Introduction
There are three forms of autophagy, including
chaperone-mediated autophagy (CMA), which is a process of
recognizing proteins with the KFERQ sequence through HSC70 and
sending them to the surface of lysosome membrane to be combined
with lysosome-associated membrane protein 2A (LAMP-2A), a specific
CMA protein receptor, to make the protein unfold and then enter the
lumen of the lysosome for lysosomal degradation (1,2). As a
rate-limiting protein of CMA, LAMP-2A is widely used as a target to
block CMA activity (3,4). Kon et al determined that the
activity of CMA in a variety of cancer cells increased, and LAMP-2A
expression was upregulated, demonstrating that CMA is a necessary
condition for malignant cell growth and tumor metastasis (5). Through the study of 593 gastric
non-cancerous foci and 173 gastric cancer tissues, LAMP-2A was
revealed to be a potential biomarker for early prediction and
prognosis of gastric cancer (6).
Tumor cells rely mainly on aerobic glycolysis to
produce ATP, which in the case of adequate oxygen supply leads to
increased glucose uptake and lactic acid production compared with
normal cells, a phenomenon known as the Warburg effect (7). Activation of oncogenes or deletion of
anti-oncogenes increase glucose uptake and lactate production.
During oncogenesis, the expression of pyruvate kinase continues to
change, which is a key glycolytic enzyme (8,9) that
catalyzes the transfer of phosphate from phosphoenolpyruvate (PEP)
to ADP, leading to the formation of pyruvate and ATP. Cancer cells
commonly express the M2 isoform of embryonic pyruvate kinase
(PKM2), which may contribute to the transition of metabolism from
oxidative phosphorylation to aerobic glycolysis and tumor formation
(10,11). Previous research has demonstrated that
PKM2 could be a potential target for tumor regulation (12). A previous study revealed that the
acetylation of PKM2 K305 promoted its lysosomal-dependent
degradation through CMA, increased the interaction between PKM2 and
CMA chaperone HSC70, and promoted cell proliferation and tumor
growth (13).
However, the specific mechanism of action of CMA and
PKM2 in RCC has yet to be reported. Based on this, the present
study investigated the effect of CMA and PKM2 on the development of
renal cancer (RC) and its possible mechanism of action.
Materials and methods
Tissues and cell lines
The tumor tissues and paired para-cancer (PC) tissue
samples (margins of tumor tissues 3 cm) were collected from 12
patients who underwent surgery for RC at the Second Affiliated
Hospital of Xi'an Jiaotong University (Xi'an, China) from January
to December 2019. The inclusion criteria was as follows: Patients
who were diagnosed with renal cancer by abdominal CT and
postoperative pathological examination; in line with the
indications of surgical treatment; complete clinical data; patients
and their family members were informed of this study and signed
informed consent. The exclusion criteria was as follows:
hypertension, diabetes mellitus, kidney disease, adrenal disease
and hepatic and renal insufficiency; people with coagulation
dysfunction; confused patients; poor compliance; women who were
lactating or pregnant. The study included 9 male and 3 female
patients, with a mean age of 54.7 years (range from 42 to 71
years). All of the included patients were informed of the study and
signed an informed consent. Our study was approved by the Ethics
Committee of the Second Affiliated Hospital of Xi'an Jiaotong
University, Xi'an, China.
Human normal renal cells HK-2 and human RCC cell
lines A498, GRC-1, 786-O and ACHN were purchased from the Cell Bank
of the Shanghai Institute of Biochemistry and Cell Biology, Chinese
Academy of Sciences. These cells were cultured in RPMI-1640 medium
(Thermo Fisher Scientific, Inc.) supplemented with 10% fetal bovine
serum (FBS; Gibco; Thermo Fisher Scientific, Inc.) in an incubator
containing 5% CO2 at 37°C.
Cell transfection
The cDNA of LAMP-2A and PKM2 were amplified using
RNA extracted from 293T cells (Cell Bank of the Shanghai Institute
of Biochemistry and Cell Biology, Chinese Academy of Sciences).
After the cells were cultured at 37°C to the logarithmic growth
phase, the cells were inoculated into a 12-well plate, and the cell
density was adjusted to 3×105 cells/well. A minimum of
two independent shRNAs for LAMP-2A or PKM2 were designed,
synthesized and packaged by Guangzhou RiboBio Co., Ltd. shRNAs were
cloned into a pLKO.1 puro vector (Addgene, Inc.) according to the
manufacturer's protocol. The shRNA sequences used are presented in
Table I. The second generation of 80%
confluent cells were transfected with lentiviral particles
(MOI=16), with 15 µg pLKO.1-shRNA-LAMP-2A/PKM2 plasmid or a
scrambled sequence using Lipofectamine 3000 (Invitrogen; Thermo
Fisher Scientific, Inc.) for 10 min at 20°C. A total of 72 h after
transfection, the cells were harvested. The cells were divided into
four groups and transfected with empty control vector (Control),
sh-LAMP-2A vector [LAMP-2A(−)], sh-PKM2 vector [PKM2(−)], and
sh-LAMP-2A + sh-PKM2 vectors [LAMP-2A(−) PKM2(−)].
 | Table I.Sequences of shRNA used in the
experiments. |
Table I.
Sequences of shRNA used in the
experiments.
| Gene | Sequence |
|---|
| LAMP-2A |
5′-CACCGCACCATCATGCTGGATATGACGAATCATATCCAGCATGATGGTGC-3′ |
|
|
3′-CGTGGTAGTACGACCTATACTGCTTAGTATAGGTCGTACTACCACGAAAA-5′ |
| PKM2 |
5′-CCGGCGGGTGAACTTTGCCATGAATTTCAAGAGAATTCATGGCAAAGTTCACCCGTTTTTGGTACC-3′ |
| Scrambled |
3′-GCCCACTTGAAACGGTACTTAAAGTTCTCTTAAGTACCGTTTCACGTGGGCAAAAAACCATGGTTAA-5′ |
Reverse transcription-quantitative
(RT-q) PCR
RT-qPCR was performed using the SYBR-Green PCR
Master Mix according to the manufacturer's instructions with an ABI
7300HT RT-PCR system (Applied Biosystems; Thermo Fisher Scientific,
Inc.) to determine the expression level of related mRNA. After
extracting the total RNA from cells or tissue with the TRizol
reagent (Invitrogen; Thermo Fisher Scientific, Inc.), the cDNA was
synthesized by reverse transcription experiment using a reverse
transcription synthesis kit (PrimeScript™ RT reagent kit; Takara
Bio, Inc.) with Oligo dT primer, Prime Script Buffer and dNTP
according to the manufacturer's instructions. GAPDH was used as an
internal reference. The primer sequences used are presented in
Table II. The thermocycling
conditions were as follows: 95°C for 8 min, 1 cycle; 95°C for 25
sec, 64°C for 20 sec; 72°C for 20 sec, 10 cycles; 93°C for 25 sec,
60°C for 35 sec, and 72°C for 20 sec, 35 cycles. The relative
expression level of related genes was calculated using the
2−ΔΔCq method (14).
ΔΔCq=[Cq target gene (sample)-Cq GAPDH (sample)]-[Cq target gene
(calibration sample)-Cq GAPDH (calibration sample)].
 | Table II.Primer sequences used in the reverse
transcription- quantitative PCR. |
Table II.
Primer sequences used in the reverse
transcription- quantitative PCR.
| Gene | Sequence (5′-3′) |
|---|
| LAMP-2A | F:
ACAGCTCAAGACTGCAGTGC |
|
| R:
ATGATGGTGCTTGAGACCAAT |
| PKM2 | F:
CCTTGCAATTATTTGAGGAACTCCGC |
|
| R:
CACGGTACAGGTGGGCCTGAC |
| PCNA | F:
GACTCGTCTCATGTCTCTTTGGTG |
|
| R:
GTATTTTGGACATGCTGGTGAGG |
| VEGF | F:
GAAGTTCATGGATGTCTATCAGCG |
|
| R:
ACTCCGTCAGAACTATCAAAGCTGC |
| HIF-1 | F:
TAAAGGAATTTCAATATTTGATGGG |
|
| R:
AAAGGGTAAAGAACAAAACACACAG |
| p21 | F:
GCCACTGTGATGCGCTAATG |
|
| R:
AGAAGATCAGCCGGCTAATG |
| Bcl-2 | F:
GTGGAGGAGCTCTTCAGGGA |
|
| R:
AGGCACCCAGGGTGATGCAA |
| Bax | F:
GGCC-CACCAGCTCTGAGCAGA |
|
| R:
GCCACGTGGGGGTCCCAAAGT |
| Cleaved | F:
CATGGAAGCGAATCAATGGACT |
| caspase-3 | R:
CTGTACCAGACCGAGATGTCA |
| GAPDH | F:
AAGGAGGCGGAGAAGAGGAC |
|
| R:
CGTCGTTACGAGTCACTTCAGG |
Western blot analysis
Cells or tissues were collected and lysed with RIPA
cleavage buffer (Thermo Fisher Scientific, Inc.). The protein
concentration was quantified with BCA reagent (Thermo Fisher
Scientific, Inc.). Then, 250 µg proteins were separated by
electrophoresis in 10% SDS-PAGE and transferred to nitrocellulose
membranes (EMD Millipore). The membranes were blocked in 5% BSA at
37°C for 2 h and then incubated with a primary antibody at 4°C
overnight. After washing with TBST (1% Tween-20), the membranes
were incubated with a secondary antibody (goat anti-rabbit IgG HRP;
1:3,000 dilution; product code ab205718; Abcam) for 2 h at room
temperature. The membranes were visualized with an ECL kit (Thermo
Fisher Scientific, Inc.), and finally the band density on the
membrane was scanned and analyzed using an image analyzer (model
no. MSD910) with Biomaster analysis software (both from Beijing
Maisiqi High Technology Co., Ltd.).
The following primary antibodies were used:
anti-LAMP-2A (1:1,000 dilution; product code ab125068; Abcam),
anti-PKM2 (1:1,000 dilution; product no. 4053; Cell Signaling
Technology, Inc.), anti-proliferating cell nuclear antigen (PCNA)
(1:1,000 dilution; product code ab92552), anti-hypoxia inducible
factor (HIF)-1 (1:1,000 dilution; product code ab179483), anti-p21
(1:1,000 dilution; product code ab109199), anti-vascular
endothelial growth factor (VEGF) (1:1,000 dilution; product code
ab150375), anti-cleaved caspase-3 (1:1,000 dilution; product code
ab32042), anti-Bcl-2 (1:1,000 dilution; product code ab32124),
anti-Bax (1:1,000 dilution; product code ab182733) and anti-GAPDH
(1:2,000 dilution; product code ab8245; all from Abcam).
Cell proliferation assay
The proliferation level of A498 cells was detected
by CCK-8 kit (Beyotime Biotechnology), and the experiment was
carried out according to the manufacturer's instructions. Briefly,
the cells were inoculated in 96-well plates with an inoculation
density of 5×103 cells/well. The cells were cultured at
37°C for 48 h and 10 µl of CCK-8 reagent was added to each well.
After incubation for 1 h, the absorbance at 450 nm was measured
with a microplate reader. The results were representative of three
independent experiments.
Cell cycle and apoptosis analysis
After transfection, cells (1×105 per
well) were collected, prepared into single-cell suspension, washed
twice with PBS, and fixed with 70% ethanol at 4°C overnight.
According to the manufacturer's instructions, 1 µl propidium iodide
(PI; Thermo Fisher Scientific, Inc.) was added and placed in the
dark at 37°C for 30 min. The cell cycle of each group was detected
by flow cytometry (FACSCanto II; BD Biosciences) with FlowJo VX 10
software (FlowJo, LLC), and the proportion of cells in the G1, S
and G2 phases was counted.
Cells were collected by the same method without
ethanol fixation. A total of 2.5 µl Annexin V-fluorescein
isothiocyanate (FITC)/propidium iodide (PI) were added into the
cell suspension according to the manufacturer's instructions, and
placed in the dark at 37°C for 15 min. Cell apoptosis was detected
by flow cytometry in each group, and early apoptotic cells, late
apoptotic cells and dead cells were counted.
Transwell migration assay
Matrigel was diluted with 0.5% FBS in DMEM (Gibco;
Thermo Fisher Scientific, Inc.) medium at 1:5 and seeded on the
upper surface of the bottom membrane of a 24-mm Transwell chamber
(8-µm pore size; Corning, Inc.) for 16 h at 37°C. Then the cells
were incubated for 4 h at 37°C in an incubator with 5%
CO2. Cells were further cultured in 5% FBS medium for 24
h at 37°C. After cell counting, the cells were seeded on the upper
chamber at 3×104 cells/well. Transwell chambers were
fixed with 4% methanol at room temperature for 30 min and stained
with 0.1% crystal violet for 20 min at room temperature. A total of
700 µl of culture medium containing 20% calf serum (Gibco; Thermo
Fisher Scientific, Inc.) was added in the lower chamber, and the
cells were cultured in an incubator at 37°C in an atmosphere
containing 5% CO2 for 72 h. Five fields were randomly
selected under a low-power microscope (magnification ×10; Olympus
Corporation) for each group to compare the differences in the
number of transmembrane cells between the groups.
Immunofluorescence
Cells were fixed with 4% methanol at room
temperature for 30 min and permeabilized with 0.1% Triton X-100 in
PBS for 20 min. The cells were incubated with anti-PKM2 (1:1,000
dilution; product no. 4053; Cell Signaling Technology, Inc.) and
anti-HSC70 (1:1,000 dilution; product code ab223356; Abcam) primary
antibodies overnight at 4°C, and then incubated in
fluorochrome-conjugated secondary antibodies goat anti-rabbit IgG
(cat. no. A-31556) and donkey anti-mouse IgG (cat. no. Q22082; both
1:2,000 dilution; both Thermo Fisher Scientific, Inc.) for 2 h at
room temperature. DAPI was used to counterstain the cell nuclei at
37°C for 30 min. Images were captured with a laser confocal
microscope (Olympus Corporation) in five different fields for each
sample.
Co-immunoprecipitation analysis
Approximately 5×106 cells were collected
48 h after transfection and lysed in 0.5 ml RIPA buffer (Beyotime
Biotechnology) and centrifuged at 1,000 × g at 4°C for 10 min.
Total cell proteins were extracted as Input controls. The
precleared lysates (500 µg) were immunoprecipitated against the
epitopes with 20 µl of protein A/G-agarose beads (Santa Cruz
Biotechnology, Inc.) and incubated with anti-PKM2 antibody (1:1,000
dilution; product no. 4053; Cell Signaling Technology, Inc.) at 4°C
overnight. The resultant mixtures were centrifuged at 250 × g at
4°C for 5 min and the supernatants were removed. The pellets were
washed three times with PBS and western blotting was performed
using an anti-HSC70 antibody (1:1,000 dilution; product code
ab223356; Abcam) according to the manufacturer's instructions.
Statistical analysis
Each sample was assessed 3 times and all experiments
were performed in triplicate. All data were statistically analyzed
and plotted using GraphPad software 8 (GraphPad Software, Inc.).
The data were expressed as the mean ± standard deviation (SD). The
comparisons between two groups were performed by unpaired Student's
t-test or Mann-Whitney U test. Comparisons among more than two
groups were performed by ANOVA with Fisher's Least Significant
Difference (LSD). P<0.05 was considered to indicate a
statistically significant difference.
Results
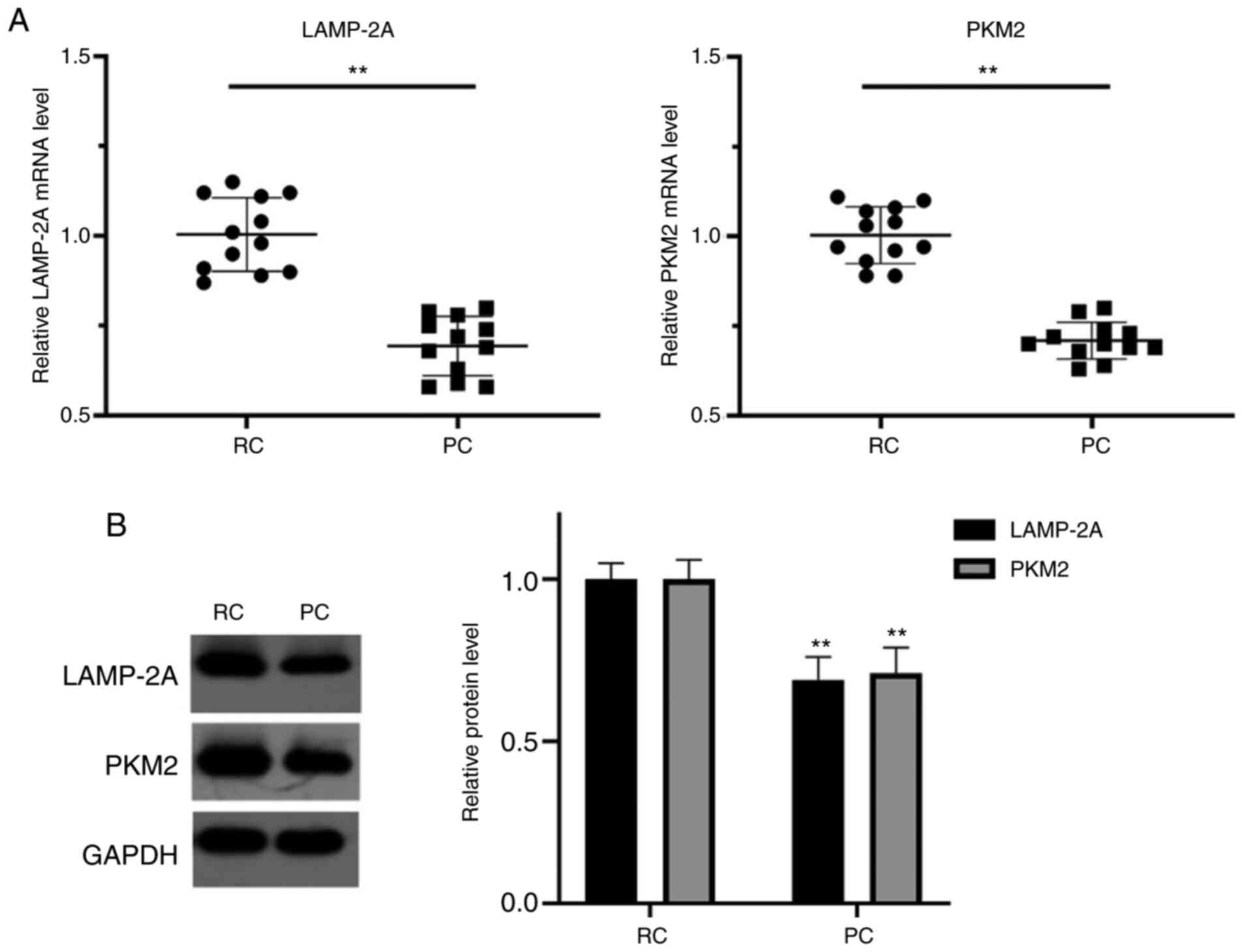
LAMP-2A and PKM2 expression levels are
significantly increased in RCC tissues
In the collected tissues of patients that underwent
RC surgery at our hospital, the mRNA levels of LAMP-2A and PKM2
were significantly higher in the RC tissues compared with their
paired PC tissues (P<0.01), respectively. In addition, the
protein levels of LAMP-2A and PKM2 in RC tissues were significantly
higher than those in their paired PC tissues (P<0.01),
respectively (Fig. 1).
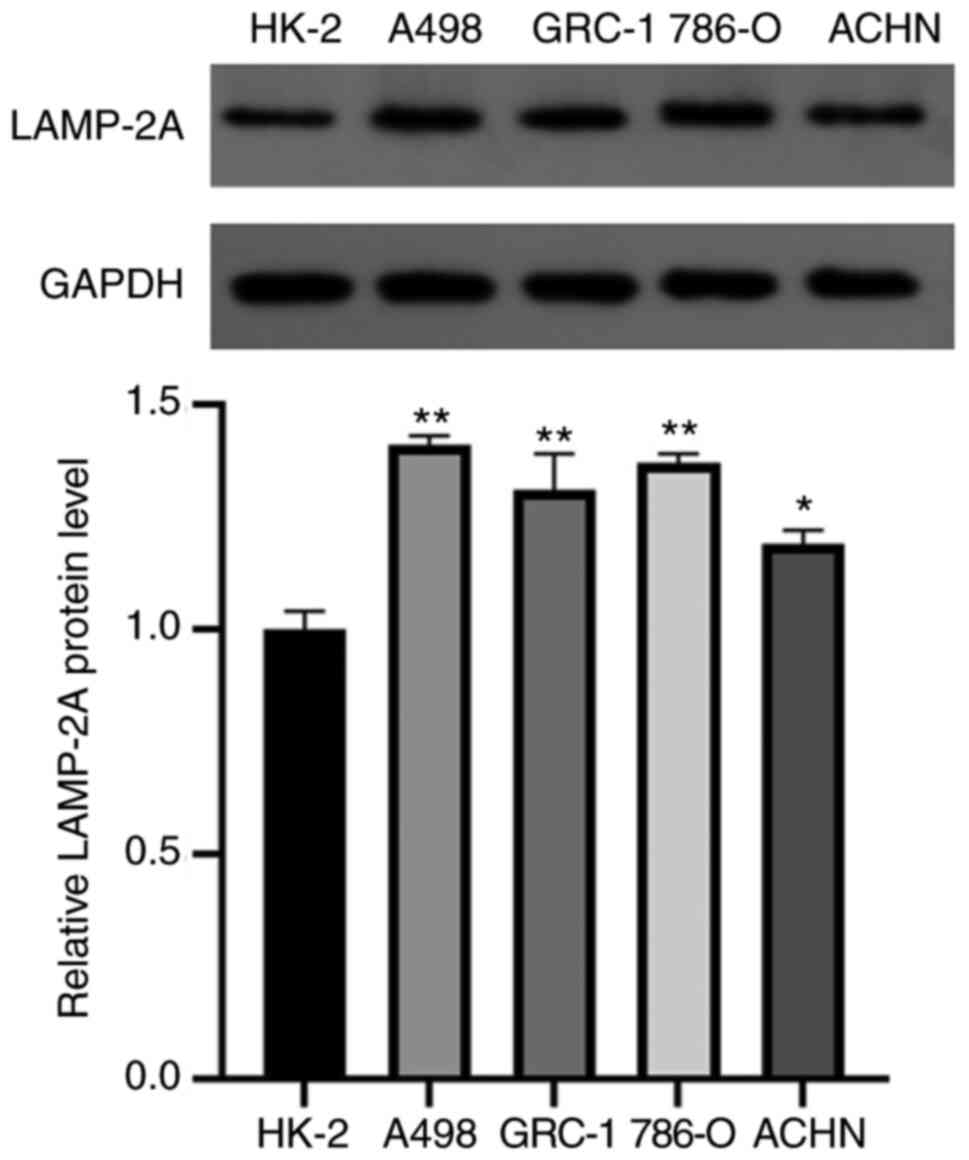
Level of LAMP-2A is significantly
increased in RCC cell lines
The protein levels of LAMP-2A were assessed in human
normal renal cells HK-2 and four RC cell lines: A498, GRC-1, 786-O
and ACHN. LAMP-2A expression levels in different cells are revealed
in Fig. 2. LAMP-2A expression levels
were significantly increased in all four types of RC cells compared
with normal renal cells (P<0.05 or P<0.01). Among them,
LAMP-2A had the highest relative expression level in A498 cells,
and was therefore selected for subsequent experiments.
LAMP-2A silencing increases PKM2
expression level in A498 cells
A498 cells were transfected with shRNAs to knock
down the expression levels of PKM2 and LAMP-2A, and the mRNA levels
revealed that shRNAs effectively knocked down the expression levels
of both genes in A498 cells compared with the scrambled sequences
(Fig. 3A). The mRNA and protein
levels of LAMP-2A and PKM2 in each group were assessed, and the
results are demonstrated in Fig. 3B and
C.
LAMP-2A mRNA and protein levels in the LAMP-2A(−)
group and LAMP-2A(−) PKM2(−) group were significantly decreased
(P<0.01) compared with the Control group, and there was no
significant difference between the PKM2(−) group and the Control
group (P>0.05).
The mRNA and protein levels of PKM2 in the PKM2(−)
group and LAMP-2A(−) PKM2(−) group were significantly decreased
(P<0.01) compared with the Control group. It is worth noting
that the mRNA and protein levels of PKM2 were increased in the
LAMP-2A(−) group compared with the Control group and the LAMP-2A(−)
PKM2(−) group compared with the PKM2(−) group (P<0.05 or
P<0.01), indicating that LAMP-2A silencing could reduce the
degradation of PKM2.
LAMP-2A and PKM2 silencing suppresses
the proliferation and invasion and induces the apoptosis of RCC
cells
The proliferation, cell cycle, apoptosis, and
invasion of four groups of cells were examined as revealed in
Fig. 4. Both LAMP-2A and PKM2
silencing significantly reduced the proliferation rate of A498 and
786-O cells (P<0.01), and the inhibition was greater with their
combination. Furthermore, both LAMP-2A and PKM2 silencing
significantly promoted the apoptosis of A498 and 786-O cells
(P<0.01) and blocked the cell cycle at G2/M phase. In addition,
both LAMP-2A and PKM2 silencing significantly inhibited the
invasive ability of A498 and 786-O cells (P<0.01), and their
combined inhibitory effect was stronger (P<0.05).
LAMP-2A and PKM2 silencing affects the
levels of related genes
The levels of mRNA and protein of genes that may be
related to CMA and PKM2 were detected (Fig. 5). Compared with the Control group, the
mRNA and protein levels of PCNA, VEGF, HIF-1 and the ratio of Bcl-2
to Bax were significantly decreased in the other three groups
(P<0.05 or P<0.01), and the levels of the LAMP-2A(−) PKM2(−)
group were significantly lower than that of the PKM2(−) group
(P<0.05). The mRNA and protein expression levels of cleaved
caspase-3 were significantly higher in the other three groups
compared with the Control group (P<0.01). Compared with the
Control group, the mRNA and protein expression level of p21 was
significantly increased in the other three groups, and the level of
the LAMP-2A(−) PKM2(−) group was significantly higher than that of
the PKM2(−) group (P<0.05).
Co-immunoprecipitation reveals the
interaction between PKM2 and HSC70
The co-immunoprecipitation of PKM2 and HSC70 in each
group was also detected, and the results are revealed in Fig. 6B. Immunofluorescence results (Fig. 6A) revealed that PKM2 and HSC70 could
co-locate in A498 cells. The interaction between PKM2 and HSC70 was
significantly higher in LAMP-2A(−) group than that in PKM2(−) and
LAMP-2A(−) PKM2(−) group, with no significant difference from the
Control group.
Discussion
CMA has been revealed to be associated with cancer
progression in recent years (15),
but its mechanisms have not been fully explored. PKM2 has been
reported to be a substrate for CMA in non-small cell lung cancer
(13). Knockout of LAMP-2A gene in
lung cancer cells indicated that CMA was necessary for
proliferation of lung cancer cells (5). In the present study, the mechanism of
CMA and PKM2 in the proliferation regulation of RCC was mainly
investigated.
In our findings, LAMP-2A and PKM2 expression levels
were significantly increased in RCC tissues and cell lines. CMA is
considered to play a critical role in a variety of tumors, and PKM2
is also recognized as a regulatory tumor target (16). The results of the present study
confirmed that both of them played an equally vital role in the
development of RCC.
The acetylation of PKM2 and CMA can promote the
degradation of PKM2 (17). The
present study revealed that the deletion of LAMP-2A could increase
the expression level of PKM2 and reduced the degradation of PKM2 in
A498 cells, which is consistent with previous results (17).
Both LAMP-2A and PKM2 silencing significantly
reduced the proliferation rate of A498 cells, and this inhibition
was greater with their combination. Correspondingly, the expression
levels of PCNA, VEGF and HIF-1 were also significantly decreased.
The downregulation of PKM2 has been reported to significantly
reduce the levels of PCNA, cyclin D1 and p27 in primary astrocytes
(18). PKM2 knockdown can lead to
impaired cell proliferation and enhanced apoptosis in vitro
and inhibit tumor growth and decrease angiogenesis in vivo.
In addition, PKM2 deficiency has been revealed to negatively affect
the proliferation chain accumulation and promoter activity of
hypoxia-induced HIF-1α, and lead to decreased VEGF secretion
(19). The expression and function of
PKM2 were closely related to HIF-1 in the feed-forward loop, and
HIF-1 was identified as a target gene of the PKM2/STAT3 pathway,
and the expression of PKM2 was in turn regulated by the HIF-1
transcription complex (20).
Experimental results revealed that inhibition of CMA inhibited the
proliferation of colon carcinoma CT26 cells (21). Both CMA blocking and PKM2 knockdown
inhibited cell proliferation, and similar results were obtained in
the present study with regard to RCC.
Silencing of LAMP-2A and PKM2 promoted apoptosis of
A498 cells and blocked the cells in the G2/M phase. Concurrently,
the levels of p21 and cleaved caspase-3 were increased while the
ratio of Bcl-2/Bax was decreased. In fact, PKM2 has been reported
to translocate to the mitochondria, interact with Bcl-2 at
threonine 69 site and phosphorylate Bcl-2 to prevent Cul3-based E3
ligase from binding to Bcl-2 and subsequent Bcl-2 degradation, thus
directly inhibiting the apoptosis of glioma cells under oxidative
stress (22). PKM2 has been revealed
to accelerate malignant behavior of colorectal cancer cells,
increase oxaliplatin resistance and reduce cell apoptosis (23). A previous study indicated that
knockdown of PKM2 blocked the non-small lung cancer cells in the
G2/M phase, which was associated with the effect of the expression
of p21 (24). PKM2 knockdown
activated spindle assembly checkpoints and blocked cell processes
during metaphase to anaphase mitosis, leading to apoptosis.
CMA is a substrate protein autophagy substitution
pathway, mediated by HSC70 and LAMP-2A. HSC70 recognizes and
targets substrate proteins with the KFERQ-like motifs for transport
to lysosomal membranes. LAMP-2A assists substrate protein
translocation to lysosomes for degradation (25). Co-immunoprecipitation revealed the
interaction between PKM2 and HSC70. Acetylation of PKM2 increases
its interaction with CMA chaperone HSC70 and accelerates its
binding to lysosomes (13). In RCC,
CMA and PKM2 may also function in this way.
A limitation of the present study was that in
vivo experiments were not conducted to verify our conclusions.
Therefore, the specific interaction mechanism between CMA and PKM2
still requires further investigation.
In conclusion, the data of the present study
supported the hypothesis that CMA affected the proliferation and
apoptosis of RCC cells through PKM2, and blockade of CMA or
knockdown of PKM2 may be a new treatment strategy for RCC.
Acknowledgements
Not applicable.
Funding
The present study was supported by Xi'an Science and
Technology Planning Project (grant no. 201805096YX4SF30) and the
Traditional Chinese Medicine Scientific Research Project of Shaanxi
Province (grant no. JCSM038).
Availability of data and materials
The datasets generated and/or analyzed during the
present study are available from the corresponding author on
reasonable request.
Authors' contributions
SX and TC contributed to the conception and design
of the study. GX and ZW performed the experiments, collected and
analyzed data. SX and TC wrote the manuscript. All authors reviewed
and approved the final version of the manuscript.
Ethics approval and consent to
participate
The study protocol was approved by the Ethics
Committee of the Second Affiliated Hospital of Xi'an Jiaotong
University (Xi'an, China). Written informed consent was obtained
from all the study subjects before enrollment.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Cuervo AM: Chaperone-mediated autophagy:
Selectivity pays off. Trends Endocrinol Metab. 21:142–150. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Agarraberes FA, Terlecky SR and Dice JF:
An intralysosomal hsp70 is required for a selective pathway of
lysosomal protein degradation. J Cell Biol. 137:825–834. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Park C, Suh Y and Cuervo AM: Regulated
degradation of Chk1 by chaperone-mediated autophagy in response to
DNA damage. Nat Commun. 6:68232015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Saha T: LAMP2A overexpression in breast
tumors promotes cancer cell survival via chaperone-mediated
autophagy. Autophagy. 8:1643–1656. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kon M, Kiffin R, Koga H, Chapochnick J,
Macian F, Varticovski L and Cuervo AM: Chaperone-mediated autophagy
is required for tumor growth. Sci Transl Med. 3:109ra1172011.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhou J, Yang J, Fan X, Hu S, Zhou F, Dong
J, Zhang S, Shang Y, Jiang X, Guo H, et al: Chaperone-mediated
autophagy regulates proliferation by targeting RND3 in gastric
cancer. Autophagy. 12:515–528. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Warburg O: On the origin of cancer cells.
Science. 123:309–314. 1956. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Altenberg B and Greulich KO: Genes of
glycolysis are ubiquitously overexpressed in 24 cancer classes.
Genomics. 84:1014–1020. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Majumder PK, Febbo PG, Bikoff R, Berger R,
Xue Q, McMahon LM, Manola J, Brugarolas J, McDonnell TJ, Golub TR,
et al: mTOR inhibition reverses Akt-dependent prostate
intraepithelial neoplasia through regulation of apoptotic and
HIF-1-dependent pathways. Nat Med. 10:594–601. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mazurek S, Boschek CB, Hugo F and
Eigenbrodt E: Pyruvate kinase type M2 and its role in tumor growth
and spreading. Semin Cancer Biol. 15:300–308. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Dombrauckas JD, Santarsiero BD and Mesecar
AD: Structural basis for tumor pyruvate kinase M2 allosteric
regulation and catalysis. Biochemistry. 44:9417–9429. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li YH, Li XF, Liu JT, Wang H, Fan LL, Li J
and Sun GP: PKM2, a potential target for regulating cancer. Gene.
668:48–53. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lv L, Li D, Zhao D, Lin R, Chu Y, Zhang H,
Zha Z, Liu Y, Li Z, Xu Y, et al: Acetylation targets the M2 isoform
of pyruvate kinase for degradation through chaperone-mediated
autophagy and promotes tumor growth. Mol Cell. 42:719–730. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang S, Hu B, You Y, Yang Z, Liu L, Tang
H, Bao W, Guan Y and Shen X: Sorting nexin 10 acts as a tumor
suppressor in tumorigenesis and progression of colorectal cancer
through regulating chaperone mediated autophagy degradation of
p21Cip1/WAF1. Cancer Lett. 419:116–127. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Arias E and Cuervo AM: Pros and cons of
chaperone-mediated autophagy in cancer biology. Trends Endocrinol
Metab. 31:53–66. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang Z, Deng X, Liu Y, Liu Y, Sun L and
Chen F: PKM2, function and expression and regulation. Cell Biosci.
9:522019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang J, Feng G, Bao G, Xu G, Sun Y, Li W,
Wang L, Chen J, Jin H and Cui Z: Nuclear translocation of PKM2
modulates astrocyte proliferation via p27 and -catenin pathway
after spinal cord injury. Cell Cycle. 14:2609–2618. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Azoitei N, Becher A, Steinestel K, Rouhi
A, Diepold K, Genze F, Simmet T and Seufferlein T: PKM2 promotes
tumor angiogenesis by regulating HIF-1α through NF-κB activation.
Mol Cancer. 15:32016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
de Wit RH, Mujić-Delić A, van Senten JR,
Fraile-Ramos A, Siderius M and Smit MJ: Human cytomegalovirus
encoded chemokine receptor US28 activates the HIF-1α/PKM2 axis in
glioblastoma cells. Oncotarget. 7:67966–67985. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Peng JQ, Han SM, Chen ZH, Yang J, Pei YQ,
Bao C, Qiao L, Chen WQ and Liu B: Chaperone-mediated autophagy
regulates apoptosis and the proliferation of colon carcinoma cells.
Biochem Biophys Res Commun. 522:348–354. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liang J, Cao R, Wang X, Zhang Y, Wang P,
Gao H, Li C, Yang F, Zeng R, Wei P, et al: Mitochondrial PKM2
regulates oxidative stress-induced apoptosis by stabilizing Bcl2.
Cell Res. 27:329–351. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lu WQ, Hu YY, Lin XP and Fan W: Knockdown
of PKM2 and GLS1 expression can significantly reverse
oxaliplatin-resistance in colorectal cancer cells. Oncotarget.
8:44171–44185. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yuan S, Qiao T, Zhuang X, Chen W, Xing N
and Zhang Q: Knockdown of the M2 isoform of pyruvate kinase (PKM2)
with shRNA enhances the effect of docetaxel in human NSCLC cell
lines in vitro. Yonsei Med J. 57:1312–1323. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gorantla NV and Chinnathambi S: Autophagic
pathways to clear the tau aggregates in Alzheimer's disease. Cell
Mol Neurobiol. 41:1175–1181. 2021. View Article : Google Scholar : PubMed/NCBI
|