Introduction
Breast cancer is one of the most prevalent malignant
diseases and the primary cause of cancer-related deaths in women
worldwide, accounting for 23% of the total cancer cases and 14% of
the cancer-related deaths (1).
Intensive research efforts have revealed that this cancer is a
heterogeneous and complex disease characterized by distinct
morphological, clinical, and molecular features. Although great
advances in diagnosis and treatment have been made, including the
most comprehensive portfolio of targeted treatments for any cancer,
the recurrence and metastasis rates of breast cancer remain
unacceptably high. Therefore, it remains important to develop new
targeted therapeutics for patients who are not aided by current
treatments (2).
The cell surface is surrounded by glycans, and most
proteins carry glycan modifications, which play an essential role
in regulating a range of biological processes including immune
regulation and cell-cell interactions. Notably, malignant
transformation is often associated with aberrant glycosylation
(3). Among the different forms of
glycosylation, sialylation has been linked with multiple malignant
characteristics, such as growth, migration, and invasion. The
primary mediators of sialylation are the sialyltransferases (STs),
a family of anabolic enzymes that have been closely linked to
cancer progression (4,5). Specifically, increasing evidence has
associated the function of the α-2,8-sialyltransferase (ST8SIA
I–VI) family with several human tumors. For example, ST8SIA4 was
revealed to participate in breast cancer progression by modifying
the sialylation profile of breast cancer cells (6).
Long non-coding (lnc) RNAs, are a type of non-coding
RNAs defined by lengths of more than 200 nucleotides. An increasing
number of studies have demonstrated that lncRNAs play essential
roles in both general biological processes as well as tumorigenesis
where they can act either as oncogenes or tumor suppressors
(7,8).
For instance, the expression of the lncRNA GAS5 was revealed to be
decreased in hepatocellular carcinoma and predicted a poor clinical
outcome for these patients (9). The
number of lncRNAs aberrantly expressed in human tumors continues to
grow with extensive research efforts demonstrating that these
function as signal mediators, molecular decoys and scaffolds or
enhancers of transcription. In addition, a large group of lncRNAs
can serve as molecular sponges that regulate functional gene
expression (10).
The metastasis-associated lung adenocarcinoma
transcript 1 (MALAT1) is one of the most well-characterized
lncRNAs, that is commonly upregulated in numerous cancer types
(11). For instance, Zhang et
al revealed that MALAT1 was upregulated in renal cell carcinoma
tissues, and knockdown of MALAT1 in renal cell carcinomas decreased
their proliferation, migration and invasion (12). Sun et al verified that MALAT1
regulated human umbilical vein endothelial cell (HUVEC)
proliferation through sponging of miR-320a to regulate forkhead box
protein (FOX)M1 expression (13).
Upregulation of MALAT1 in breast cancer was revealed to induce
epithelial-mesenchymal transition (EMT) and decrease trastuzumab
sensitivity in human epidermal growth factor receptor
(HER)2+ breast cancer (14). However, the role of MALAT1 in breast
cancer development remains controversial, and an improved
understanding of the functional relevance of MALAT1 to this disease
is urgently needed.
MicroRNAs (miRNAs) are a class of small nucleotide
molecules with a length of 20–22 nucleotides (15). Mechanistically, miRNAs suppress mRNA
transcription by directly binding to the 3′-untranslated regions
(3′-UTRs) (16). Previously, miRNAs
have been reported to play a multifaceted role in tumorigenesis.
For example, Chen et al reported that miR-495 was
downregulated in breast cancer cells (17). In gastric cancer cells, miR-92b-3p was
overexpressed and accelerated cell malignant progression via
upregulation of MMP-2/9 expression (18). Intriguingly, numerous lncRNAs function
as competing endogenous RNAs (ceRNAs), acting as molecular sponges
for miRNAs and altering their availability and regulation of their
coding gene targets (REF) (19,20).
Our previous study revealed that miR-26a/26b
regulated breast cancer progression by targeting ST8SIA4 (6). As a logical extension of this work, our
present study sought to provide an improved definition of this
mechanism. Herein, the potential associations between miR-26a/26b,
ST8SIA4 and the lncRNA MALAT1 were investigated. The present study
provided new insights into the underlying mechanisms regulated by
MALAT1 in breast cancer and indicated that the
MALAT1/miR-26a/26b/ST8SIA4 axis holds great promise as a new
therapeutic target for breast cancer.
Materials and methods
Human tumor tissues
A total of 36 pairs of breast cancer and adjacent
tissues were removed from the patients (age, 28–76 years; median,
45 years) with breast cancer who received surgeries at the First
Affiliated Hospital of Dalian Medical University (Dalian, China)
between June 2015 to August 2017. The subjects were pathologically
confirmed as breast cancer, and they had hardly underwent any
antitumor therapies before their tissues were obtained. All samples
were stored in liquid nitrogen before analysis. The study was
approved by the Ethics Committee of The First Affiliated Hospital
of Dalian Medical University (approval no. YJ-KY-FB-2017-32) with
all enrolled patients providing written informed consent.
Cell lines and cell culture
The breast cancer cell lines MDA-MB-231, MCF-7 and
human epithelial breast cell line MCF-10A were obtained from the
American Type Culture Collection (ATCC). All cells were cultured in
RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.) in a 5%
CO2 incubator at 37°C. All media were supplemented with
10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.)
and replaced every 2 days.
RNA isolation and reverse
transcription-quantitative (RT-q)PCR
Total RNA from tissues and cells was extracted by
TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.) and reverse
transcribed into cDNA using SuperScript Reverse Transcriptase III
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions. Quantitative PCR reactions were
carried out on an ABI 7500 PCR System using SYBR-Green Supermix
(Applied Biosystems; Thermo Fisher Scientific, Inc.) with the
following settings: Initial denaturation at 95°C for 30 sec,
followed by 40 cycles of denaturation at 95°C for 5 sec, annealing
at 60°C for 30 sec, and melting curve analysis. Fluorescent signals
were detected after each cycle. Relative mRNA expression levels
were calculated with normalization to GAPDH or U6 using the
2−ΔΔCq method (21). The
sequences of the primers were as follows: MALAT1 forward,
5′-CTCTCCCCTCCCTTGGTCTT-3′ and reverse,
5′-TCCCAATCCCCACATTTAAAAT-3′; ST8SIA4 forward,
5′-AGAGGTGGACGATCTGCACAATAA−3′ and reverse,
5′-TGACAAGTGACCGACTCAAAGACA−3′; GAPDH forward,
5′-CATGTTCGTCATGGGTGTGAA−3′ and reverse,
5′-GGCATGGACTGTGGTCATGAG−3′; miR-26a forward,
5′-GGGGGTTCAAGTAATCCAGGATA-3′ and reverse,
5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAGCCTA-3′; miR-26b
forward, 5′-GGGGGTTCAAGTAATTCAGGAT-3′ and reverse,
5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACACCTAT-3′; U6
forward, 5′-CGCTTCGGCAGCACATATACTA-3′ and reverse,
5′-GGAACGCTTCACGAATTTGC-3′. All experiments were performed in
triplicate.
Cell transfection
The MALAT1 cDNA was subcloned into the pmirGLO
vector (Promega Corporation). miR-26a/26b mimic, inhibitor, miR-NC,
and small interfering (si)RNA targeting MALAT1 and scramble siRNA
were synthesized by Shanghai GenePharma Co., Ltd. MALAT1 siRNA
sequence was: 5′-GATCCATAATCGGTTTCAA-3′, and the sequence of the
scramble siRNA was: 5′-UUCUCCGAACGUGUCACGUTT−3′. Lipofectamine 3000
(Invitrogen; Thermo Fisher Scientific, Inc.) was used for the
transfection according to the manufacturer's instructions.
Mock-transfected cells were used as the negative control. Cells
were harvested 48 h after transfection for further studies. First,
5 µg siRNA was dissolved in 125 µl Opti-MEM culture medium with
free FBS. Then, 3.75 µl Lipofectamine 3000 was dissolved in 125 µl
Opti-MEM culture medium for 5 min at room temperature. Finally, the
aforementioned two solutions were mixed while resting for 15 min at
room temperature; then 250 µl mixed solution was added to each well
in the 6-well plates, and the transfection medium was changed to
cultured medium containing 10% FBS after 4–6 h at 37°C. RT-qPCR was
used to determine the transfection efficiency.
Cell Counting Kit-8 (CCK-8) assay
Cell viability (1×103 cells/well) was
assessed every 24 h after the indicated treatments. Briefly, 10 µl
CCK-8 reagent was added and then the plates were cultured for
another 1–2 h at 37°C in the dark. The density of each well was
measured at 450 nm using an automated microplate reader (BioTek
China). There were 3–5 multiple wells in each group. The experiment
was performed in triplicate.
Colony formation assay
A total of ~300 cells were seeded into each well of
6-well plates and were further cultured for 2 weeks. When the
macroscopic colonies (>50 cells) were evidently observed, the
cells were fixed with 4% paraformaldehyde for 10 min at room
temperature and then stained with Giemsa (1X) for another 20 min at
room temperature. Finally, images of the visibly stained colonies
were captured and colonies were counted with the naked eye. The
experiment was performed in triplicate.
Wound healing assay
Cells (2×105) were seeded into 12-well
plates and cultured. Subsequently, after a 70–80% confluent culture
was obtained, the cell monolayer was scraped with a 200-µl pipette
tip. Then, the cells were washed and recultured with a serum-free
medium. Cell wounds were observed and images were captured under an
inverted microscope at 0 and 24 h and the degree of wound healing
was expressed as the width change between the two-time points. At
least eight fields for each condition were selected randomly in
each well with a magnification of ×100. The experiment was
performed in triplicate.
Cell migration and invasion
assays
The detection of cell migration and invasion
abilities was carried out by using a 24-well Transwell chamber
(Corning, Inc.) with Matrigel (BD). The upper chamber of Transwell
culture plates was coated with 0.4 mg/ml Matrigel (BD Biosciences)
and incubated for 24 h at 4°C. In brief, 3×104 cells in
200 µl serum-free medium were added into the upper chambers of each
Transwell insert (8-µm pore size), and 600 µl complete culture
medium containing 10% FBS was added to the lower chamber. After 24
h of incubation at 37°C, cells were fixed with methanol (99.5%) for
30 min and stained for 30 min at room temperature with 0.1% crystal
violet after fixation with methanol. Images of the cells on the
apical chamber membrane were captured and cells selected from five
random microscopic fields were counted at a magnification of ×400
with an inverted microscope.
Immunofluorescence analysis
Ki67 immunofluorescence analysis was also performed
to observe cell proliferation. Cells were cultured in 24-well
plates containing a sterile glass coverslip. After treatment, the
cells were fixed with cold 4% paraformaldehyde for 15 min at room
temperature and permeabilized in 0.2% Triton X-100 for 20 min at
room temperature. Thereafter, the cells were blocked with 10% goat
serum (Invitrogen; Thermo Fisher Scientific, Inc.) in
phosphate-buffered saline (PBS; Gibco; Thermo Fisher Scientific,
Inc.) for 30 min at room temperature, before the addition of
anti-Ki67 antibody (product code ab15580; 1:500; Abcam) overnight
at 4°C. After washing twice with PBS, the cells were incubated with
an FITC-anti-rabbit IgG (cat. no. sc-2012; 1:1,000; Santa Cruz
Biotechnology, Inc.) for 2 h at room temperature and then
counterstained with DAPI (0.1 µg/ml; Invitrogen; Thermo Fisher
Scientific, Inc.) for 5 min in the dark at room temperature. Cell
staining was visualized using fluorescence microscopy with images
captured at a magnification of ×200.
Fluorescence in situ hybridization
(FISH)
Specific probes targeting MALAT1 or miR-26a/26b
probes were synthesized by Shanghai GenePharma Co., Ltd. The RNA
FISH assay was performed using a FISH kit (Shanghai GenePharma Co.,
Ltd.) according to the manufacturer's protocols. Cells were fixed
with 4% paraformaldehyde at room temperature and incubated with
0.5% Triton X-100 in PBS on ice for 10 min. Then, cells were
pre-hybridized for 30 min at 37°C before being hybridized with 1 µM
Cy3-labeled MALAT1 and 1 µM FITC-labeled miR-26a/26b probes
(Shanghai GenePharma Co., Ltd.) at 37°C overnight. The cells were
then counterstained with DAPI (1 µg/ml) for 10 min at room
temperature before visualization using a fluorescence microscope at
a magnification of ×400.
Dual-luciferase reporter assay
The pGL3-based luciferase reporter plasmids
(Invitrogen; Thermo Fisher Scientific, Inc.) containing the 3′-UTR
region of wild-type (WT) MALAT1 or mutant (MUT) MALAT1 mutated with
altered miR-26a/26b binding sites were designed and constructed.
Firefly luciferase and Renilla luciferase were used for
normalization. A total of 5×104 cells were then
co-transfected with pGL3-MALAT1 WT or MUT plasmid in combination
with miR-26a/26b mimics using Lipofectamine 3000 (Invitrogen;
Thermo Fisher Scientific, Inc.). After 24 h, the Dual-Luciferase
Reporter Assay System (Promega Corporation) was used to evaluate
reporter activity.
Western blotting
Cell lysates were prepared using RIPA lysis buffer
(25 mM Tris•HCl pH 7.6, 150 mM NaCl, 1% NP-40, 1% sodium
deoxycholate, 0.1% SDS) by following the manufacturer's protocol.
The protein concentration was measured with a BCA Protein Assay Kit
(Pierce; Thermo Fisher Scientific, Inc.). Protein samples (20–40
µg) were resolved on 10% SDS-PAGE for separation and transferred
onto polyvinylidene difluoride membranes, followed by incubation
with blocking buffer [5% non-fat dry milk in PBS containing 0.1%
Tween-20 (PBST)] for 1 h at room temperature. Thereafter, the blots
were incubated with an anti-ST8SIA4 monoclonal antibody (cat. no.
AP9771C; 1:1,000; Abcepta) at 4°C overnight. Next, a secondary
anti-rabbit HRP-conjugated IgG (cat. no. sc-2357; 1:1,000; Santa
Cruz Biotechnology, Inc.) was added and incubated for 1 h at room
temperature. After washing, the immuno-reactive bands and images
were developed using an ECL kit (Amersham; Cytiva). All bands were
determined and analyzed by LabWorks (TM ver4.6, UVP, BioImaging
Systems). Blotting against GAPDH (cat. no. sc-47724; 1:1,000; Santa
Cruz Biotechnology, Inc.) was used as the loading control and
internal reference.
RNA immunoprecipitation (RIP)
assay
The binding relationship between MALAT1 and
miR-26a/26b was explored using the RIP assay kit (cat. no. 17-700;
EMD Millipore) following the recommended guidelines from the
manufacturer. Then, 2×107 cells were lysed in the
presence of RNase inhibitor and mixed with magnetic beads
pre-incubated with anti-argonaute (Ago)2 antibody (cat. no.
04-642-I; 1:1,000; EMD Millipore) at 4°C overnight to capture
endogenous miR-26a/26b. The levels of coprecipitated MALAT1 were
determined using RT-qPCR. IgG antibody (cat. no. AP124; 1:1,000;
EMD Millipore) precipitates were used as the negative control.
Bioinformatics analysis
StarBase software v2.0 (http://starbase.sysu.edu.cn) was used to predict the
binding sites shared by miRNAs and lncRNAs.
Statistical analysis
All analyses were performed using SPSS 17.0
statistical packages (SPSS, Inc.). The data were presented as the
mean ± SD. Unpaired t-tests were used to compare two groups.
Comparisons between multiple groups were performed by one-way ANOVA
analysis and a post hoc pairwise analysis was performed using
Tukey's post hoc test. Correlation between gene expression was
performed using Spearman's correlation analysis. The Kaplan-Meier
method was used to construct the survival curves and the difference
was assessed by a log-rank test. P<0.05 was considered to
indicate a statistically significant difference.
Results
Expression of MALAT1 is associated
with breast cancer metastasis and overall survival
In the present study, MALAT1 expression levels in a
total of 36 pairs of cancer and adjacent non-tumor tissues from
breast cancer patients were detected by RT-qPCR. After normalizing
against GAPDH, it was revealed that the expression of MALAT1 was
significantly increased in cancer tissues compared with paired
non-tumor tissues (Fig. 1A). Notably,
cancer tissues from breast cancer patients with metastasis
demonstrated higher MALAT1 levels than those without metastasis
(Fig. 1B). MALAT1 levels were also
evaluated in breast cancer cells by RT-qPCR (Fig. 1C). Breast cancer cells expressed
higher MALAT1 levels than the normal MCF-10A cells, especially in
MDA-MB-231 cells which displayed higher levels than MCF-7 cells.
Moreover, survival analysis indicated that relatively high MALAT1
expression predicted a poor prognosis in breast cancer patients
(Fig. 1D). These results indicated
that MALAT1 upregulation may play a critical role in breast cancer
tumorigenesis.
MALAT1 functions to promote breast
cancer cell aggressiveness
To further investigate the biological function of
MALAT1 in breast cancer cells, MALAT1 was overexpressed in the
MCF-7 cell line (Fig. S1A). As
anticipated, MALAT1-overexpressing MCF-7 cells exhibited
significantly increased cell proliferation compared with parental
MCF-7 cells as determined by using CCK-8 assays (Fig. 2A). Consistently, the proportion of
cells expressing Ki67 was also increased in MALAT1-overexpressing
cells (Fig. 2B). Furthermore,
assessment of the motility of MALAT1-overexpressing cells revealed
shorter wound distances in wound healing assays relative to the
negative control (mock) cells. The enhanced aggressiveness of MCF-7
cells with MALAT1 overexpression was also evident in the Transwell
assays (Fig. 2D). Conversely,
knockdown of MALAT1 in MDA-MB-231 cells indicated reductions in
cell proliferation when assessed by CCK-8 and Ki67
immunofluorescence assays (Fig. 2E and
F). Moreover, siMALAT1-transfected MDA-MB-231 cells
demonstrated reduced motility in the wound healing assay relative
to control cells (Fig. 2G), and more
importantly, MALAT1 knockdown inhibited the migration and invasion
of MDA-MB-231 cells (Fig. 2H). Thus,
these results collectively indicated that MALAT1 promoted
proliferation, motility and invasion in breast cancer cells.
MALAT1 directly regulates
miR-26a/26b
Accumulating evidence has revealed that lncRNAs can
target and regulate miRNAs by ‘sponge-like’ effects, which
subsequently inhibit miRNA-mediated functions (22,23). To
provide improved comprehension of the molecular mechanism involving
MALAT1 in breast cancer, RT-qPCR was performed to examine the
expression of MALAT1 and miR-26a/26b in breast cancer tissues. The
results revealed that there was a negative relationship between
MALAT1 and miR-26a/26b (Fig. 3A).
Moreover, significant decreases of miR-26a/26b expression were
observed in MALAT1-overexpressing MCF-7 cells (Fig. 3B). Consistently, knockdown of MALAT1
increased the expression level of miR-26a/26b in MDA-MB-231 cells
(Fig. 3C). Furthermore, ectopic
expression of MALAT1 in MCF-7 cells significantly upregulated the
expression of ST8SIA4, which is a major target of miR-26a/26b
(Fig. 3D). Conversely, MALAT1
downregulation reduced the abundance of ST8SIA4 in MDA-MB-231
cells. Collectively, MALAT1 directly regulated the expression of
the miR-26a/26b targeting ST8SIA4.
MALAT1 serves as a miR-26a/26b sponge
in breast cancer cells
The underlying interaction mechanism by which MALAT1
and miR-26a/26b are involved in breast cancer was further
investigated. Bioinformatics analysis (http://starbase.sysu.edu.cn) predicted that MALAT1
directly binds to miR-26a/26b with the complementary binding sites
revealed in Fig. 4A and B.
Dual-luciferase reporter assay revealed that co-transfection of
miR-26a/26b mimics and WT MALAT1 led to decreased reporter
activity. Instructively, there was no significant change in
luciferase activity when MUT MALAT1 and miR-26a/26b mimics were
cotransfected indicating a specific interaction through the
identified binding sites. Consistently, the co-location of MALAT1
and miR-26a/26b in the cytoplasm of breast cancer cells was
verified by the FISH assay, which indicated their functional
crosstalk (Fig. 4C and D). Moreover,
both MALAT1 (Fig. 4E) and miR-26a/26b
(Fig. 4F) were enriched in RIP assays
conducted against the Ago2 protein, indicating that Ago2 directly
bound to MALAT1 and miR-26a/26b in breast cancer cells.
Collectively, these data indicated that MALAT1 acted as a molecular
sponge for miR-26a/26b.
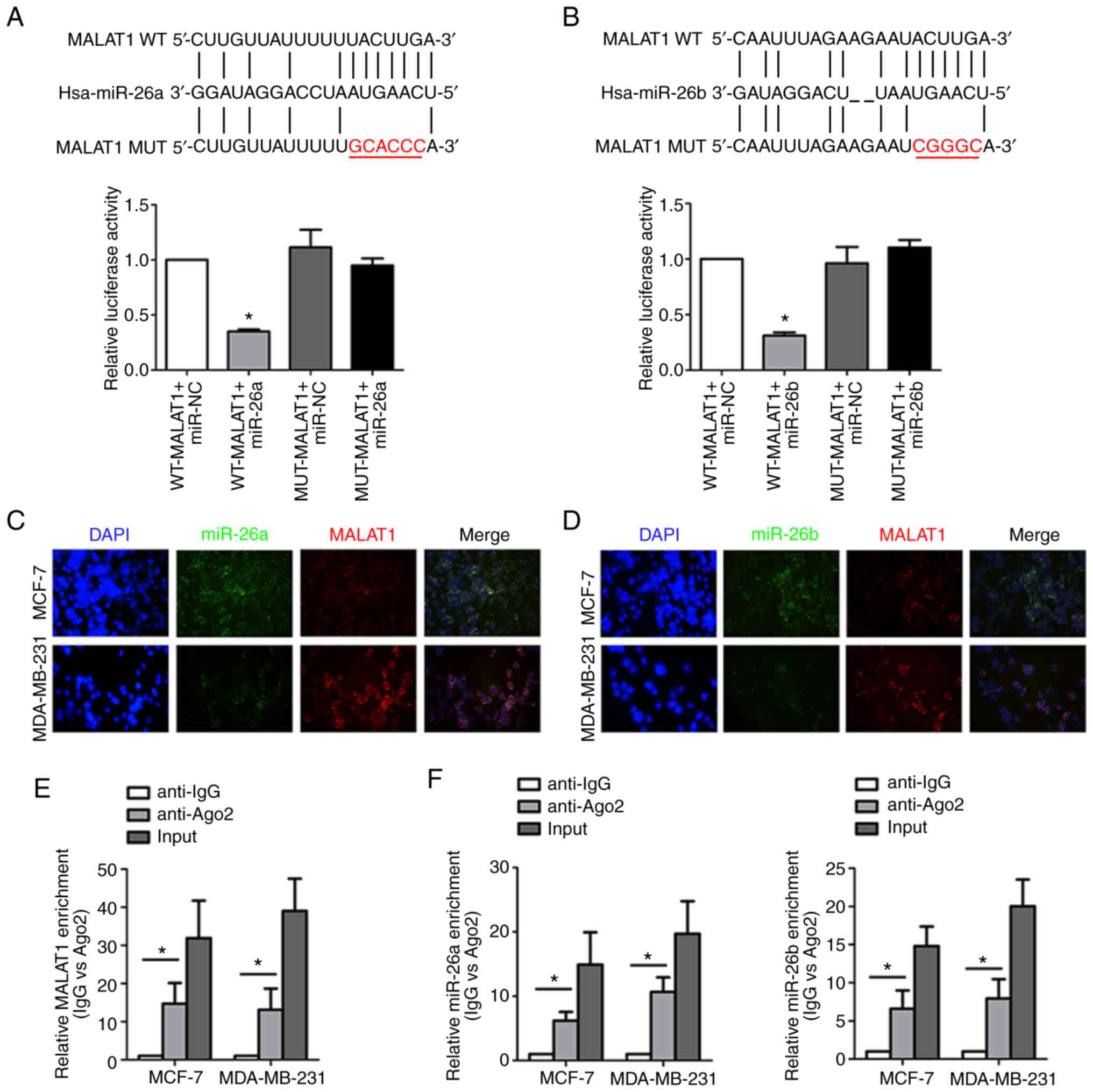 | Figure 4.MALAT1 acts as a molecular sponge for
miR-26a/26b. (A) The predicted binding sites between MALAT1 and
miR-26a. In addition, luciferase activities were assessed to
analyze the direct binding of MALAT1 and miR-26a. (B) The predicted
binding sites between MALAT1 and miR-26b. Luciferase activity was
also assessed to analyze the direct binding of MALAT1 and miR-26b.
(C) MALAT1 and miR-26a colocalization was detected by FISH assay in
MDA-MB-231 cells and MCF-7 cells. Red fluorescence, MALAT1; green
fluorescence, miR-26a; DAPI was used to stain nuclear DNA. (D)
MALAT1 and miR-26b colocalization were detected by FISH assay in
MDA-MB-231 cells and MCF-7 cells. Red fluorescence, MALAT1; green
fluorescence, miR-26b; DAPI was used to stain nuclear DNA. (E)
Coprecipitated RNA was detected by RIP assay. MALAT1 levels were
presented as fold enrichment in the Ago2 pellet relative to the IgG
immunoprecipitate. (F) Enrichment of miR-26a/26b was revealed by
the RIP assay. *P<0.05. MALAT1, metastasis-associated lung
adenocarcinoma transcript 1; miR, microRNA; WT, wild-type; MUT,
mutant; FISH, fluorescence in situ hybridization; RIP, RNA
immunoprecipitation; Ago, argonaute; IgG, immunoglobulin G. |
MALAT1/miR-26a/26b/ST8SIA4 axis
contributes to breast cancer progression
A previous study demonstrated that miR-26a/26b
suppressed breast cancer progression by downregulating the levels
of ST8SIA4 (6). To confirm the
effects of MALAT1 on miR-26a/26b and ST8SIA4 during breast cancer
progression, further functional experiments were carried out. In
fact, the modulatory effects of downregulating miR-26a/26b and
MALAT1 on ST8SIA4 were increased in MDA-MB-231 cells (Figs. 5A, S1B and
C). In comparison with the controls, the miR-26a/26b inhibitors
resulted in increased ST8SIA4 expression, while silencing MALAT1
decreased ST8SIA4 levels. In addition, silencing of MALAT1 in
miR-26a/26b-inhibited cells partially reversed the increases in
ST8SIA4. Similar trends were evident in the western blot assays
where knockdown of MALAT1 suppressed the increased levels of
ST8SIA4 protein induced by miR-26a/26b inhibitors (Fig. 5B), consistent with a model wherein
MALAT1 regulates ST8SIA4 via miR-26a/26b.
Functional assays also revealed the likely
significance of MALAT1 and miR-26a/26b during breast cancer
progression. Colony formation assays along with Ki67 staining
assessing the proportion of proliferating cells demonstrated that
breast cancer cells transfected with miR-26a/26b mimics exhibited a
higher proliferative activity than control cells, while cells
transfected with siMALAT1 displayed comparatively lower
proliferative activity. Interestingly, cotransfection of
miR-26a/26b mimics and siMALAT1 reversed the altered proliferation
which was induced by miR-26a/26b mimics alone (Fig. 5C and D). Scratch and Transwell assays
also revealed that miR-26a/26b inhibition increased the ability of
migration and invasion in MDA-MB-231 cells. Moreover, relative to
transfection of miR-26a/26b inhibitors only, these cells exhibited
further decreases in migration and invasion abilities after
cotransfection of siMALAT1 with the miR-26a/26b inhibitors
(Fig. 5E-H).
In concert with the aforementioned experiments using
MDA-MB-231 cells, the effects of miR-26a/26b and MALAT1 were
investigated in MCF-7 cells (Figs. 6A and
B, S1B and C). Transfection of
MALAT1 or miR-26a/26b into MCF-7 cells revealed that overexpression
of miR-26a/26b suppressed cell viability, migration, and invasion,
whereas ectopically expressed MALAT1 reversed these properties
(Fig. 6C-H). Collectively, our
findings indicated that MALAT1 promoted the progression of breast
cancer through a miR-26a/26b/ST8SIA4 axis.
Discussion
Breast cancer carcinogenesis is a complex biological
process involving the contribution of different genomic mutations
interacting with various biological signaling networks. Mounting
evidence has indicated that the genesis of breast cancer involves
epigenetic modifications (e.g., DNA methylation), modulation of
tumor-susceptible genes (e.g., BRCA1 and BRCA2), and the
dysregulation of lncRNAs, with tumorigenesis occurring when the
balance of oncogenes and anti-oncogenes are altered (24,25).
Consequently, in-depth investigations of the underlying mechanisms
promoting breast cancer could provide valuable insights into novel
diagnostic markers as well as the identification of therapeutic
targets. In the present study, the functional effects of the lncRNA
MALAT1 were identified and characterized in breast cancer
progression, revealing a novel mechanism by which MALAT1 regulated
ST8SIA4 through miR-26a/26b.
Advances in sequencing technology along with careful
mechanistic analyses have revealed that numerous lncRNAs
participate in tissue homeostasis together with disease
pathogenesis by exerting regulatory effects on gene expression at
the transcriptional, post-transcriptional and epigenetic levels
(26). Numerous lncRNAs are
aberrantly expressed in tumors, and in specific cases these have
been revealed to exert powerful effects on cancer initiation,
progression and metastasis. For example, a study by Liu et
al reported that the lncRNA HOTAIR regulated colorectal cancer
malignancy by inducing c-Met sialylation and activating the
JAK2/STAT3 pathway (27). Studies
performed by Guan et al in breast cancer also indicated that
lncRNA SNHG20 promoted cell proliferation, invasion, and migration
by regulating HER2 via miR-495 (28).
Moreover, Gao et al reported that lncRNA ATB/miR-200c/CDK2
signaling was responsible for intensified proliferation and
restrained apoptosis of colorectal cancer cells (29). Previous studies have revealed that
MALAT1 exhibited a close association with breast cancer, but its
precise functions in breast cancer progression remain unclear
(30,31). Our study confirmed that MALAT1 was
especially overexpressed in breast cancer tissues with high MALAT1
expression being associated with metastasis and poor overall
survival. Interestingly, a recent research also revealed that the
MALAT1 expression in serum was increased in breast cancer patients,
while its levels became downregulated after patients received
breast-conserving surgery combined with neo-adjuvant chemotherapy
(32). It was also confirmed that
MALAT1 was overexpressed in breast cancer cell lines and moreover,
provided comprehensive evidence that MALAT1 increased the migration
and invasion of breast cancer cells in vitro. Therefore,
consistent with the studies of Arun and Spector (33), our results indicated MALAT1 acted as
an essential regulator of breast cancer progression.
Mounting evidence has indicated that lncRNAs act as
competing endogenous RNAs (ceRNAs) that regulate gene expression by
sponging compatible miRNAs. For example, Li et al reported
that MALAT1 contributed to glioma autophagy and proliferation by
sponging miR-101 (34). Functional
correlations between MALAT1 and miR-30a-5p were also recorded in
hepatocellular carcinoma, indicating that MALAT1 acted as an
oncogenic lncRNA in HCC that promoted cell migration and invasion
(35). In the present study, focus
was placed on the relationship between MALAT1 and miR-26a/26b in
breast cancer. miR-26a and miR-26b are members of the miR-26
family; they are transcribed from three genomic loci, miR-26a-1,
miR-26a-2 and miR-26b, which reside in the introns of genes coding
for the CTDSPL, CTDSP2 and CTDSP1 proteins, respectively (36). Previous studies have noted that
miR-26a/26b are tumor-suppressor genes that considerably suppress
the invasion process (37,38). In this study, it was revealed that the
expression of MALAT1 was negatively correlated with miR-26a/26b in
breast cancer tissues and that MALAT1 could inhibit miR-26a/26b
expression. Furthermore, bioinformatics analysis highlighted a
possible interaction between these two molecules with FISH assays
demonstrating that MALAT1 and miR-26a/26b colocalized in breast
cancer cells. Further substantiating this theory, luciferase
reporter and RIP assays identified that MALAT1 acted as a molecular
sponge for miR-26a/26b.
The malignant process of tumors is frequently
accompanied by changes in the structure and expression of
oligosaccharide proteins and glycolipids on the cell surface. It
has been revealed that the expression of sialylated glycoconjugates
are altered during cell differentiation, disease progression and
oncogenic transformation (39). ST
expression patterns are significantly different between cancer and
normal tissues (40). It was
previously reported that ST8SIA4 was upregulated in breast cancer
cells and tissues and acted to promote malignant progression with
evidence revealing that ST8SIA4 was a target gene of miR-26a/26b
(6). Extending the aforementioned
work here, a positive correlation was revealed between MALAT1 and
ST8SIA4 and functional evidence for their association was provided.
For example, it was demonstrated that MALAT1 silencing in
miR-26a/26b-inhibited cells partially reversed the increases in
ST8SIA4 levels. Therefore, a ceRNA role of MALAT1 appeared to be a
reasonable hypothesis to explain these results. In fact, the
mechanism whereby MALAT1 acted as a molecular sponge for
miR-26a/26b, suppressing cell progression in breast cancer by
targeting ST8SIA4, was analyzed. Moreover, MALAT1 overexpression
partially rescued the suppression of cell proliferation, migration
and invasion induced by miR-26a/26b mimics. In summary, our results
demonstrated a crucial role for miR-26a/26b as a direct target of
MALAT1 in regulating breast cancer progression with clear evidence
of crosstalk between MALAT1, miR-26a/26b and ST8SIA4.
In conclusion, the present work revealed that MALAT1
was overexpressed in breast cancer where it functioned as an
oncogene. Notably, high MALAT1 expression in breast cancer patient
tissues was positively associated with tumor metastasis and poor
clinical outcomes. It was also established that MALAT1 acted as a
ceRNA for miR-26a/26b in breast cancer cells and the inhibition of
miR-26a/26b thereby weakened its inhibitory effects on ST8SIA4
expression. The present study identified a
MALAT1/miR-26a/26b/ST8SIA4 axis which crucially contributed to the
cell proliferation, invasion and migration of breast cancer cells.
This discovery also provided new insights into the underlying
mechanisms regulated by MALAT1 in breast cancer and indicated that
the MALAT1/miR-26a/26b/ST8SIA4 axis holds great promise as a new
therapeutic target for breast cancer. However, lncRNA MALAT1 and
miR-26a/26b could, respectively, possess targets of diverse miRNAs
and mRNAs, and discovery of multiple lncRNA/miRNA/genes may be
conducive to thorough suppression of breast cancer cells. This
shortcoming of the present study should be improved in the
future.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by grants (grant
nos. 20180550476 and 20180550729) from the Natural Science
Foundation of Liaoning Province (China).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XM and HY conceived and supervised the study. NW and
SC designed and performed the experiments. XW, LZ and NW analyzed
the data. XM, NW, SC and HY wrote the manuscript. All authors read
and approved the final version of the manuscript.
Ethics approval and consent to
participate
The Ethics Committee of The First Affiliated
Hospital of Dalian Medical University approved the present study
and written informed consents were acquired from all enrolled
patients.
Patient consent for publication
Not applicable.
Competing interests
The author declare that they have no competing
interests.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ruiterkamp J, Voogd AC, Tjan-Heijnen VC,
Bosscha K, van der Linden YM, Rutgers EJ, Boven E, van der Sangen
MJ and Ernst MF; Dutch Breast Cancer Trialists' Group (BOOG), :
SUBMIT: Systemic therapy with or without upfront surgery of the
primary tumor in breast cancer patients with distant metastases at
initial presentation. BMC Surg. 12:52012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kölbl AC, Andergassen U and Jeschke U: The
role of glycosylation in breast cancer metastasis and cancer
control. Front Oncol. 5:2192015. View Article : Google Scholar
|
|
4
|
Vajaria BN, Patel KR, Begum R and Patel
PS: Sialylation: An avenue to target cancer cells. Pathol Oncol
Res. 22:443–447. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Petretti T, Kemmner W, Schulze B and
Schlag PM: Altered mRNA expression of glycosyltransferases in human
colorectal carcinomas and liver metastases. Gut. 46:359–366. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ma XL, Dong WJ, Su Z, Zhao LF, Miao Y, Li
NN, Zhou HM and Li J: Functional roles of sialylation in breast
cancer progression through miR-26a/26b targeting ST8SIA4. Cell
Death Dis. 7:e25612016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bach DH and Lee SK: Long noncoding RNAs in
cancer cells. Cancer Lett. 419:152–166. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bartonicek N, Maag JL and Dinger ME: Long
noncoding RNAs in cancer: Mechanisms of action and technological
advancements. Mol Cancer. 15:432016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tu ZQ, Li RJ, Mei JZ and Li XH:
Down-regulation of long non-coding RNA GAS5 is associated with the
prognosis of hepatocellular carcinoma. Int J Clin Exp Pathol.
7:4303–4309. 2014.PubMed/NCBI
|
|
10
|
Hombach S and Kretz M: Non-coding RNAs:
Classification, biology and functioning. Adv Exp Med Biol.
937:3–17. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li ZX, Zhu QZ, Zhang HB, Hu Y, Wang G and
Zhu YS: MALAT1: A potential biomarker in cancer. Cancer Manag Res.
10:6757–6768. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang H, Yang F, Chen SJ, Che JP and Zheng
JH: Upregulation of long non-coding RNA MALAT1 correlates with
tumor progression and poor prognosis in clear cell renal cell
carcinoma. Tumor Biol. 36:2947–2955. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sun JY, Zhao ZW, Li WM, Yang G, Jing PY,
Li P, Dang HZ, Chen Z, Zhou YA and Li XF: Knockdown of MALAT1
expression inhibits HUVEC proliferation by upregulation of miR-320a
and downregulation of expression. Oncotarget. 8:61449–61509.
2017.
|
|
14
|
Wu Y, Sarkissyan M, Ogah O, Kim J and
Vadgama JV: Expression of MALAT1 promotes trastuzumab resistance in
HER2 overexpressing breast cancers. Cancers (Basel). 12:19182020.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Browne G, Dragon JA, Hong D, Messier TL,
Gordon JA, Farina NH, Boyd JR, VanOudenhove JJ, Perez AW, Zaidi SK,
et al: MicroRNA-378-mediated suppression of Runx1 alleviates the
aggressive phenotype of triple-negative MDA-MB-231 human breast
cancer cells. Tumour Biol. 37:8825–8839. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chen Y, Luo DL, Tian WG, Li ZR and Zhang
XH: Demethylation of miR-495 inhibits cell proliferation, migration
and promotes apoptosis by targeting STAT-3 in breast cancer. Oncol
Rep. 37:3581–3589. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li C, Huo B, Wang Y and Cheng C:
Downregulation of microRNA-92b-3p suppresses proliferation,
migration, and invasion of gastric cancer SGC-7901 cells by
targeting Homeobox D10. J Cell Biochem. 120:17405–17412. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chan JJ and Tay Y: Noncoding RNA: RNA
regulatory networks in cancer. Int J Mol Sci. 19:13102018.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tay Y, Rinn J and Pandolfi PP: The
multilayered complexity of ceRNA crosstalk and competition. Nature.
505:344–352. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang Y, Zhu Z, Huang S, Zhao Q, Huang C,
Tang Y, Sun C, Zhang Z, Wang L, Chen H, et al: LncRNA XIST
regulates proliferation and migration of hepatocellular carcinoma
cells by acting as miR-497-5p molecular sponge and targeting PDCD4.
Cancer Cell Int. 19:1982019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhao Z, Sun W, Guo Z, Zhang J, Yu H and
Liu B: Mechanisms of lncRNA/microRNA interactions in angiogenesis.
Life Sci. 254:1169002020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang X, Teer JK, Tousignant RN, Levin AM,
Boulware D, Chitale DA, Shaw BM, Chen Z, Zhang Y, Blakeley JO, et
al: Breast cancer risk and germline genomic profiling of women with
neurofibromatosis type 1 who developed breast cancer. Genes
Chromosomes Cancer. 57:19–27. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Amelio I, Bernassola F and Candi E:
Emerging roles of long non-coding RNAs in breast cancer biology and
management. Semin Cancer Biol. 72:36–45. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Dimartino D, Colantoni A, Ballarino M,
Martone J, Mariani D, Danner J, Bruckmann A, Meister G, Morlando M
and Bozzoni I: The long non-coding RNA lnc-31 interacts with Rock1
mRNA and mediates its YB-1-dependent translation. Cell Rep.
23:733–740. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liu B, Liu Q, Pan S, Huang Y, Qi Y, Li S,
Xiao Y and Jia L: The HOTAIR/miR-214/ST6GAL1 crosstalk modulates
colorectal cancer procession through mediating sialylated c-Met via
JAK2/STAT3 cascade. J Exp Clin Cancer Res. 38:4552019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Guan YX, Zhang MZ, Chen XZ, Zhang Q, Liu
SZ and Zhang YL: Lnc RNA SNHG20 participated in proliferation,
invasion, and migration of breast cancer cells via miR-495. J Cell
Biochem. 119:7971–7981. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Gao Z, Zhou H, Wang Y, Chen J and Ou Y:
Regulatory effects of lncRNA ATB targeting miR-200c on
proliferation and apoptosis of colorectal cancer cells. J Cell
Biochem. 121:332–343. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Xiao Y, Pan J, Geng Q and Wang G: LncRNA
MALAT1 increases the stemness of gastric cancer cells via enhancing
SOX2 mRNA stability. FEBS Open Bio. 9:1212–1222. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zuo Y, Li Y, Zhou Z, Ma M and Fu K: Long
non-coding RNA MALAT1 promotes proliferation and invasion via
targeting miR-129-5p in triple-negative breast cancer. Biomed
Pharmacother. 95:922–928. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Sun Z and Liu J and Liu J: The expression
of lncRNA-MALAT1 in breast cancer patients and its influences on
prognosis. Cell Mol Biol (Noisy-le-grand). 66:72–78. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Arun G and Spector DL: MALAT1 long
non-coding RNA and breast cancer. RNA Biol. 16:860–863. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Li Z, Xu C, Ding B, Gao M, Wei X and Ji N:
Long non-coding RNA MALAT1 promotes proliferation and suppresses
apoptosis of glioma cells through derepressing Rap1B by sponging
miR-101. J Neurooncol. 134:19–28. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Pan Y, Tong S, Cui R, Fan J, Liu C, Lin Y,
Tang J, Xie H, Lin P, Zheng T, et al: Long non-coding MALAT1
functions as a competing endogenous RNA to regulate vimentin
expression by sponging miR-30a-5p in hepatocellular carcinoma. Cell
Physiol Biochem. 50:108–120. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li Y, Sun Z, Liu B, Shan Y, Zhao L and Jia
L: Tumor-suppressive miR-26a and miR-26b inhibit cell
aggressiveness by regulating FUT4 in colorectal cancer. Cell Death
Dis. 8:e28922017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhu Y, Lu Y, Zhang Q, Liu JJ, Li TJ, Yang
JR, Zeng C and Zhuang SM: MicroRNA-26a/b and their host genes
cooperate to inhibit the G1/S transition by activating the pRb
protein. Nucleic Acids Res. 40:4615–4625. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kato M, Goto Y, Matsushita R, Kurozumi A,
Fukumoto I, Nishikawa R, Sakamoto S, Enokida H, Nakagawa M,
Ichikawa T and Seki N: MicroRNA-26a/b directly regulate La-related
protein 1 and inhibit cancer cell invasion in prostate cancer. Int
J Oncol. 47:710–718. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Li F and Ding J: Sialylation is involved
in cell fate decision during development, reprogramming and cancer
progression. Protein Cell. 10:550–565. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Garnham R, Scott E, Livermore KE and
Munkley J: ST6GAL1: A key player in cancer. Oncol Lett. 18:983–989.
2019.PubMed/NCBI
|




















