the cells that adhered to the lower surface of the filter were
stained with 1% crystal violet for 10 min at room temperature.
Then, the quantity of invading or migrating cells was determined
under an Olympus IX73-FL-PH inverted microscope (Olympus
Corporation). All experiments were repeated three times.
Wound healing assay
A549 and H1299 cells were seeded in a 6-well plate
using medium with FBS-free for 24 h after transfection and covered
70–80% of the plate. A straight line was scratched the plate with a
10-µl pipette tip and every line was spaced at 0.5 cm. PBS was used
to removed cell debris. The cell healing was filmed at 0 and 24 h
using an Olympus IX73-FL-PH inverted microscope (Olympus
Corporation). The distance of lines was measured using ImageJ 1.8.0
(National Institutes of Health). All experiments were repeated
three times.
Western blotting
Total proteins were extracted from cells with RIPA
lysis buffer (Beyotime Institute of Biotechnology), and the
concentrations of the proteins were measured using a BCA protein
assay kit (Beyotime Institute of Biotechnology). The samples were
mixed with loading buffer, were electrophoresed on 10% SDS-PAGE
gels and transferred to PVDF membranes (MilliporeSigma). The
membranes were blocked with 5% non-fat milk (Beyotime Institute of
Biotechnology) for 2 h at room temperature and incubated overnight
at 4°C with primary antibodies [anti-TRIP13, anti-β-actin,
anti-vimentin, anti-E-cadherin, anti-snail family transcriptional
repressor 1 (Snail)]. The membranes were completely washed three
times with TBS-Tween-20 (0.1%) (Beyotime Institute of
Biotechnology) and further incubated with a specific HRP-conjugated
secondary antibody (1:2,000; cat. no. ab205718; Abcam) for 1 h at
room temperature. The densitometry was measured using ImageJ 1.8.0.
All experiments were repeated three times. The primary antibodies
were anti-TRIP13 (1:1,000; cat. no. ab204331; Abcam), anti-β-actin
(1:2,000; cat. no. ab8227; Abcam), anti-vimentin (1:1,000; cat. no.
5741S; Cell Signaling Technology, Inc.), anti-E-cadherin (1:1,000;
cat. no. 3195S; Cell Signaling Technology, Inc.) and anti-Snail
(1:1,000; cat. no. NBP2-27293; Novus Biologicals, Inc.).
Statistical analysis
All the data are presented as the mean ± SD. Most of
the differences between two groups were determined using an
unpaired t-test. The expression level of TRIP13 between NSCLC
tissues and adjacent normal tissues detected via RT-qPCR was
compared using a paired Student's t-test. Multiple comparison
between the group was performed using one-way ANOVA and Tukey's
test. P<0.05 was considered to indicate a statistically
significant difference. All the data statistical analyses were
conducted using GraphPad Prism 8.0 (GraphPad Software Inc.).
Results
TRIP13 is upregulated in NSCLC
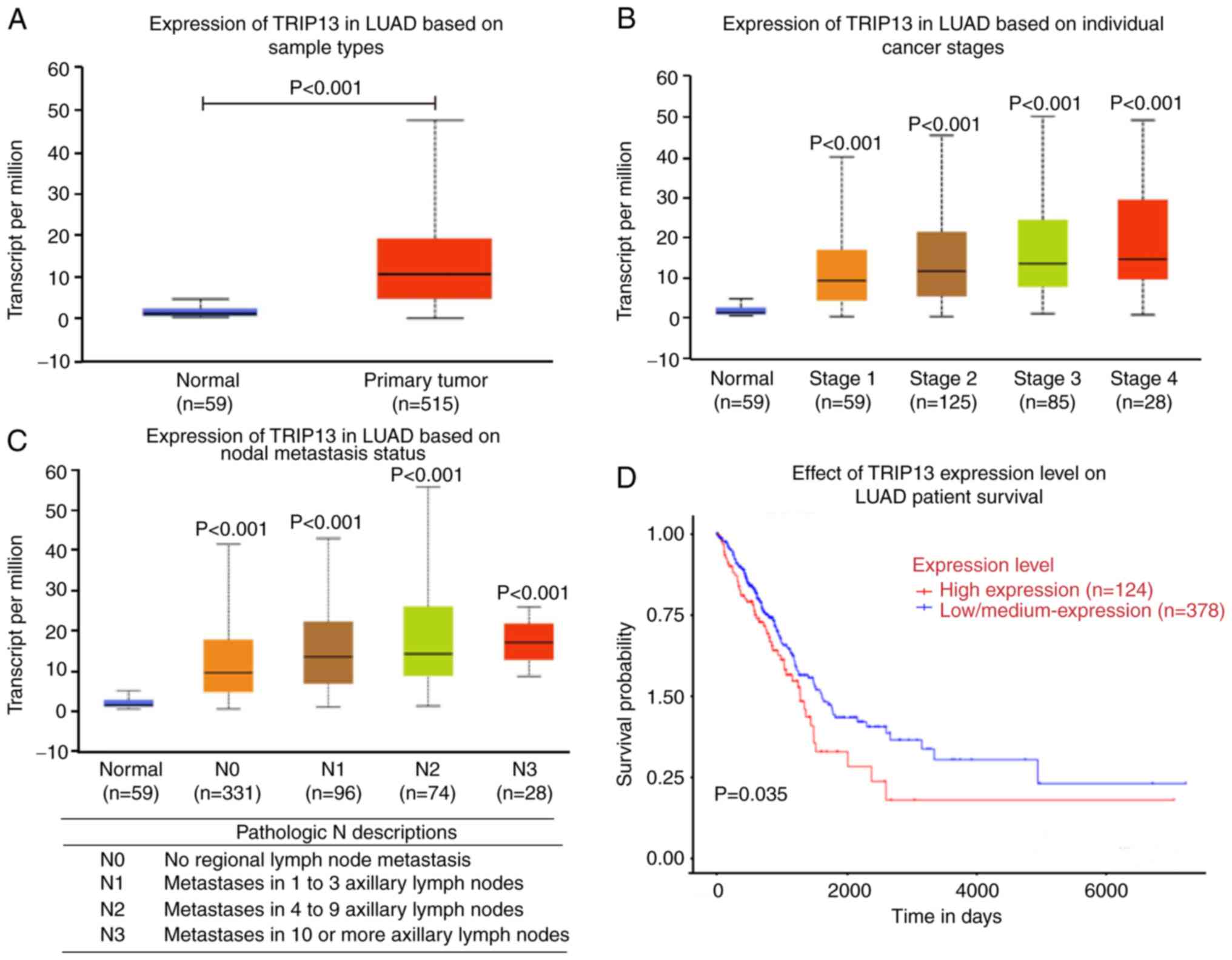
To study the expression of TRIP13 in NSCLC, clinical
data from TCGA, including data from 59 normal samples and 515
primary tumor samples, were analyzed. It was found that TRIP13 was
upregulated in the NSCLC tissues compared with the adjacent normal
tissues (Fig. 1A). The data
demonstrated that the expression level of TRIP13 was more
significantly increased with higher stages of NSCLC, and each stage
was statistically significant compared with the normal group
(Fig. 1B).
To further evaluate the effect of TRIP13 expression
on prognosis, four cancer stages were identified based on the
number of axillary lymph nodes affected. As the number of affected
nodes increased, the expression level of TRIP13 was significantly
upregulated in the NSCLC tissues compared with the normal tissues,
and increased TRIP13 expression predicted poor prognosis (Fig. 1C). Furthermore, the survival analysis
revealed that patients with high TRIP13 levels (n=124) had poorer
prognosis compared with those with low TRIP13 levels (n=378)
(Fig. 1D).
IHC analysis identified that the nuclei were stained
yellow brown or dark brown and that TRIP13 was located in the
nucleus in 10 NSCLC tissues (Fig.
2A). The results of TRIP13 positive cells were assessed by two
pathologists who did not know the tumor grade. In total, 10
high-power fields were observed and the positive percentage was
calculated. The quantitative data are shown in Fig. 2B. To determine the biological function
of TRIP13 in NSCLC cells, three cell lines, namely, A549, H1299 and
H661 cell lines, were selected for the experiment (Fig. 2C). The expression level of TRIP13 was
low in A549 cells, but its expression was high in H1299 cells.
Knockdown of TRIP13 inhibits the
proliferation, invasion and migration and increases apoptosis of
H1299 cells
si1 and si2 were used to knockdown the expression of
TRIP13 in H1299 cells, which had a higher endogenous TRIP13
expression than the other cell lines. The RT-qPCR results revealed
that the knockdown efficiency of si1 and si2 was substantial, and
that the expression of TRIP13 was knocked down (Fig. 3A). To further study the cellular
biological functions, a CCK-8 assay was used to examine the
proliferative abilities of the cells at 0, 24, 48 and 72 h. The
knockdown of TRIP13 in H1299 cells significantly suppressed
proliferation compared with the NC group (Fig. 3B) from 24 to 72 h. Moreover, the
knockdown of TRIP13 inhibited the H1299 cell migratory and invasive
abilities, as evaluated using the Transwell assay (Fig. 4A). The wound healing experiment
effectively detected lateral cell migration. The migratory ability
of the knockdown group was significantly decreased compared with
the NC group, as determined by comparing the gaps in the wound
healing assay (Fig. 4B).
Flow cytometry accurately detected cellular
apoptosis and the cell cycle progression. The change in the
apoptotic rate could be one of the mechanisms that affect the
malignant progression of NSCLC. The results indicated that
knockdown of TRIP13 increased the rate of apoptosis in H1299 cells
compared with the NC at 48 h after transfection (Fig. 5A). In addition, the distribution of
the cell cycle reflected the stage of DNA synthesis in cancer
cells. In the TRIP13 knockdown group, there was a significant
decrease in the number of cells in the G1 phase; on the
other hand, TRIP13 knockdown induced H1299 cell cycle arrest in the
S phase and caused no changes in the number of cells in the
G2 phase (Fig. 5B).
TRIP13 overexpression inhibits
apoptosis and promotes proliferation, invasion and migration of
A549 cells
The aforementioned experimental results demonstrated
that knockdown of TRIP13 significantly affected the function of
NSCLC cells. In order to prove the role of TRIP13 in promoting
cancer progression more effectively, the changes in abilities of
cell apoptosis, proliferation, invasion and migration after
overexpression of TRIP3 were detected. The transfection efficiency
was determined via RT-qPCR, and it was found that TRIP13 was
significantly overexpressed (Fig.
6A). The TRIP13 overexpression group had increased cell
proliferation compared with the vector group (Fig. 6B). The Transwell assay was used for
detecting the H1299 cell migratory and invasive abilities. The
results demonstrated that these abilities were significantly
increased in the TRIP13 overexpression group compared with the
vector group (Fig. 7A). Moreover, the
wound healing experiment identified that TRIP13 overexpression
significantly promoted lateral cell migration (Fig. 7B).
In contrast to TRIP13 knockdown, TRIP13
overexpression in A549 cells consistently reduced the apoptotic
rate at 48 h after transfection (Fig.
8A). In addition, the TRIP13 overexpression group showed a
significant decrease in the number of cells in the S phase.
However, the number of cells in the G1 and G2
phase were markedly increased (Fig.
8B).
TRIP13 induces migration and invasion
via EMT in NSCLC cells
A large number of studies have reported that EMT was
an important biological process which epithelial-derived malignant
tumor cells acquire the ability to migrate and invade (23–25).
Western blotting was used to analyze the expression levels of
several EMT-related factors, such as the epithelial marker
E-cadherin and the mesenchymal markers vimentin and Snail, after
transfection (Fig. 9A). The results
demonstrated that TRIP13 knockdown could increase E-cadherin
expression and decrease vimentin and Snail expression in H1299
cells. However, TRIP13 overexpression led to the opposite results.
TRIP13 overexpression decreased the protein expression level of
E-cadherin and enhanced the protein expression levels of vimentin
and Snail (Fig. 9). The gray scale
ratio of the three proteins to the internal reference was used to
reflect the differences between each group (Fig. 9B and C).
Discussion
China has a high incidence of lung cancer, and due
to its poor prognosis, effective diagnostic criteria and treatment
options are important for patients. The present study considered
TRIP13 to be a potential biomarker worthy of further
investigation.
Accumulating evidence has suggested that TRIP13
serves a carcinogenic role in numerous cancer types. For example,
Zhang et al (26) reported
that TRIP13 silencing could suppress the function of glioblastoma
cells by regulating the expression of F-box and WD repeat domain
containing 7 (FBXW7). c-Myc is a proto-oncogene that can affect the
development of several tumor cells, and it has been shown that the
TRIP13/FBXW7/c-Myc pathway may provide a reliable target for
glioblastoma treatment. In hepatocellular carcinoma, TRIP13 was
positively correlated with actinin α 4 and activated the Akt/mTOR
pathway to accelerate the malignant progression of hepatocellular
carcinoma (27). The mitotic
checkpoint complex (MCC) coordinates with the SAC to maintain
genomic stability and inhibits the expression of certain complexes,
such as anaphase promoting complex (28). TRIP13 bound to p31comet to transform
activate closed mitotic arrest deficient 2 like 1 (MAD2) into open
MAD2, which was not active and could cause the decomposition of
MCC. TRIP13 was further shown to be a AAA+ ATPase that binds to
related proteins (29–31).
As the use of chemotherapeutic drugs has increased
in the treatment of tumors, cancer cells have become more
resistant. A previous study reported that upregulation of TRIP13
could enhance the progression of bladder cancer cells. These
authors revealed that the proliferation of tumor cells was
significantly decreased after knockdown of TRIP13. Moreover, the
apoptotic rate was also significantly reduced in these
drug-resistant cells, as shown by flow cytometry (18). To further investigate the mechanism
via which TRIP13 participates in DNA repair, the researchers
detected the expression level of H2AX phosphorylated at serine 139
(γH2AX), which is a typical marker of DNA damage, and RAD50 double
strand break repair protein (RAD50), which is a typical marker of
DNA repair. The results showed that overexpression of TRIP13 could
inhibit the expression of γH2AX and enhance the expression of RAD50
after treatment with cisplatin. Thus, these researchers suggested
that TRIP13 may promote DNA repair in tumor cells (32). Therefore, TRIP13 could be considered a
novel potential target for the diagnosis and treatment of
tumors.
In the present study, a high expression of TRIP13 in
NSCLC was identified. TRIP13 was differentially expressed in NSCLC
of different grades, based on the database results, and indicated a
poor prognosis. Furthermore, knockdown of TRIP13 in H1299 cells
increased apoptosis and inhibited proliferation, invasion and
migration. Consistently, overexpression of TRIP13 inhibited
apoptosis and promoted proliferation, invasion and migration of
A549 cells.
EMT is closely associated with the aggressive
behavior of cancer cells (33).
E-cadherin is responsible for cell adhesion and cell cytoskeleton
organization. The loss of E-cadherin is a key step in the EMT
process, leading to the transition of epithelial cells to
aggressive mesenchymal cells. It has also been confirmed that
E-cadherin promotes cell proliferation and polarization to
accelerate the metastasis of tumors (34). Moreover, changes in vimentin
expression are closely associated with the malignant progression of
tumors (35). As an important
component of the signaling pathway, Snail promotes the progression
of NSCLC (36). Previous studies have
reported that TRIP13 induced changes in the expression levels of
several EMT markers to promote the progression of several cancer
types (32,37,38).
However, there are some limitations in the present
study. It has been proved that TRIP13 could affect the metastasis
of NSCLC via EMT pathways. However, the other pathways that TRIP13
could affect metastasis are yet to be further determined. The
relationship between TRIP13 and prognosis of patients has not been
evaluated. These were the major points for the future work.
In conclusion, the present study demonstrated that
upregulated TRIP13 could be a novel and potential biomarker for the
diagnosis and treatment of NSCLC and served an important role in
the malignant progression of NSCLC.
Acknowledgements
Not applicable.
Funding
This study was supported by the Health Bureau of
Nantong City (grant no. WKZL2018054) and the Nantong Science and
Technology Bureau (grant no. MSZ18008).
Availability of data and materials
The datasets generated during the current study are
available from The Cancer Genome Atlas dataset (https://cancer.gov/).
Authors' contributions
JS was devoted to the conception and design of this
study. RL and QZ performed the experiments. LJ, LC and FW analyzed
the experimental data and presented statistical results. RL, QZ and
FW collected the tissues. RL, QZ and JS wrote the manuscript. JS
contributed to revise this manuscript and approved the final
version to be published. RL, QZ and JS were responsible for
confirming the authenticity of the raw data. All authors read and
agreed to be accountable for all aspects of the work in ensuring
that questions related to the accuracy or integrity of any part of
the work are appropriately investigated and resolved.
Ethics approval and consent to
participate
Informed consent was obtained from each patient.
This study was approved by the Ethics Committee of The Third
People's Hospital of Nantong Affiliated to Nantong University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
TRIP13
|
thyroid hormone receptor-interacting
protein 13
|
|
NSCLC
|
non-small cell lung cancer
|
|
TCGA
|
The Cancer Genome Atlas
|
|
RT-qPCR
|
reverse transcription-quantitative
PCR
|
|
CCK-8
|
Cell Counting Kit-8
|
|
IHC
|
immunohistochemistry
|
|
SAC
|
spindle assembly checkpoint
|
|
EMT
|
epithelial-mesenchymal
transformation
|
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bade B and Dela Cruz C: Lung cancer 2020:
Epidemiology, etiology, and prevention. Clin Chest Med. 41:1–24.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Schuller HM: The impact of smoking and the
influence of other factors on lung cancer. Expert Rev Respir Med.
13:761–769. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rivera GA and Wakelee H: Lung cancer in
never smokers. Adv Exp Med Biol. 893:43–57. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jones GS and Baldwin DR: Recent advances
in the management of lung cancer. Clin Med (Lond). 18 (Suppl
2):S41–S46. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Steven A, Fisher SA and Robinson BW:
Immunotherapy for lung cancer. Respirology. 21:821–833. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kim D, Lee YS, Kim DH and Bae SC: Lung
cancer staging and associated genetic and epigenetic events. Mol
Cells. 43:1–9. 2020.PubMed/NCBI
|
|
8
|
Skřičková J, Kadlec B, Venclíček O and
Merta Z: Lung cancer. Cas Lek Cesk. 157:226–236. 2018.
|
|
9
|
Wang Y, Huang J, Li B, Xue H, Tricot G, Hu
L, Xu Z, Sun X, Chang S, Gao L, et al: A small-molecule inhibitor
targeting TRIP13 suppresses multiple myeloma progression. Cancer
Res. 80:536–548. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ye Q, Rosenberg S, Moeller A, Speir J, Su
T and Corbett K: TRIP13 is a protein-remodeling AAA+ ATPase that
catalyzes MAD2 conformation switching. Elife. 4:e073672015.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Clairmont CS, Sarangi P, Ponnienselvan K,
Galli LD, Csete I, Moreau L, Adelmant G, Chowdhury D, Marto JA and
D'Andrea AD: TRIP13 regulates DNA repair pathway choice through
REV7 conformational change. Nat Cell Biol. 22:87–96. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yost S, de Wolf B, Hanks S, Zachariou A,
Marcozzi C, Clarke M, de Voer R, Etemad B, Uijttewaal E, Ramsay E,
et al: Biallelic TRIP13 mutations predispose to Wilms tumor and
chromosome missegregation. Nat Genet. 49:1148–1151. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ma HT and Poon RYC: TRIP13 regulates both
the activation and inactivation of the spindle-assembly checkpoint.
Cell Rep. 14:1086–1099. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tao Y, Yang G, Yang H, Song D, Hu L, Xie
B, Wang H, Gao L, Gao M, Xu H, et al: TRIP13 impairs mitotic
checkpoint surveillance and is associated with poor prognosis in
multiple myeloma. Oncotarget. 8:26718–26731. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yao J, Zhang X, Li J, Zhao D, Gao B, Zhou
H, Gao S and Zhang L: Silencing TRIP13 inhibits cell growth and
metastasis of hepatocellular carcinoma by activating of
TGF-β1/smad3. Cancer Cell Int. 18:2082018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ju L, Li X, Shao J, Lu R, Wang Y and Bian
Z: Upregulation of thyroid hormone receptor interactor 13 is
associated with human hepatocellular carcinoma. Oncol Rep.
40:3794–3802. 2018.PubMed/NCBI
|
|
17
|
Sheng N, Yan L, Wu K, You W, Gong J, Hu L,
Tan G, Chen H and Wang Z: TRIP13 promotes tumor growth and is
associated with poor prognosis in colorectal cancer. Cell Death
Dis. 9:4022018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gao Y, Liu S, Guo Q, Zhang S, Zhao Y, Wang
H, Li T, Gong Y, Wang Y, Zhang T, et al: Increased expression of
TRIP13 drives the tumorigenesis of bladder cancer in association
with the EGFR signaling pathway. Int J Biol Sci. 15:1488–1499.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Dong L, Ding H, Li Y, Xue D, Li Z, Liu Y,
Zhang T, Zhou J and Wang P: TRIP13 is a predictor for poor
prognosis and regulates cell proliferation, migration and invasion
in prostate cancer. Int J Biol Macromol. 121:200–206. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang Q, Dong Y, Hao S, Tong Y, Luo Q and
Aerxiding P: The oncogenic role of TRIP13 in regulating
proliferation, invasion, and cell cycle checkpoint in NSCLC cells.
Int J Clin Exp Pathol. 12:3357–3366. 2019.PubMed/NCBI
|
|
21
|
Mittal V: Epithelial mesenchymal
transition in tumor metastasis. Annu Rev Pathol. 13:395–412. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Pastushenko I and Blanpain C: EMT
transition states during tumor progression and metastasis. Trends
Cell Biol. 29:212–226. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yeung KT and Yang J:
Epithelial-mesenchymal transition in tumor metastasis. Mol Oncol.
11:28–39. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Saitoh M: Involvement of partial EMT in
cancer progression. J Biochem. 164:257–264. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhang G, Zhu Q, Fu G, Hou J, Hu X, Cao J,
Peng W, Wang X, Chen F and Cui H: TRIP13 promotes the cell
proliferation, migration and invasion of glioblastoma through the
FBXW7/c-MYC axis. Br J Cancer. 121:1069–1078. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhu MX, Wei CY, Zhang PF, Gao DM, Chen J,
Zhao Y, Dong SS and Liu BB: Elevated TRIP13 drives the AKT/mTOR
pathway to induce the progression of hepatocellular carcinoma via
interacting with ACTN4. J Exp Clin Cancer Res. 38:4092019.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Alfieri C, Chang L, Zhang Z, Yang J,
Maslen S, Skehel M and Barford D: Molecular basis of APC/C
regulation by the spindle assembly checkpoint. Nature. 536:431–436.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Alfieri C, Chang L and Barford D:
Mechanism for remodelling of the cell cycle checkpoint protein MAD2
by the ATPase TRIP13. Nature. 559:274–278. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ye Q, Kim D, Dereli I, Rosenberg SC,
Hagemann G, Herzog F, Tóth A, Cleveland DW and Corbett KD: The AAA+
ATPase TRIP13 remodels HORMA domains through N-terminal engagement
and unfolding. EMBO J. 36:2419–2434. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lu S, Qian J, Guo M, Gu C and Yang Y:
Insights into a crucial role of TRIP13 in human cancer. Comput
Struct Biotechnol J. 17:854–861. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lu S, Guo M, Fan Z, Chen Y, Shi X, Gu C
and Yang Y: Elevated TRIP13 drives cell proliferation and drug
resistance in bladder cancer. Am J Transl Res. 11:4397–4410.
2019.PubMed/NCBI
|
|
33
|
Lu W and Kang Y: Epithelial-mesenchymal
plasticity in cancer progression and metastasis. Dev Cell.
49:361–374. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Venhuizen J, Jacobs F, Span P and Zegers
M: P120 and E-cadherin: Double-edged swords in tumor metastasis.
Semin Cancer Biol. 60:107–120. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Strouhalova K, Přechová M, Gandalovičová
A, Brábek J, Gregor M and Rosel D: Vimentin intermediate filaments
as potential target for cancer treatment. Cancers (Basel).
12:1842020. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Xie Q, Zhu Z, He Y, Zhang Z, Zhang Y, Wang
Y, Luo J, Peng T, Cheng F, Gao J, et al: A lactate-induced
Snail/STAT3 pathway drives GPR81 expression in lung cancer cells.
Biochim Biophys Acta Mol Basis Dis. 1866:1655762020. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Agarwal S, Behring M, Kim HG,
Chandrashekar DS, Chakravarthi BVSK, Gupta N, Bajpai P, Elkholy A,
Al Diffalha S, Datta PK, et al: TRIP13 promotes metastasis of
colorectal cancer regardless of p53 and microsatellite instability
status. Mol Oncol. 14:3007–3029. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kurita K, Maeda M, Mansour MA, Kokuryo T,
Uehara K, Yokoyama Y, Nagino M, Hamaguchi M and Senga T: TRIP13 is
expressed in colorectal cancer and promotes cancer cell invasion.
Oncol Lett. 12:5240–5246. 2016. View Article : Google Scholar : PubMed/NCBI
|