Introduction
Colorectal cancer (CRC) was the 3rd most common
malignancy in Australia in 2017 overall, with estimated all-age
incidence rates of 67.3 per 100,000 in males and 49.4 per 100,000
in females. Emergent evidence has suggested that the incidence of
colorectal cancer in people under 50 years of age (‘early-onset
colorectal cancer’) is rising in high-income countries (1). Among the various molecular drivers of
CRC, genetic and epigenetic alterations are the main cause of the
hereditary and sporadic forms (2).
Factors involved in CRC include the characteristics and properties
of tumors that provide information about the prognosis. Prognostic
and predictive biomarkers are selection factors that facilitate
treatment decision making and treatment response prediction,
respectively, in patients (3).
Previous studies have revealed that noncoding RNAs
directly act as tumor suppressors or oncogenes at the
transcriptional or post-transcriptional level to regulate the
occurrence and development of tumors (4,5). Long
noncoding RNAs (lncRNAs) are an interesting class of transcripts
longer than 200 nucleotides that have functions other than coding
proteins (6,7). Emerging evidence has revealed that
lncRNAs play a role in a variety of human cancers and can even be
used as predictive biomarkers or therapeutic targets for cancer
treatment (8,9). LncRNA CDKN2B-antisense RNA 1 (AS1) has
already been implicated in the advancement of various tumors
(8,9).
The lncRNA CDKN2B-AS1/miR-141/cyclin D network has been revealed to
regulate the progression and metastasis of renal cell carcinoma
(8). The lncRNA
CDKN2B-AS1/miR-324-5p/ROCK1 axis has been revealed to regulate cell
cycle progression in laryngeal squamous cell cancer (10). MicroRNAs (miRNAs) are small noncoding
RNAs 18–25 nucleotides in length that function by regulating target
genes. Studies have demonstrated that the differential expression
of miRNAs has an important relationship with the occurrence,
development and prognosis of CRC (10–12).
LncRNAs act as sponges/competing endogenous RNAs (ceRNAs) to
regulate the post-transcriptional regulation of miRNAs and are
factors in the etiology of numerous diseases (12). Thus, this study focused on the
differential expression of CDKN2B-AS1 and related miRNAs in
CRC.
CDKN2B-AS1 may play different roles during
carcinogenesis in cancers (8–10). In the present study, the biological
role and functions of CDKN2B-AS1 were investigated in CRC. The
expression of CDKN2B-AS1 was detected in CRC tissues and cancer
cell lines. Moreover, functional experiments were designed to
reveal the effects of CDKN2B-AS1 on the malignant behavior of CRC
cells. Furthermore, the relevant underlying mechanisms of
CDKN2B-AS1 in CRC were discussed in depth. Collectively, it was
determined that CDKN2B-AS1 may be involved in the progression of
CRC through the regulation of proliferation.
Materials and methods
CRC patients and tissue samples
The 69 collected CRC tissues and their corresponding
adjacent non-tumor tissue samples (sampled at more than 5 cm from
the tumor, 48 males and 21 females; age range, 30–65 years) were
obtained from Qingdao Municipal Hospital (Qingdao, China) between
May 2018 and May 2019 and stored at −80°C for further study. The
inclusion criteria was as follows: The patients had to be diagnosed
with colon cancer by pathological diagnosis and had to be aged
between 20 and 75 years. Patients were excluded if they suffered
from other malignant tumors or if they had been treated with
radiotherapy or chemotherapy within 3 months before enrolment. The
study was performed in accordance with clinical study protocols and
the principles of the Declaration of Helsinki (modified 2018). All
patients provided informed consent for research purposes. The
procedures in the present study were approved (approval no.
2018-R-32) by the Qingdao Municipal Hospital Ethics Review
Committee.
Cell lines and culture conditions
Human CRC cell lines (LOVO, HCT-116, DLD-1, SW480,
and RKO), a human normal cell line (NCM460) and 293T cells were
purchased from the Institute of Biochemistry and Cell Biology of
the Chinese Academy of Sciences. The cells were cultured in Roswell
Park Memorial Institute (RPMI-1640; Gibco; Thermo Fisher
Scientific, Inc.) with 10% fetal bovine serum (FBS, Gibco; Thermo
Fisher), 100 U/ml penicillin, and 100 mg/ml streptomycin (Gibco;
Thermo Fisher Scientific, Inc.) at 37°C in a 5% CO2
atmosphere.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from CRC tissues and CRC
cell lines using TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc.). The PrimeScript RT Reagent kit (Takara
Biotechnology Co., Ltd.) was used to reverse transcribe mRNA to
cDNA according to the manufacturer's instructions (temperature
protocol: 42°C for 60 min, 70°C for 5 min and then maintained at
4°C). The relative expression level of CDKN2B-AS1 was detected by a
SYBR-Green PCR Master Mix kit (Takara, Biotechnology Co., Ltd.)
according to the manufacturer's instructions. RT-qPCR was performed
at 95°C for 1 min, 40 cycles at 95°C for 10 sec and 58°C for 40
sec. The sequences of the specific primers were as follows:
CDKN2B-AS1 forward, 5′-GACTTCTGTTTTCTGGCCACC and reverse,
TCGGGAAAGGATTCCAGCAC-3′; upregulator of cell proliferation (URGCP)
forward, 5′-CGCAATCATCTCCTTCCATT-3′ and reverse,
5′-GATTTGGGAGAAGTAGCCCC-3′; GAPDH forward,
5′-GGTGCTGAGTATGTCGTGGAGTCTA-3′ and reverse,
5′-TCTTGAGGGAGTTGTCATATTTCTC-3′. GAPDH expression was used as an
endogenous control, and the 2−ΔΔCq method (13) was used to calculate the results.
Bioinformatics analysis
lncRNASNP2 (http://bioinfo.life.hust.edu.cn/lncRNASNP#!/mirna) and
TargetScan 7.2 (http://www.targetscan.org/) were used to predict the
putative target genes of CDKN2B-AS1 and miR-28-5p.
Knockdown of CDKN2B-AS1
expression
Two si-CDKN2B-AS1 small interfering (si)RNAs (50 nM)
targeting CDKN2B-AS1 (si#1 and si#2) were synthesized by Guangzhou
RiboBio Co., Ltd. All cells were transfected with siRNAs at room
temperature according to the manufacturer's instructions for
Lipofectamine 3000 Transfection Reagent (Thermo Fisher Scientific,
Inc.) for 24-h transfection. miR-28-5p mimics (50 nM) and
inhibitors (100 nM) were synthesized by Shanghai GenePharma Co.,
Ltd. miR-28-5p (50 nM) was transfected into cells at room
temperature according to the instructions for Lipofectamine 3000
Transfection Reagent (Thermo Fisher Scientific, Inc.) for 24-h
transfection. The siRNAs, miR-28-5p mimic, inhibitor and control
sequences are provided in Table SI,
and si-negative control (NC) (50 nM) used in the present study was
scrambled.
Dual-luciferase reporter plasmid
transfection
pmiR-RB-REPORT™ plasmids [CDKN2B-AS1-wild-type (WT)
and URGCP-WT] and their related mutant plasmids [CDKN2B-AS1-mutant
(MUT) and URGCP-MUT] were synthesized by Guangzhou RiboBio Co.,
Ltd. Cells (5×104) were seeded into 24-well plates and
co-transfected with the reporter plasmids alongside miR-28-5p mimic
(50 nM) or NC mimic (50 nM) applying Lipofectamine® 3000
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocols. Subsequently, 48 h later, the transfected
cells were lysed to detect their luciferase activity by employing a
dual-luciferase reporter assay system (Promega Corporation). The
dual-luciferase reporter assay system was used to assess firefly
luciferase activity which was normalized to Renilla
luciferase activity.
Cell Counting Kit-8 (CCK-8) assay
The proliferation of CRC cells was assessed by CCK-8
assay (Meilun Biotechnology Co., Ltd.). After transfection with
siRNA, the cells (2×103) were plated and cultured at
37°C in 96-well plates for 24, 48, 72, and 96 h, and then 20 µl
CCK-8 solution was added to the wells. To estimate cell
proliferation, the absorbance of the cell medium at 490 nm was
measured using a multifunctional microplate reader (SpectraMax
M5).
Colony formation assay
The cells (1,000/well) were seeded and cultured at
37°C for 10–14 days after transfection with siRNAs. Subsequently,
they were fixed with 4% paraformaldehyde at room temperature for 30
min and stained with 0.5% crystal violet at room temperature for 30
min, and the number of colonies was counted under an inverted light
microscope at a magnification of ×100 (D850; Nikon
Corporation).
In situ hybridization
DIG-labeled LNA-CDKN2B-AS1 probes were synthesized
following the manufacturer's instructions by Guangzhou RiboBio Co.,
Ltd. and the probe sequences are available upon request. In brief,
a 5-mm section of paraffin-embedded tissue was fixed with 4%
formaldehyde solution, washed, and permeabilized with Triton X-100
solution for 5 min at room temperature. The DIG-labeled
LNA-CDKN2B-AS1 probe was incubated with the tissues for
hybridization at 37°C, overnight. Then, the expression was
determined using diaminobenzidine solution (1:900; Boster
Biological Technology) for 3 min at room temperature, and the
staining intensity was observed using a BX51 light microscope
(Olympus Corporation). The staining was quantified by counting the
number of positive cells at a magnification of ×400.
Subcellular fractionation
A nuclear and cytoplasmic protein extraction kit
(Beyotime Institute of Biotechnology) and TRIzol reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) were used to extract
cytoplasmic and nuclear RNA according to the manufacturer's
instructions. CDKN2B-AS1 expression in the cytoplasmic and nuclear
fractions was assessed by qPCR. The thermocycling conditions were
as follows: 95°C for 30 sec, 95°C for 3 sec and 60°C for 30 sec,
for 40 cycles; 95°C for 15 sec, 60°C for 60 sec and 95°C for 15
sec. GAPDH and U6 were the cytoplasmic and nuclear controls,
respectively. The primers of U6 were as follows: forward,
5′-CTCGCTTCGGCAGCACA-3′ and reverse,
5′-AACGCTTCACGAATTTGCGT-3′.
Protein extraction and western
blotting
RIPA buffer (Thermo Fisher Scientific, Inc.)
containing protease inhibitor cocktail (Roche Diagnostics) was used
to extract total protein from CRC tissues and CRC cell lines. The
quantification of total protein was conducted with a BCA Protein
Assay kit (Sangon Biotech Co., Ltd.). Equal amounts of proteins (20
µg) were separated by 10% SDS-PAGE gels, followed by transferring
onto PVDF membranes (Sigma-Aldrich; Merck KGaA). After being
blocked with 5% fat-free milk at room temperature for 2 h, the
blots were probed with primary antibodies. Primary antibodies
anti-cyclin D1 (1:1,000; product no. 55506), cyclin-dependent
kinase (CDK)4 (1:1,000; product no. 12790), Rb (1:1,000; product
no. 9309), phosphorylated (p)-Rb (1:1,000; product no. 8516),
caspase-9 (1:1,000; product no. 9508), cleaved caspase-9 (1:1,000;
product no. 9505), caspase-3 (1:1,000; product no. 9662), cleaved
caspase-3 (1:1,000; product no. 9664; all from Cell Signaling
Technology, Inc.), URGCP (1:3,000; cat. no. PA5-44534; Thermo
Fisher Scientific, Inc.), and β-actin (1:1,000; product no. 4970;
Cell Signaling Technology, Inc.) were incubated with the PVDF
membranes overnight at 4°C. The PVDF membranes were then incubated
with HRP-labeled secondary antibody (1:1,000 dilution; product no.
7074; Cell Signaling Technology, Inc.) for 1 h at room temperature.
An enhanced chemiluminescence kit (EMD Millipore) was used to
expose the membrane. Images were captured through autoradiography
with X-ray film. ImageJ 1.8.0 software (National Institutes of
Health) was used to quantify the density of the target bands.
Statistical analysis
The data are presented as the mean ± standard
deviation and were analyzed using the statistical software package
SAS 8.0 for Windows (SAS Institute, Inc.). The paired Student's
t-test was used to analyze the CDKN2B-AS1 levels in 69
paired CRC and control tissues. The unpaired Student's t-test was
applied for two-group comparisons of the other assays, and one-way
ANOVA with Tukey's post hoc test was used for multiple group
comparisons. Two-tailed Pearson's correlation was used to analyze
the correlation between CDKN2B-AS1 and miR-28-5p levels. P<0.05
was considered to indicate a statistically significant
difference.
Results
CDKN2B-AS1 is overexpressed in CRC
tissues and cells
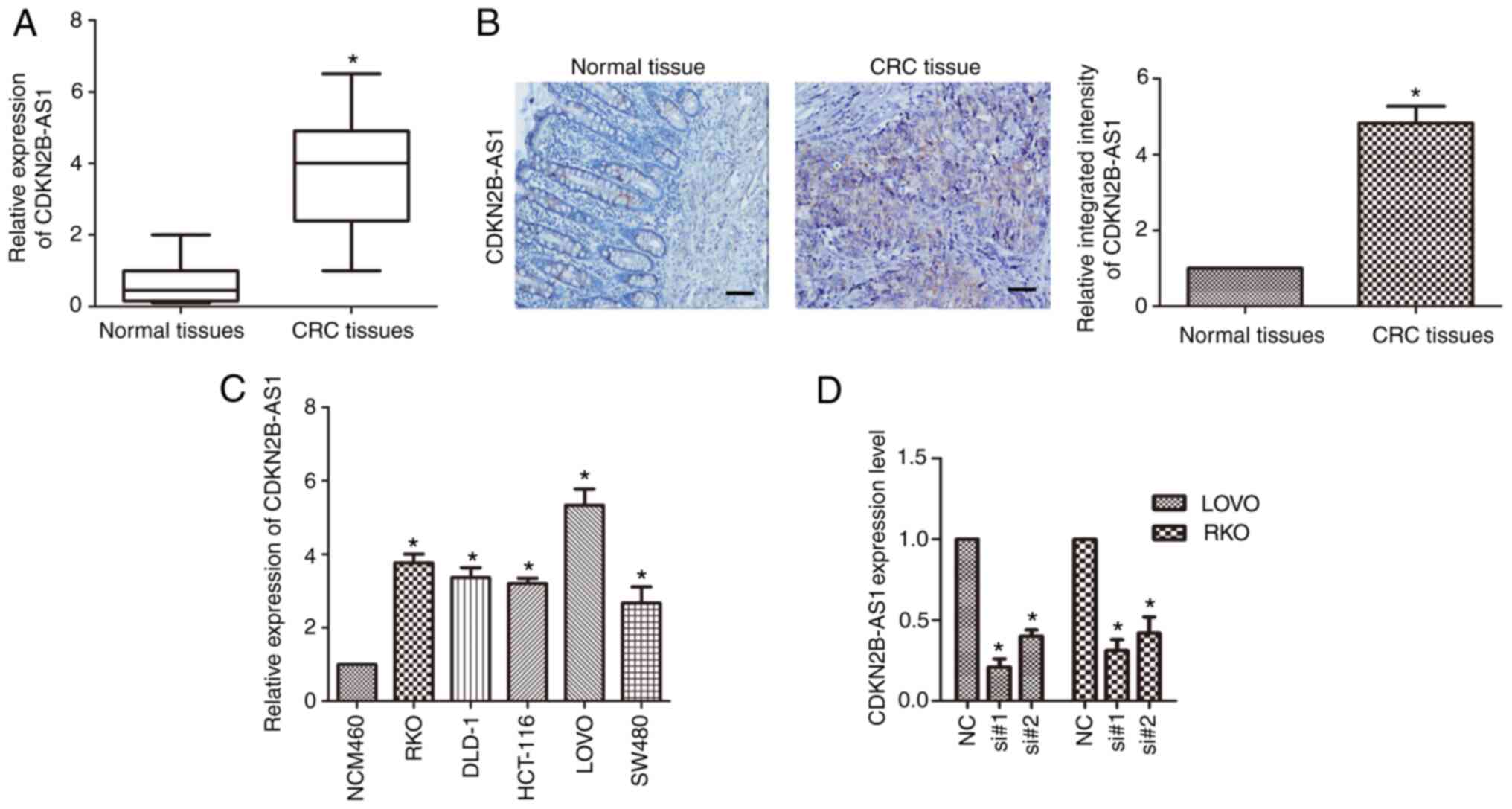
qPCR was applied to detect the expression of
CDKN2B-AS1 in CRC. The results revealed that CDKN2B-AS1 was highly
overexpressed in CRC tissue compared with matched adjacent normal
tissue (n=6; P<0.01; Fig. 1A).
Furthermore, in situ hybridization of a DIG-labeled
LNA-CDKN2B-AS1 probe revealed that CDKN2B-AS1 was mainly
distributed in the cytoplasm of CRC tissue (Fig. 1B). The expression of CDKN2B-AS1 in CRC
cell lines (LOVO, HCT-116, DLD-1, SW480 and RKO) was also detected
by qPCR. It was revealed that the expression of CDKN2B-AS1 was
upregulated in CRC cell lines compared with the normal colonic cell
line (n=6; P<0.01; Fig. 1C). In
LOVO and RKO cells, the expression of CDKN2B-AS1 was markedly
higher than that in the other cells (n=6; P<0.05; Fig. 1C); hence, the LOVO and RKO cell lines
were used for the following study. Two siRNAs (si#1 and si#2)
targeting CDKN2B-AS1 were used, and the knockdown efficiency of
these siRNAs was investigated in both LOVO and RKO cells.
CDKN2B-AS1 expression was significantly downregulated in LOVO cells
compared with the control cells (n=6, P<0.05; Fig. 1D) and in RKO cells compared with the
control cells (n=6, P<0.05; Fig.
1D).
Knockdown of CDKN2B-AS1 inhibits the
proliferation of CRC cells
As the expression of CDKN2B-AS1 was markedly higher
in both LOVO and RKO cells than in the other cells, experiments
with CDKN2B-AS1 knockdown were performed in these cells. To
determine the effect of CDKN2B-AS1 on the proliferation of both
LOVO and RKO cells, CCK-8 assays were conducted. As revealed in
Fig. 2A, the number of proliferating
cells in the si-CDKN2B-AS1 transfection group was decreased
compared with that in the NC group (n=6, P<0.05; Fig. 2A). Colony formation assays also
demonstrated that there were fewer CRC cell colonies in the
si-CDKN2B-AS1 group than in the control group (n=6, P<0.05;
Fig. 2B). To determine the molecular
mechanisms of si-CDKN2B-AS1-induced cell proliferation inhibition,
cell cycle-related proteins cyclin D1, CDK4, Rb, p-Rb and
apoptosis-related proteins (cleaved caspase-9 and cleaved
caspase-3) were examined by western blotting. The results
demonstrated that the protein levels of cyclin D1, and CDK4 were
decreased in CDKN2B-AS1-knockdown CRC cells compared with the NC
cells (n=6, P<0.05; Fig. 2C and
D). Moreover, it is illustrated that p-Rb/Rb expression levels
are increased after CDKN2B-AS1 knockdown compared with the negative
control cells (n=6, P<0.05; Fig. 2C
and D). The protein levels of cleaved caspase-9 and cleaved
caspase-3 were upregulated in the knockdown cells compared with the
NC cells (n=6, P<0.05; Fig. 2C and
D). Therefore, these results indicated that CDKN2B-AS1 promoted
CRC proliferation by regulating cell cycle-related and
apoptosis-related proteins in CRC.
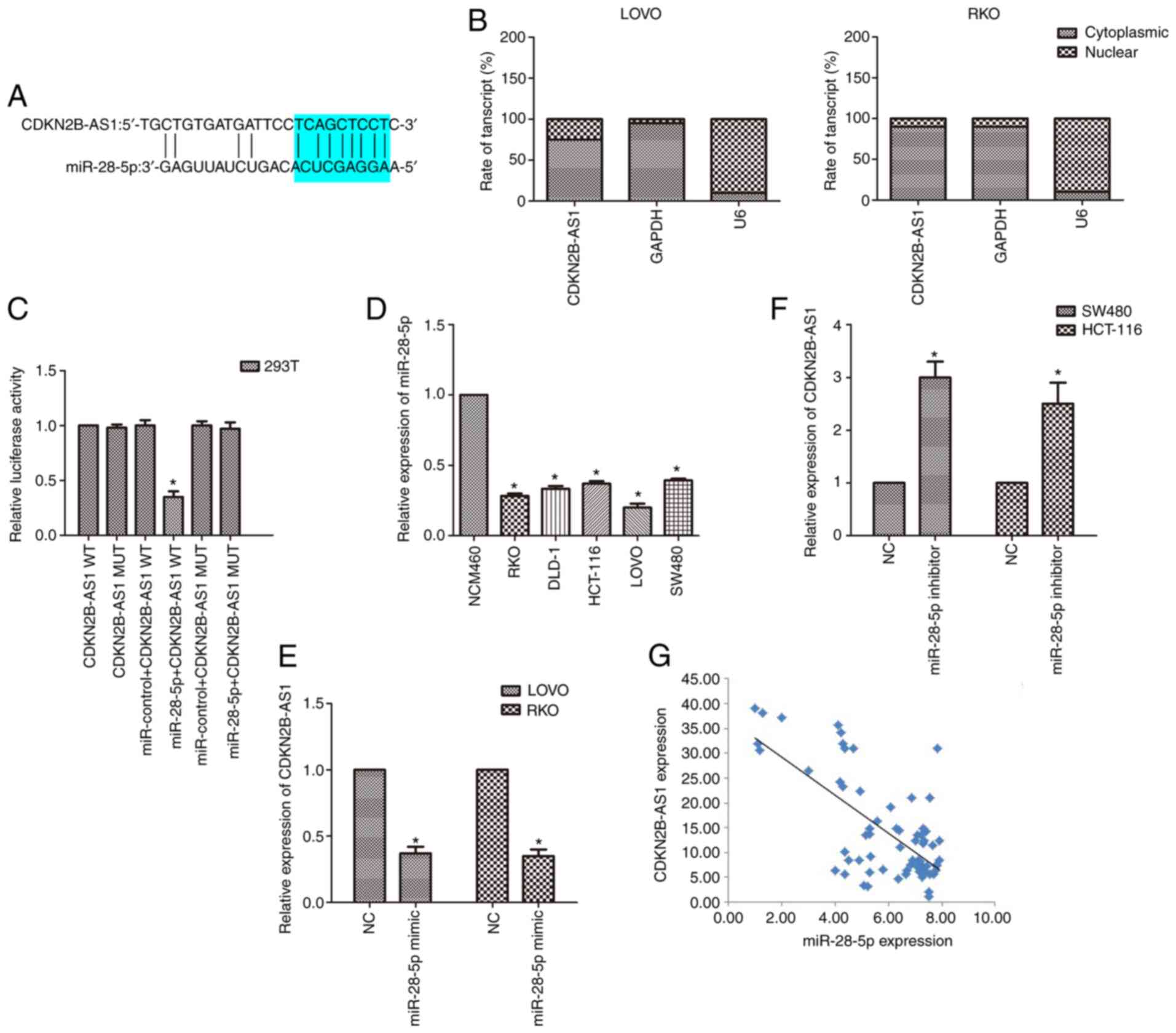
CDKN2B-AS1 sponges miR-28-5p in CRC
cells
To further explore the molecular mechanism of
CDKN2B-AS1 in the malignancy of CRC, a bioinformatics tool
(lncRNASNP2; http://bioinfo.life.hust.edu.cn/lncRNASNP#!/mirna) was
used to identify potential related miRNAs. The bioinformatics data
demonstrated that miR-28-5p is a potential target of CDKN2B-AS1
(Fig. 3A). Subcellular fractionation
revealed that CDKN2B-AS1 was principally located in the cytoplasm
of CRC cells (Fig. 3B). This binding
relationship was further validated by luciferase reporter assay.
The results revealed that the luciferase activity of cells
co-transfected with miR-28-5p and CDKN2B-AS1-WT reporter plasmids
was decreased compared with that of cells transfected with
CDKN2B-AS1-MUT alone or co-transfected with CDKN2B-AS1-MUT and
miR-28-5p mimic (n=6, P<0.05; Fig.
3C). The expression of miR-28-5p in CRC cell lines was also
detected by qPCR. It was revealed that the expression of miR-28-5p
was downregulated in CRC cell lines compared with the normal
colonic cell line (n=6; P<0.05; Fig.
3D). As the expression of miR-28-5p was markedly lower in both
LOVO and RKO cells than in the other cells, the effect of
upregulation of miR-28-5p expression was explored in these cells
(n=6; P<0.05; Fig. 3D).
Conversely, because the expression of miR-28-5p was higher in both
SW480 and HCT-116 cells than in the other cells, the effect of
downregulation of miR-28-5p expression was explored in SW480 and
HCT-116 cells (n=6; P<0.05; Fig.
3D). Moreover, upregulation of miR-28-5p expression induced a
reduction in CDKN2B-AS1 levels, whereas downregulation of miR-28-5p
caused an increase in CDKN2B-AS1 expression by qPCR results (n=6,
P<0.05; Fig. 3E and F).
Furthermore, the correlation analysis revealed a negative
relationship between miR-28-5p and CDKN2B-AS1 in CRC specimens
(P<0.01, r=−0.714; Fig. 3G).
CDKN2B-AS1 function is suppressed by
miR-28-5p
The results revealed that the binding relationship
between CDKN2B-AS1 and miR-28-5p raised the possibility that
miR-28-5p levels may be the intermediate between CDKN2B-AS1 and its
oncogenic effects. Among the examined siRNAs, siRNA-1(si#1)
exhibited the highest knockdown efficiency of CDKN2B-AS1 in cells,
thus, this siRNA was selected in this experiment. To examine this
possibility, CCK-8 and colony formation assays were performed.
After transfection with the miR-28-5p inhibitor, CCK-8 assays
revealed that the decrease in the number of live cells was partly
attenuated in CDKN2B-AS1-knockdown CRC cells (n=6, P<0.05;
Fig. 4A). The miR-28-5p inhibitor
caused a significant increase in colony formation, whereas
CDKN2B-AS1 knockdown abrogated this increase in colony formation
(n=6, P<0.05; Fig. 4B and C).
These data indicated that the CRC cell proliferation effects of
CDKN2B-AS1 were mediated by miR-28-5p.
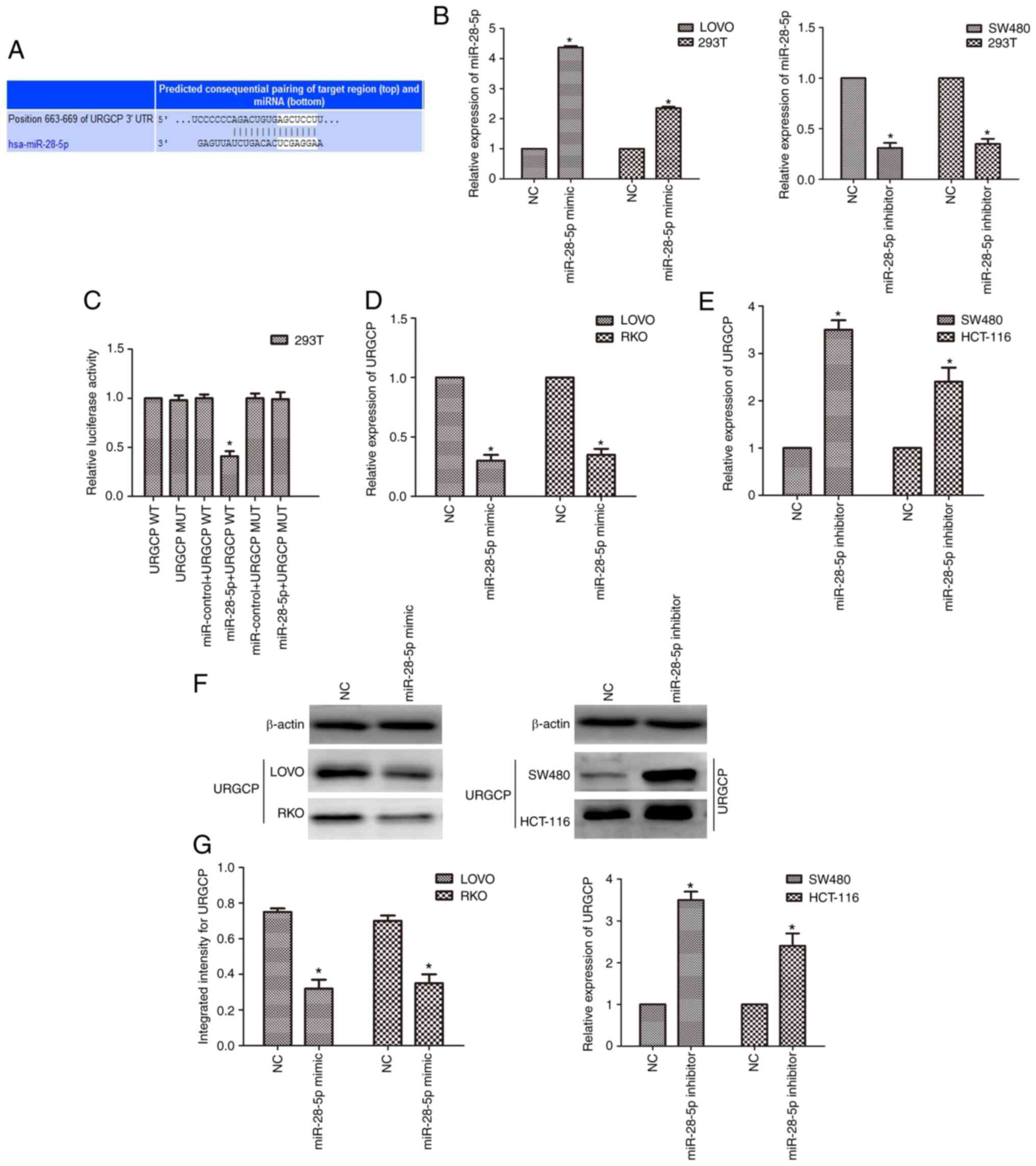
URGCP is a target of miR-28-5p
The potential targets of miR-28-5p were investigated
to reveal the mechanism underlying the effects of miR-28-5p. It was
revealed by bioinformatics tools that URGCP is a target of
miR-28-5p (http://www.targetscan.org/vert_72/; Fig. 5A). After transfection with the
miR-28-5p mimic, the expression of miR-28-5p was upregulated, while
it was downregulated after transfection of the miR-28-5p inhibitor
in both CRC cells and 293T cells (n=6, P<0.05; Fig. 5B). To verify the direct binding
relationship between miR-28-5p and URGCP, a luciferase reporter
system was used. After co-transfection of miR-28-5p mimics and
URGCP-WT, the luciferase activity was significantly reduced, while
the luciferase activity was not altered when cells were transfected
with URGCP-MUT (n=6, P<0.05; Fig.
5C). Moreover, the qPCR and western blot results revealed that
the miR-28-5p mimic suppressed the expression of URGCP in LOVO and
RKO cells, and the miR-28-5p inhibitor upregulated the expression
of URGCP in SW480 and HCT-116 cells (n=6, P<0.05; Fig. 5D-G).
CDKN2B-AS1 regulates URGCP expression
via miR-28-5p and regulates the proliferation of CRC cells
After transfection with CDKN2B-AS1-siRNAs with or
without a miR-28-5p inhibitor in CRC cells, the qPCR results
revealed that knockdown of CDKN2B-AS1 reduced the expression of
URGCP, whereas the miR-28-5p inhibitor partly abolished this
decreasing effect (n=6, P<0.05; Fig.
6A). Western blot assays revealed that the downregulation of
URGCP protein expression in CDKN2B-AS1-knockdown cells was rescued
by miR-28-5p inhibitor transfection (n=6, P<0.05; Fig. 6B and C). To further verify whether the
CDKN2B-AS1-mediated inhibition of proliferation was dependent on
the activation of the miR-28-5p targeting URGCP, western blot
analysis was used. The results demonstrated that the expression of
cyclin D1 and CDK4, which was decreased in CDKN2B-AS1-knockdown
cells, was partly rescued by miR-28-5p inhibitor transfection (n=6,
P<0.05; Fig. 6D and E). Moreover,
it is illustrated that p-Rb/Rb expression levels are increased
after CDKN2B-AS1 knockdown and decreased after miR-28-5p inhibitor
transfection. The increases in the levels of the cleaved fragments
of caspase-9 and caspase-3 in CDKN2B-AS1 knockdown cells were
partly attenuated by transfection with the miR-28-5p inhibitor
(n=6, P<0.05; Fig. 6D and E).
These data further demonstrated that the proliferation induced by
CDKN2B-AS1 depletion in CRC cells was partly mediated by the
miR-28-5p targeting URGCP.
Discussion
Accumulating evidence has indicated that lncRNAs can
be used as biological markers and therapeutic targets of cancer and
play a variety of roles in biological processes related to cancer
(14). Recent studies have reported
that CDKN2B-AS1 is involved in the occurrence and development of a
variety of tumors (15–18). CDKN2B-AS1 has been revealed to
regulate the malignancy of renal clear cell carcinoma by mediating
NUF2 transcription (16). A recent
study also revealed that CDKN2B-AS1-CDKN2B is involved in the
molecular genetics of open-angle glaucoma pathogenesis (17). In a study conducted by Akbari et
al CDKN2B-AS1 was revealed to be related to intrinsic apoptotic
genes in CRC (18). However, the
detailed functions of CDKN2B-AS1 and the mechanisms through which
it exerts its biological functions are not well understood. In the
present study, it was revealed that CDKN2B-AS1 was upregulated in
CRC tissue vs. normal tissue. In addition, the expression of
CDKN2B-AS1 in CRC cell lines was detected and it was revealed that
CDKN2B-AS1 was overexpressed in CRC cells vs. normal cells. These
results indicated that CDKN2B-AS1 could be a useful diagnostic
biomarker or therapeutic target in CRC.
The role of CDKN2B-AS1 in CRC was also examined by
assessing the biological behavior of CRC cell lines with altered
expression of this target. It was revealed by CCK-8 and colony
formation assays that the downregulation of CDKN2B-AS1
significantly inhibited the proliferation of CRC cells. Previous
studies have confirmed that cyclin D1 promotes the G1
phase transition of cells by activating CDK4, resulting in
increased Rb phosphorylation (p-Rb) (19,20).
Assessment of the mechanisms by which CDKN2B-AS1 knockdown affected
proliferation revealed that the expression of cell cycle-related
proteins (cyclin D1 and CDK4) was reduced in knockdown cell lines.
Moreover, it is illustrated that p-Rb/Rb expression levels are
increased after CDKN2B-AS1 knockdown. Emerging evidence has
indicated that the upregulation of intrinsic apoptotic signals
leads to cell death by recruiting and further activating the
promoter caspase-9 and effector caspases (caspase-3/6/9) (21). In the present study, it was revealed
that CDKN2B-AS1 knockdown induced the apoptosis of CRC cells by
activating apoptosis-related proteins (caspase-9 and
caspase-3).
In recent years, increasing evidence has
demonstrated the hypothesis that lncRNAs exert their biological
effect by acting as ceRNAs to affect the development of cancers
(22). The cytoplasmic localization
of CDKN2B-AS1 was further demonstrated by quantitative methods
using subcellular fractionation experiments, which complemented the
hybridization results. The role of CDKN2B-AS1 and miRNA in CRC was
determined by bioinformatics analysis and dual-luciferase analysis.
It was identified that miR-28-5p directly targeted CDKN2B-AS1. The
localization of miR-28-5p in the cytoplasm and its interaction with
lncRNAs have been studied in numerous cancers including CRC
(23,24). Moreover, the results revealed that the
expression of miR-28-5p was negatively correlated with the
expression of CDKN2B-AS1 in CRC tissues and cell lines. The results
indicated that CDKN2B-AS1 is a sponge of CDKN2B-AS1. Recent studies
have revealed that miR-28-5p acts as a tumor suppressor in multiple
human cancers, including CRC (25,26). In
functional experiments, CCK-8 and colony formation assays revealed
that miR-28-5p could partly reverse the effect of CDKN2B-AS1 on CRC
cells. Therefore, the effects of CDKN2B-AS1 on CRC cell
proliferation can be explained in part by its role as a molecular
sponge of miR-28-5p.
In the present study, the mechanism of miR-28-5p was
determined by bioinformatics analysis and dual-luciferase analysis.
It was identified that miR-28-5p directly targeted URGCP. URGCP,
also known as upregulated gene 4 (URG4), exhibits increased
expression in osteosarcoma (OS) tissues vs. normal tissues
(27). miR-671-5p has been revealed
to inhibit gastric cancer cell proliferation and promote cell
apoptosis by targeting URGCP (28).
URG4/URGCP has also been revealed to enhance the angiogenic
capacity of human hepatocellular carcinoma cells in vitro
via activation of the NF-κB signaling pathway (29). In the present study, it was revealed
that URGCP was upregulated in CRC. It was next investigated whether
URGCP plays a role in mediating CDKN2B-AS1-induced cellular
proliferation and apoptosis inhibition in CRC cells. It was
revealed that CDKN2B-AS1 sponges miR-28-5p to regulate
proliferation and apoptosis via URGCP. To illustrate the mechanism,
the expression of proliferation-related proteins and
apoptosis-related proteins was also assessed. Western blot analysis
demonstrated that the decrease in cycle-related proteins caused by
CDKN2B-AS1 knockdown was partly blocked by the miR-28-5p inhibitor.
Additionally, our experiments revealed that the upregulation of
apoptosis-related proteins caused by CDKN2B-AS1 knockdown was
partly suppressed after transfection with the miR-28-5p inhibitor.
These data demonstrated that CDKN2B-AS1 depletion-mediated
proliferation inhibition and apoptosis induction was partly
mediated by URGCP in CRC cells.
While it would be helpful to determine the
association of this gene with prognosis in CRC patients, samples
collected more than five years after surgery are required and the
average period of follow-up is 6 months. The samples collected for
the present study were obtained not more than one year after
surgery, and these data will be provided in future research.
Numerous previous studies on the ceRNA mechanism have used dual
luciferase quantitative methods and interaction assays instead of
RNA immunoprecipitation (RIP) studies (30,31).
Therefore, in the present study, dual luciferase quantitative
methods and interaction assays were also used. In our future
studies, RIP assays will be performed to further study the ceRNA
mechanism. In a number of studies, colocalization experiments of
miRNAs and lncRNAs were not performed (32,33), and
our study also lacks colocalization experiments of miR-28-5p and
lncRNA CDKN2B-AS1. These data will be provided in future
research.
In summary, it was revealed that the upregulated
lncRNA CDKN2B-AS1 promoted cell proliferation and inhibited cell
apoptosis in CRC. Mechanistically, CDKN2B-AS1 functioned as a
miR-28-5p sponge to positively regulate URGCP in CRC. The present
study revealed the precise role of this regulatory axis and
indicated that the CDKN2B-AS1/miR-28-5p/URGCP axis may be a
therapeutic target in CRC.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All the datasets generated and analyzed in the
present study are included in this published article.
Authors' contributions
MLM and HYZ conceived the study design. HYZ and SYZ
designed the experiments and supervised all research. MLM, SYZ and
XLY carried out the experiments and prepared the draft of the
manuscript. XLY analyzed the data. All authors read and approved
the final manuscript and agree to be accountable for all aspects of
the research in ensuring that the accuracy or integrity of any part
of the work are appropriately investigated and resolved.
Ethics approval and consent to
participate
CRC specimens were obtained following the guidelines
approved by the Ethics Review Committee of Qingdao Municipal
Hospital (Qingdao, China) and written informed consent was obtained
from patients in all cases.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
CRC
|
colorectal cancer
|
|
lncRNA
|
long noncoding RNA
|
|
miRNAs or miRs
|
microRNAs
|
|
ceRNAs
|
competing endogenous RNAs
|
|
RT-qPCR
|
reverse transcription-quantitative
polymerase chain reaction
|
|
siRNA
|
small interfering RNA
|
|
URGCP
|
upregulator of cell proliferation
|
References
|
1
|
Nindra U, Shahnam A and Mahon KL: Review
of systemic chemotherapy in unresectable colorectal peritoneal
carcinomatosis. Asia-Pac J Clin Oncol. Feb 19–2021.(Epub ahead of
print). View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Grady WM and Carethers JM: Genomic and
epigenetic instability in colorectal cancer pathogenesis.
Gastroenterology. 135:1079–1099. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Oldenhuis CN, Oosting SF, Gietema JA and
de Vries EG: Prognostic versus predictive value of biomarkers in
oncology. Eur J Cancer. 44:946–953. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Huarte M, Guttman M, Feldser D, Garber M,
Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M,
et al: A large intergenic noncoding RNA induced by p53 mediates
global gene repression in the p53 response. Cell. 142:409–419.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tang Y, Tang R, Tang M, Huang P, Liao Z,
Zhou J, Zhou L, Su M, Chen P, Jiang J, et al: LncRNA DNAJC3-AS1
regulates fatty acid synthase via the EGFR pathway to promote the
progression of colorectal cancer. Front Oncol. 10:6045342020.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Nagano T and Fraser P: No-nonsense
functions for long noncoding RNAs. Cell. 145:178–181. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Dinger ME, Pang KC, Mercer TR and Mattick
JS: Differentiating protein-coding and noncoding RNA: Challenges
and ambiguities. PLoS Comput Bbiol. 4:e10001762008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Dasgupta P, Kulkarni P, Majid S, Hashimoto
Y, Shiina M, Shahryari V, Bhat NS, Tabatabai L, Yamamura S, Saini
S, et al: LncRNA CDKN2B-AS1/miR-141/cyclin D network regulates
tumor progression and metastasis of renal cell carcinoma. Cell
Death Dis. 11:6602020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Song C, Qi Y, Zhang J, Guo C and Yuan C:
CDKN2B-AS1: An indispensable long non-coding RNA in multiple
diseases. Curr Pharm Des. 26:5335–5346. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu F, Xiao Y, Ma L and Wang J: Regulating
of cell cycle progression by the lncRNA CDKN2B-AS1/miR-324-5p/ROCK1
axis in laryngeal squamous cell cancer. Int J Biol Markers.
35:47–56. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Dos Santos IL, Penna KGBD, Dos Santos
Carneiro MA, Libera LSD, Ramos JEP and Saddi VA: Tissue micro-RNAs
associated with colorectal cancer prognosis: A systematic review.
Mol Biol Rep. 48:1853–1867. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tay Y, Rinn J and Pandolfi PP: The
multilayered complexity of ceRNA crosstalk and competition. Nature.
505:344–352. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Andrisani O: Epigenetic mechanisms in
hepatitis B virus-associated hepatocellular carcinoma. Hepatoma
Res. 7:122021.PubMed/NCBI
|
|
15
|
Zukerman R, Harris A, Vercellin AV, Siesky
B, Pasquale LR and Ciulla TA: Molecular genetics of glaucoma:
Subtype and ethnicity considerations. Genes (Basel). 12:552020.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Luo Y, Tao H, Jin L, Xiang W and Guo W:
CDKN2B-AS1 exerts oncogenic role in osteosarcoma by
promoting cell proliferation and epithelial to mesenchymal
transition. Cancer Biother Radiopharm. 35:58–65. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Rathi S, Danford I, Gudiseva HV, Verkuil
L, Pistilli M, Vishwakarma S, Kaur I, Dave T, O'Brien JM and
Chavali VRM: Molecular genetics and functional analysis implicate
CDKN2BAS1-CDKN2B involvement in POAG pathogenesis. Cells.
9:19342020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Akbari F, Peymani M, Salehzadeh A and
Ghaedi K: Integrative in silico and in vitro transcriptomics
analysis revealed new lncRNAs related to intrinsic apoptotic genes
in colorectal cancer. Cancer Cell Int. 20:5462020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chiron D, Martin P, Di Liberto M, Huang X,
Ely S, Lannutti BJ, Leonard JP, Mason CE and Chen-Kiang S:
Induction of prolonged early G1 arrest by CDK4/CDK6 inhibition
reprograms lymphoma cells for durable PI3Kδ inhibition through
PIK3IP1. Cell Cycle. 12:1892–1900. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gogolin S, Ehemann V, Becker G, Brueckner
LM, Dreidax D, Bannert S, Nolte I, Savelyeva L, Bell E and
Westermann F: CDK4 inhibition restores G(1)-S arrest in
MYCN-amplified neuroblastoma cells in the context of
doxorubicin-induced DNA damage. Cell Cycle. 12:1091–1104. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Girouard J, Lafleur MJ, Parent S, Leblanc
V and Asselin E: Involvement of Akt isoforms in chemoresistance of
endometrial carcinoma cells. Gynecol Oncol. 128:335–343. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Smillie CL, Sirey T and Ponting CP:
Complexities of post-transcriptional regulation and the modeling of
ceRNA crosstalk. Crit Rev Biochem Mol Biol. 53:231–245. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tian L, Han F, Yang J, Ming X and Chen L:
Long noncoding RNA LINC01006 exhibits oncogenic properties in
cervical cancer by functioning as a molecular sponge for
microRNA285p and increasing PAK2 expression. Int J Mol Med.
47:462021. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wu W, He K, Guo Q, Chen J, Zhang M, Huang
K, Yang D, Wu L, Deng Y, Luo X, et al: SSRP1 promotes colorectal
cancer progression and is negatively regulated by miR-28-5p. J Cell
Mol Med. 23:3118–3129. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Fazio S, Berti G, Russo F, Evangelista M,
D'Aurizio R, Mercatanti A, Pellegrini M and Rizzo M: The miR-28-5p
targetome discovery identified SREBF2 as one of the mediators of
the miR-28-5p tumor suppressor activity in prostate cancer cells.
Cells. 9:3542020. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Proença MA, Biselli JM, Succi M, Severino
FE, Berardinelli GN, Caetano A, Reis RM, Hughes DJ and Silva AE:
Relationship between Fusobacterium nucleatum, inflammatory
mediators and microRNAs in colorectal carcinogenesis. World J
Gastroenterol. 24:5351–5365. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Huang J, Zhu B, Lu L, Lian Z, Wang Y, Yang
X, Satiroglu-Tufan NL, Liu J and Luo Z: The expression of novel
gene URG4 in osteosarcoma: Correlation with patients' prognosis.
Pathology. 41:149–154. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Qiu T, Wang K, Li X and Jin J: miR-671-5p
inhibits gastric cancer cell proliferation and promotes cell
apoptosis by targeting URGCP. Exp Ther Med. 16:4753–4758.
2018.PubMed/NCBI
|
|
29
|
Xing S, Zhang B, Hua R, Tai WC, Zeng Z,
Xie B, Huang C, Xue J, Xiong S, Yang J, et al: URG4/URGCP enhances
the angiogenic capacity of human hepatocellular carcinoma cells in
vitro via activation of the NF-κB signaling pathway. BMC Cancer.
15:3682015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Shan TD, Tian ZB, Li Q, Jiang YP, Liu FG,
Sun XG, Han Y, Sun LJ and Chen L: Long intergenic noncoding RNA
00908 promotes proliferation and inhibits apoptosis of colorectal
cancer cells by regulating KLF5 expression. J Cell Physiol.
236:889–899. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liu HZ, Shan TD, Han Y and Liu XS:
Silencing long non-coding RNA CASC9 inhibits colorectal cancer cell
proliferation by acting as a competing endogenous RNA of miR-576-5p
to regulate AKT3. Cell Death Discov. 6:1152020. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Piao HY, Guo S, Jin H, Wang Y and Zhang J:
LINC00184 involved in the regulatory network of ANGPT2 via ceRNA
mediated miR-145 inhibition in gastric cancer. J Cancer.
12:2336–2350. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lee WJ, Shin CH, Ji H, Jeong SD, Park MS,
Won HH, Pandey PR, Tsitsipatis D, Gorospe M and Kim HH:
hnRNPK-regulated LINC00263 promotes malignant phenotypes through
miR-147a/CAPN2. Cell Death Dis. 12:2902021. View Article : Google Scholar : PubMed/NCBI
|