Introduction
Colorectal cancer (CRC) is the third most common
type of cancer and its mortality rate has been increasing worldwide
(1,2).
CRC is a multifactorial disease with a high risk of metastasis and
recurrence. Radical surgery is the most common treatment approach
for CRC. However, the majority of patients with CRC are unable to
undergo radical surgery, since they are already in an advanced
stage of the disease and have metastases at the time of diagnosis
(3). Therefore, identifying sensitive
diagnostic biomarkers for the early detection of CRC and novel
targets for CRC treatment are of great importance.
Circular RNAs (circRNAs/circ) are a novel class of
non-coding RNAs, characterized by covalently closed circular
structures, making them highly conserved and stable. circRNAs may
act as therapeutic targets and biomarkers for several diseases
(4,5).
Increasing evidence has indicated that circRNAs are closely
associated with the initiation and development of cancer (6–8).
Furthermore, recent studies have confirmed that circRNAs may serve
as pivotal targets for the diagnosis and treatment of CRC (9–11). It has
been reported that circRNA nuclear receptor-interacting protein 1
(NRIP1), a novel circRNA, is upregulated in several tumor types,
including gastric cancer (12),
stomach adenocarcinoma (13) and
esophageal squamous cell carcinoma (14). However, studies on the expression and
biological function of circNRIP1 in CRC are still lacking.
‘MicroRNA (miRNA/miR) sponging’, referring to the
interaction between circRNAs and miRNAs, is based on the competing
endogenous RNA (ceRNA) theory. circRNAs most commonly exert their
function via acting as ceRNAs to sequester miRNAs (15–17). Via
this mechanism of acting as miRNA sponges, circRNAs have been
confirmed to have roles in several types of cancer (18). For instance, circ-methionine
adenosyltransferase 2B was reported to promote glycolysis and
malignancy of hepatocellular carcinoma (HCC) via sponging
miR-338-3p under hypoxic stress conditions (19). In addition, circ-protein arginine
methyltransferase 5 promoted the metastasis of bladder urothelial
carcinoma via sponging miR-30c to induce epithelial-mesenchymal
transition (20). Of note, the
ability of circRNAs to act as ceRNAs to sequester miRNAs has also
been indicated to have an important role in CRC (21). Similarly, circNRIP1 was able to
promote tumor initiation and development via different miRNAs in
several types of cancer (12,14,22).
5-Fluorouracil (5-FU), a chemotherapeutic drug, is
commonly used in the treatment of CRC. It has been indicated that
the combination of 5-FU and cisplatin was able to improve clinical
efficacy (23). However, drug
resistance, caused by numerous factors, may hamper the efficacy of
5-FU (24). Emerging evidence has
also suggested that certain circRNAs have an important role in 5-FU
resistance. For instance, upregulation of circ-cerebellar
degeneration-related protein 1 (CDR1) was associated with 5-FU
resistance and inhibited the circCDR1as/miR-7/cyclin E1 axis to
enhance the sensitivity of breast cancer to 5-FU (25). In addition, another study suggested
that circ-DEAD-box helicase 17 was significantly upregulated in CRC
tissues and cells, while its silencing helped the cells overcome
5-FU resistance via regulating the miR-31-5p/KN motif and ankyrin
repeat domains 1 axis (26).
The present study aimed to examine whether circNRIP1
affects the chemotherapeutic effects of 5-FU on CRC and further
reveal its underlying potential molecular mechanisms of action. It
was hypothesized that circNRIP1 has a significant role in the
resistance of CRC to 5-FU. To verify this hypothesis, the effects
of knockdown on 5-FU sensitivity and cell proliferation, migration,
invasion and apoptosis of CRC cells were evaluated.
Mechanistically, the present study aimed to predict whether
circNRIP1 is able to act as a ceRNA to sponge miR-532-3p.
Materials and methods
Tissue specimens
A total of 72 pairs of tumor and adjacent normal
tissues (collected >5 cm away from the tumor tissues and these
specimens were histologically confirmed to be non-cancerous) were
collected from patients with CRC (39 male and 33 female patients;
age range, 43–77 years) who underwent surgical resection at the
Zhengzhou Central Hospital Affiliated to Zhengzhou University
(Zhengzhou, China) and the Shaanxi Provincial Cancer Hospital
(Xi'an, China) between March 2015 and January 2016. None of the
patients received any radiotherapy or chemotherapy prior to
surgery. All subjects provided written informed consent prior to
the collection and use of their specimens. The histopathological
type of CRC was diagnosed by two clinical pathologists. The study
protocol was approved by the Committee for the Protection of Human
Subjects of Zhengzhou Central Hospital (Zhengzhou, China). The
Cancer Genome Atlas (TCGA) (http://gepia.cancer-pku.cn/index.html) was used for
analyzing miR-532-3p levels in CRC.
Cell culture and transfection
The human CRC cell lines SW620, HT-29, HCT116 and
SW480, the human normal colon epithelial cell line NCM460 and 293T
cells were obtained from the Cell Bank of the Chinese Academy of
Sciences. Authentication of all cell lines was performed by short
tandem repeat profiling. 293T cells were cultured in DMEM (Gibco;
Thermo Fisher Scientific, Inc.) and the other cell lines in
RPMI-1640 (Gibco; Thermo Fisher Scientific, Inc.) medium
supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.)
and penicillin-streptomycin solution (Gibco; Thermo Fisher
Scientific, Inc.). Cells were incubated at 37°C in a humidified
atmosphere with 5% CO2.
Cells were transfected with the following small
interfering RNAs (siRNAs) or miRNA: circNRIP1-specific si-RNA1
(si-cNRIP1-1), 5′-ACGCACAAAGAAAGAAGUGUU-3′; circNRIP1-specific
si-RNA2 (si-cNRIP1-2), 5′-UUAAAUGCAAAUAUCAGUGUU-3′, and siRNA
negative control (si-NC), 5′-UUCUCCGAACGUGUCACGUTT-3′; miR-532-3p
mimics, 5′-CCUCCCACACCCAAGGCUUGCA-3′ and NC mimics,
5′-UUCUCCGAACGUGUCACGUTT-3′; miR-532-3p inhibitors,
5′-UGCAAGCCUUGGGUGUGGGAGG-3′ and NC inhibitors,
5′-CAGUACUUUUGUGUAGUACAA-3′ (all from Shanghai GenePharma Co.,
Ltd.). Between both circNRIP1-specific siRNAs, si-cNRIP1-1 was used
for subsequent experiments and was then referred to as si-cNRIP1.
Cells were transfected with 50 nM siRNAs and/or 50 nM miR-532-3p
mimics or inhibitor using Lipofectamine® 3000
(Invitrogen; Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocol. All transfection experiments were
performed for a total of three times. The transfection efficiency
was confirmed by reverse transcription-quantitative PCR (RT-qPCR)
analysis.
RNA extraction and RT-qPCR
TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) was used to extract total RNA from tissues
and cells. Subsequently, the RNA samples were treated with DNase I
and their concentration was measured using a NanoDrop®
2000 spectrophotometer (Thermo Fisher. Scientific, Inc.). Total RNA
was then reverse transcribed into cDNA using the RevertAid First
Strand cDNA Synthesis kit (Thermo Fisher. Scientific, Inc.)
according to the manufacturer's protocol. The target genes were
amplified by qPCR using a SYBR Green reaction mix (Solarbio Life
Science, Beijing, China) on the ABI 7500 Fast RT-qPCR system
(Applied Biosystems; Thermo Fisher. Scientific, Inc.) according to
the manufacturer's protocol. GAPDH and U6 were used as reference
genes. The thermocycling conditions for qPCR were as follows: 95°C
for 2 min, followed by 40 cycles of 95°C for 10 sec and 60°C for 34
sec. The primer sequences used for qPCR were as follows: circNRIP1
forward, 5′-GGATCAGGTACTGCCGTTGAC-3′ and reverse,
5′-TGGACCATTACTTTGACAGGTG-3′; GAPDH, forward,
5′-GGGAGCCAAAAGGGTCAT-3′ and reverse, 5′-GAGTCCTTCCACGATACCAA-3′;
miR-532-3p forward, 5′-ACACTCCCCTCCCACACCCAAGG-3′ and reverse,
5′-GTGCAGGGTCCGAGGT-3′; and U6 forward, 5′-CTCGCTTCGGCAGCACA-3′ and
reverse, 5′-CACAGCTTCTCTTTGATGTCAC-3′. The relative gene expression
levels were calculated using the 2−ΔΔCq method (27).
Cell viability assay
The proliferation rate of CRC cells was determined
using a Cell Counting Kit-8 (CCK-8; Beyotime Institute of
Biotechnology). In brief, cells were seeded into 96-well plates at
5×103 cells in 100 µl medium per well. Following cell
incubation at 37°C overnight, the medium was replaced with fresh
medium containing different concentrations of 5-FU (1.25, 2.5, 5,
10, 20, 30 or 40 µM). Following incubation for 0, 24, 48 or 72 h,
10 µl CCK-8 solution was added to the designated wells and cells
were incubated for an additional 2 h. The optical density (OD) in
each well was measured at a wavelength of 450 nm with an automatic
microplate reader (SpectraMax M2; Molecular Devices, LLC). The cell
viability was calculated using the following formula: Cell
viability (%) = (average OD value of the experimental group -
average OD value of the blank control group)/(average OD value of
the control group - average OD value of the blank control group)
×100%.
Cell apoptosis assay
Cell apoptosis was assessed using an Annexin V-FITC
double staining assay and analyzed using the CytoFLEX flow
cytometer (Beckman Coulter, Inc.; CytExpert 2.3). In brief,
following treatment with 5-FU for 24 h, 2×106 cells were
harvested and washed with PBS. The cells were then resuspended in
400 µl 1X binding buffer and incubated with 5 µl Annexin V-FITC at
room temperature for 15 min, followed by incubation with 10 µl
propidium iodide in an ice bath for 5 min. Cell apoptosis was then
determined with a flow cytometer and the percentage of early + late
apoptotic cells was determined.
Cell migration and invasion
assays
Cell migration and invasion were assessed using
Transwell assays and matching Transwell upper chamber inserts (pore
size, 8 µm; Corning, Inc.). In brief, cells were harvested and
resuspended in serum-free medium. Only the Transwell inserts in the
upper chamber for the invasion assays, but not those for the
migration assays, were pre-coated with Matrigel® (BD
Biosciences). Subsequently, 5×104 cells were seeded into
the upper chamber with FBS-free RPMI-1640, while the lower one was
filled with 600 µl RPMI-1640 supplemented with 10% FBS. Cells were
incubated for 24 h at 37°C. Migrated or invaded cells were fixed in
4% paraformaldehyde, stained with 0.1% crystal violet
(Sigma-Aldrich; Merck KGaA) and observed under a microscope
(Olympus Corporation).
Luciferase reporter assay
The interaction between circNRIP1 and miR-532-3p was
predicted using the StarBase database (http://starbase.sysu.edu.cn/). The sequences of
circNRIP1 encompassing the wild-type (WT) or mutant (MUT) binding
sites of miR-532-3p were inserted into the pmirGLO luciferase
reporter vector (Promega Corporation) to generate the corresponding
luciferase reporter plasmids, namely WT-cNRIP1 and MUT-cNRIP1,
respectively. The above reporter plasmids were co-transfected with
miR-532-3p or miR-NC mimics into 293T cells. Following incubation
for 48 h, the luciferase activity was measured using a luciferase
reporter assay kit (Promega Corporation) according to the
manufacturer's protocol.
Statistical analysis
All statistical analyses were performed using SPSS
19.0 (IBM Corp.) The differences between two groups were compared
using paired or unpaired Student's t-test, while those among ≥3
groups were compared using one-way ANOVA followed by Tukey's post
hoc test. The association between circNRIP1 expression and
clinicopathological features was analyzed using a χ2
test. The correlation between two variables was determined using
the Pearson correlation test. The overall survival of patients was
assessed by the Kaplan-Meier method and a log-rank test. The
diagnostic value for CRC were assessed by receiver operating
characteristic analysis. P<0.05 was considered to indicate a
statistically significant difference.
Results
circNRIP1 is upregulated in CRC and is
associated with CRC progression
In the present study, the expression levels of
circNRIP1 were determined in 72 paired CRC and normal tissues
samples. circNRIP1 was significantly upregulated in CRC tissues
compared with normal samples (Fig.
1A). Furthermore, the prognostic value of circNRIP1 was
evaluated. Kaplan-Meier survival analysis indicated that CRC cases
with high circNRIP1 expression levels exhibited poor overall
survival compared with those with low expression (median survival,
31 vs. 46 months; log-rank=4.614; P=0.0317; Fig. 1B). To investigate the association
between the expression levels of circNRIP1 and the
clinicopathological features of patients, patients with CRC were
divided into the high (n=36) and low (n=36) expression groups,
based on the median circNRIP1 expression levels. A significant
association was observed between the expression of circNRIP1 and
lymph node metastasis, distant metastasis and TNM stage (P=0.029,
0.012 and 0.001, respectively; Table
I). No other significant associations were obtained between the
expression of circNRIP1 and other clinicopathological parameters
such as tumor size, sex, age, smoking history and alcohol
consumption.
 | Table I.Associations between circNRIP1
expression and clinicopathological characteristics of patients with
colorectal cancer. |
Table I.
Associations between circNRIP1
expression and clinicopathological characteristics of patients with
colorectal cancer.
|
|
| circNRIP1
expression |
|
|---|
|
|
|
|
|
|---|
| Variables | Cases | Low | High | P-value |
|---|
| Sex |
|
|
| 0.637 |
|
Male | 39 | 21 | 18 |
|
|
Female | 33 | 15 | 18 |
|
| Age, years |
|
|
| 0.155 |
|
<60 | 33 | 20 | 13 |
|
|
≥60 | 39 | 16 | 23 |
|
| Smoking
history |
|
|
| >0.999 |
|
Yes | 37 | 18 | 19 |
|
| No | 35 | 18 | 17 |
|
| Alcohol
consumption |
|
|
| 0.809 |
|
Yes | 44 | 21 | 23 |
|
| No | 28 | 15 | 13 |
|
| Tumor size, cm |
|
|
| 0.153 |
| ≥5 | 41 | 17 | 24 |
|
|
<5 | 31 | 19 | 12 |
|
| Lymph node
metastasis |
|
|
| 0.029 |
|
Yes | 46 | 19 | 27 |
|
| No | 26 | 18 | 8 |
|
| Distant
metastasis |
|
|
| 0.012 |
|
Yes | 13 | 2 | 11 |
|
| No | 59 | 34 | 25 |
|
| TNM stage |
|
|
| 0.001 |
|
I/II | 44 | 29 | 15 |
|
|
III/IV | 28 | 21 | 7 |
|
Overexpression of circNRIP1 attenuates
the 5-FU-mediated inhibition of viability of HCT116 and SW480
cells
To investigate the biological effect of circNRIP1 on
CRC, the expression of circNRIP1 was first determined in CRC cell
lines and normal NCM460 colon epithelial cells. The results
indicated that circNRIP1 was markedly upregulated in both CRC cell
lines compared with NCM460A cells (Fig.
2A). To further investigate the roles of circNIP1 in CRC, two
circNRIP1 siRNAs were synthesized to knockdown circNRIP1
expression. RT-qPCR results demonstrated that the expression of
circNRIP1 was significantly decreased in HCT116 and SW480 cells
transfected with si-cNRIP1 (Fig.
2B).
 | Figure 2.Overexpression of circNRIP1 in CRC
attenuates 5-FU-mediated inhibition of HCT116 and SW480 cell
viability. (A) Expression levels of circNRIP1 in normal and CRC
cell lines. (B) circNRIP1 expression in CRC cells transfected with
si-cNRIP1 or si-NC. (C-G) Effects of circNRIP1 knockdown on the
proliferation of (C) HCT116 and (D) SW480 cells, and on (E)
apoptosis, (F) invasion and (G) migration of CRC cells (scale bars,
100 µm). (H and I) Effect of circNRIP1 knockdown on the sensitivity
of (H) HCT116 and (I) SW480 cells to 5-Fu. *P<0.05, **P<0.01
and ***P<0.001 vs. the NCM460 group or the si-NC group.
circNRIP1, circRNA nuclear receptor-interacting protein 1; CRC,
colorectal cancer; 5-FU, 5-fluorouracil; si, small interfering RNA;
NC, negative control; OD, optical density; PI, propidium iodide; Q,
quadrant; LR, lower right; LL, lower left; UL, upper left; UR,
upper right. |
Subsequently, the effects of circNRIP1 on the
biological function of CRC cells were explored. CCK-8 assays
revealed that circNRIP1 knockdown markedly attenuated HCT116 and
SW480 cell proliferation (Fig. 2C and
D). However, circNRIP1 knockdown significantly promoted HCT116
and SW480 cell apoptosis (Fig. 2E).
In addition, Transwell assays indicated that circNRIP1 knockdown
significantly decreased the invasive and migratory abilities of
HCT116 and SW480 cells (Fig. 2F and
G). All of these results suggested that circNRIP1 acts as an
oncogene in CRC.
To determine whether circNRIP1 affects the
5-FU-mediated inhibition of HCT116 and SW480 cell viability, cells
transfected with si-cNRIP1 or si-NC were treated with different
concentrations of 5-FU Compared with that of si-NC-transfected
cells, the 5-FU-mediated inhibition of the cell viability of HCT116
and SW480 was significantly enhanced following transfection with
si-cNRIP1. Furthermore, the present study also evaluated the
effects of si-cNRIP1 silencing on the IC50 of 5-FU in
HCT116 and SW480 cells. Compared with those in the si-NC group, the
5-FU IC50 values were markedly reduced in HCT116 and
SW480 cells transfected with si-cNRIP1 (decreased by 33.74 and
34.47% in HCT116 cells; and 37.91 and 40.79% in SW480 cells;
Fig. 2H and I).
circNRIP1 sponges miR-532-3p in CRC
cells
To explore the mechanisms underlying the effect of
circ-NRIP1 to regulate the resistance of CRC cells to 5-FU, the
potential miRNA targets of circNRIP1 were predicted using the
StarBase database. The bioinformatics analysis predicted that
miR-532-3p may be directly targeted by circNRIP1 (Fig. 3A). In addition, the expression profile
of miR-532-3p in CRC was extracted from the TCGA database. The
analysis indicated that miR-532-3p was downregulated in tumor
tissues compared with normal ones (Fig.
3B). The expression of miR-532-3p was also evaluated in the 72
paired CRC and normal tissue samples. RT-qPCR analysis demonstrated
that the expression of miR-532-3p was reduced in CRC tissues
compared with that in adjacent normal tissues (Fig. 3C). Subsequently, the correlation
between miR-532-3p and circNRIP1 was analyzed and the results
suggested that their expression levels were negatively correlated
in CRC tissues (Fig. 3D). The
dual-luciferase reporter assay demonstrated that co-transfection of
cells with WT-circNRIP1 and miR-532-3p mimics significantly
decreased the luciferase activity in 293T cells (Fig. 3E). The expression levels of miR-532-3p
were also detected in cell lines. Consistent with the results
obtained in tumor tissues, miR-532-3p was significantly
downregulated in CRC cell lines compared with NCM460 cells
(Fig. 3F). Cells transfected with
miR-532-3p mimics exhibited significantly upregulated miR-532-3p
expression compared with the NC mimics group and those transfected
with miR-532-3p inhibitor exhibited significantly downregulated
miR-532-3p expression compared with the NC inhibitor group
(Fig. 3G), while circNRIP1 silencing
markedly enhanced the expression of miR-532-3p compared with the
control group (Fig. 3H).
Roles of miR-532-3p in CRC cells
To further investigate the effects of miR-532-3p
expression on CRC, cell proliferation and apoptosis in CRC cells
were evaluated. The results suggested that overexpression of
miR-532-3p significantly inhibited proliferation and promoted
apoptosis in HCT116 and SW480 cells (Fig.
4A-C). The 5-FU chemosensitivity was also determined in CRC
cells overexpressing miR-532-3p. Compared with the miR-NC group,
miR-532-3p overexpression significantly enhanced the 5-FU-induced
inhibition of cell viability and decreased the IC50
values of 5-FU in HCT116 and SW480 cells (Fig. 4D and E).
 | Figure 4.Roles of miR-532-3p in CRC cells. (A
and B) Effect of miR-532-3p overexpression on the proliferation of
(A) HCT116 and (B) SW480 cells. (C-E) Influence of miR-532-3p on
the toxicity of 5-FU on CRC cells. (C) Apoptosis of CRC cells and
viability of (D) HCT116 and (E) SW480 cells treated with increasing
concentrations of 5-FU. *P<0.05, **P<0.01 and ***P<0.001
vs. the miR-NC group. miR-532-3p, microRNA-532-3p; CRC, colorectal
cancer; IC50, half maximal inhibitory concentration; NC,
negative control; 5-FU, 5-fluorouracil; OD, optical density; PI,
propidium iodide; Q, quadrant; LR, lower right; LL, lower left; UL,
upper left; UR, upper right. |
circNRIP1 reduces the sensitivity of
CRC cells to 5-FU via regulating the expression of miR-532-3p
The aforementioned results suggested that circNRIP1
knockdown and miR-532-3p overexpression significantly enhanced the
sensitivity of CRC cells to 5-FU at least in part through the
binding of circNRIP1 to miR-532-3p to inhibit its expression.
Therefore, it was hypothesized that circNRIP1 reduces the
sensitivity of CRC cells to 5-FU via regulating the expression of
miR-532-3p. To verify this hypothesis, CRC cells were
co-transfected with si-cNRIP1 and miR-532-3p inhibitor and treated
with 5-FU. The results demonstrated that cell co-transfection with
si-cNRIP1 and miR-532-3p inhibitor significantly promoted cell
proliferation and inhibited apoptosis compared with the si-cNRIP1 +
miR-NC group (Fig. 5A-C). In
addition, compared with the si-cNRIP1 + miR-NC group,
co-transfection with si-cNRIP1 and miR-532-3p inhibitor
significantly attenuated the 5-FU-mediated inhibition of cell
viability and increased the IC50 values of 5-FU in
HCT116 and SW480 cells (Fig. 5D and
E).
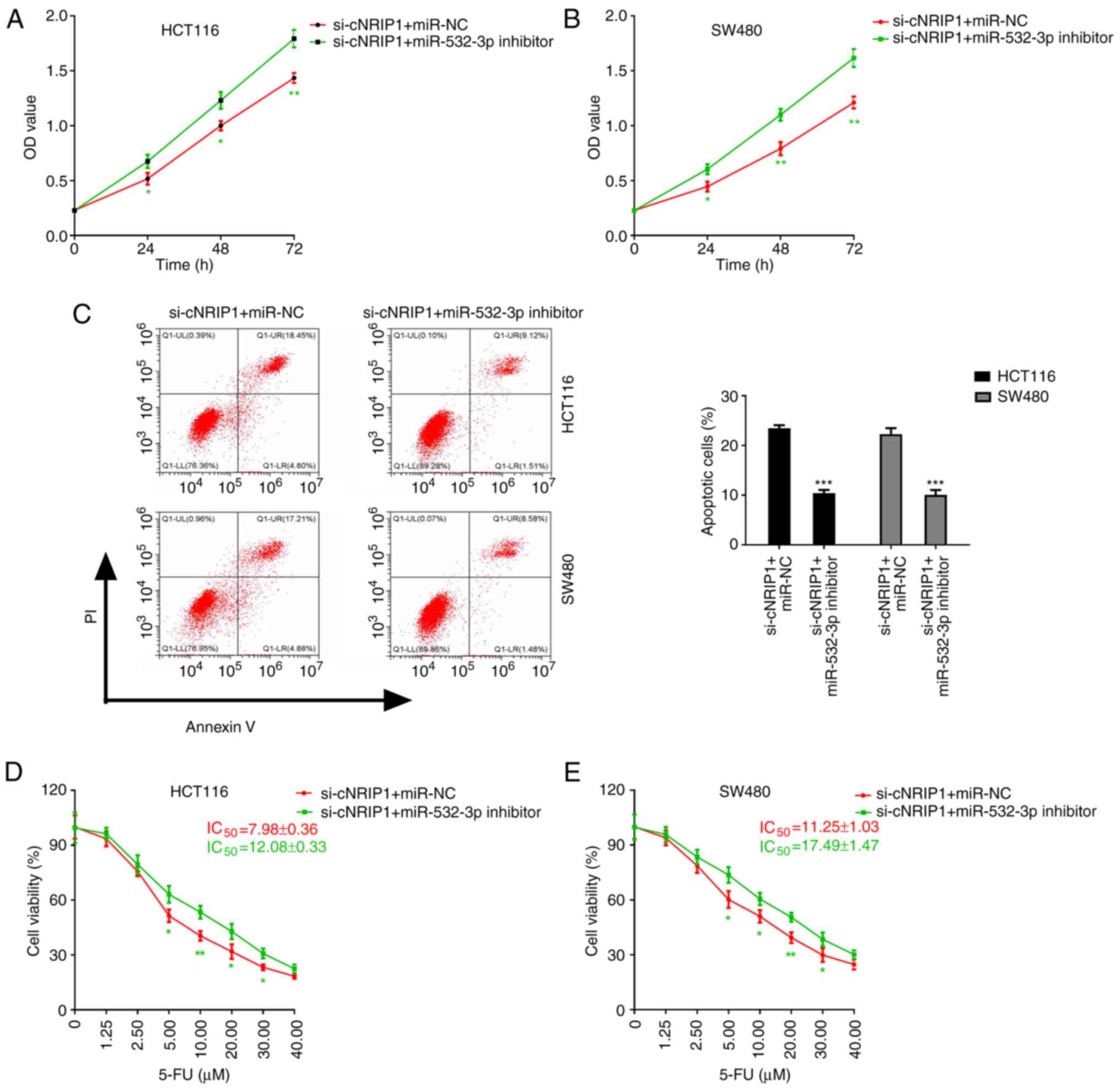 | Figure 5.circNRIP1 attenuates the sensitivity
of CRC cells to 5-FU via regulating the expression of miR-532-3p.
(A and B) Effect of cell co-transfection with si-cNRIP1 and
miR-532-3p inhibitor on the proliferation of (A) HCT116 and (B)
SW480 cells. (C-E) Influence of co-transfection with si-cNRIP1 and
miR-532-3p inhibitor on the effect of 5-FU on CRC cells. (C)
Apoptosis of CRC cells and viability of (D) HCT116 and (E) SW480
cells treated with increasing concentrations of 5-FU. *P<0.05,
**P<0.01 and ***P<0.001 vs. the si-cNRIP1 + miR-NC group.
circ-NRIP1, circRNA nuclear receptor-interacting protein 1; CRC,
colorectal cancer; miR-532-3p, microRNA-532-3p; 5-FU,
5-fluorouracil; si, small interfering RNA; IC50, half
maximal inhibitory concentration; NC, negative control; OD, optical
density; PI, propidium iodide; Q, quadrant; LR, lower right; LL,
lower left; UL, upper left; UR, upper right. |
Diagnostic value of circNRIP1 and
miR-532-3p in CRC
Subsequently, the association between circNRIP1 and
miR-532-3p with the clinicopathological features of patients and
their diagnostic value for CRC were investigated using receiver
operating characteristic curves (Fig.
6). The results indicated that both circNRIP1 and miR-532-3p
exhibited good predictive value regarding lymph node metastasis
[area under the curve (AUC), 0.814 and 0.701, respectively;
P<0.001 and P=0.009, respectively; Fig. 6A and B], distant metastasis (AUC,
0.863 and 0.768, respectively; P<0.001 and P=0.002,
respectively, Fig. 6D and E) and TNM
stage (AUC, 0.747 and 0.760, respectively; P<0.001 for each;
Fig. 6G and H). In addition, the
combination of the two indicators also exhibited a good predictive
value for lymph node metastasis (AUC=0.849; P<0.001; Fig. 6C), distant metastasis (AUC=0.906;
P<0.001; Fig. 6F) and TNM stage
(AUC=0.802; P<0.001; Fig. 6I).
These results indicated the combination of circNRIP1 and miR-532-3p
may be used as a biomarker for the diagnosis of CRC.
Discussion
Emerging evidence suggested that circRNAs have a
crucial role in resistance to chemotherapeutics and serve as key
targets for the diagnosis and treatment of CRC (9–11).
circNRIP1 was reported to be abnormally expressed in numerous tumor
types and may have an important role in promoting malignant
features (12–14). However, the expression and biological
function of circNRIP1 in CRC has remained elusive. The present
study also demonstrated that circNRIP1 was abnormally upregulated
in CRC specimens and cells. In terms of clinical value, the
expression of circNRIP1 was associated with the clinical
progression of CRC. Increased expression of circNRIP1 was indicated
to promote tumor metastasis and was associated with TNM stage and
poor prognosis of patients with CRC. Hence, circNRIP1 may be used
as a biomarker for the diagnosis of CRC.
As a type of circRNA, circNRIP1 may affect the
biological properties of tumor cells. Li et al (22) reported that circNRIP1 promoted cell
migration and invasion in cervical cancer. In addition, Dong et
al (28) demonstrated that
circNRIP1 silencing attenuated cell viability, migration and colony
formation and promoted apoptosis in renal carcinoma cell lines.
Furthermore, Zhang et al (29)
revealed that circNRIP1 knockdown inhibited cell proliferation,
migration and invasion and downregulate AKT1 in gastric cancer
cells. The functional experiments of the present study also
demonstrated that circNRIP1 promoted CRC cell proliferation,
migration and invasion, while inhibiting apoptosis. These results
suggested that circNRIP1 may also exert an oncogenic role in
CRC.
Of note, the biological functions of circRNAs are
triggered by miRNAs. Recent studies have suggested that circNRIP1
promotes the progression of tumors via sponging miRNAs. For
instance, a study indicated that circNRIP1 has an oncogenic role in
a renal carcinoma cell line via targeting miR-505 (28). Another study revealed that circNRIP1
acts as an miR-149-5p sponge to promote gastric cancer progression
via the AKT1/mTOR signaling pathway (29). Hence, the present study aimed to
investigate whether circNRIP1 is able to promote the progression of
CRC via sponging miRNAs. miR-532-3p was predicted as a target miRNA
of circNRIP1, which was confirmed by dual-luciferase reporter
assays. Of note, miR-532-3p has been recognized as a tumor
suppressor gene in several types of human cancer (30–32).
Analysis of the TCGA dataset and in vitro results of the
present study indicated that miR-532-3p was significantly
downregulated in CRC tumor tissues and cell lines. Clinical value
analysis and functional experiments also demonstrated that
miR-532-3p acts as a tumor suppressor in CRC. In addition, the
results of co-transfection experiments of CRC cells with si-cNRIP1
and miR-532-3p inhibitor suggested that miR-532-3p silencing
abrogated the defects caused by circNRIP1 depletion. These findings
indicated that knockdown of miR-532-3p reversed the circNRIP1
depletion-mediated effects in CRC cells. Consistent with the
results of the present study, previous studies suggested that
certain circRNAs affect tumor progression via acting on miR-532-3p.
Furthermore, Ouyang et al (30) demonstrated that loss of androgen
receptors promoted HCC invasion and metastasis via activating the
circ-leucyl and cystinyl aminopeptidase/miR-532-3p/ras-related
protein rab-9a signaling pathway under hypoxia. In addition, Dai
et al (33) reported that
circ-FYVE, RhoGEF and PH domain containing 4 attenuated gastric
cancer progression via modulating the miR-532-3p/adenomatous
polyposis coli/β-catenin signaling pathway.
It has also been reported that the differential
expression of circRNAs, including that of circNRIP1, may affect the
sensitivity of cancers to chemotherapy with 5-FU. Lin et al
(34) reported that circNRIP1 is able
to regulate the resistance of nasopharyngeal carcinoma cells to
5-FU and cisplatin via the miR-515-5p/IL-25 axis. In addition, Xu
et al (35) demonstrated that
circNRIP1 was able to maintain the hypoxia-induced resistance of
gastric carcinoma cells to 5-FU through hypoxia-inducible
factor-1α-dependent glucose metabolism via sponging miR-138-5p. The
present study indicated that circNRIP1 knockdown significantly
enhanced the sensitivity of CRC cells to 5-FU. Furthermore, the
results confirmed that circNRIP1 promoted the progression of CRC
via sponging miR-532-3p. Hence, in the present study, it was
hypothesized that circNRIP1 promoted the resistance of CRC cells to
5-FU via sponging miR-532-3p. First, the present study verified
that increased expression of circNRIP1 significantly suppressed the
expression of miR-532-3p. In addition, co-transfection experiments
revealed that miR-532-3p silencing reversed the effects of
circNRIP1-depletion on the chemotherapy sensitivity of CRC cells to
5-FU. However, the absence of an experiment of overexpression of
circNRIP1 is a limitation of the present study.
In conclusion, the present study demonstrated that
circNRIP upregulation promoted the progression of CRC, while
circNRIP1 silencing sensitized CRC cells to 5-FU via sponging
miR-532-3p. These results suggested that circ-NRIP1 and miR-532-3p
may be considered as potential sensitizing targets and biomarkers
for the diagnosis of CRC.
Acknowledgements
Not applicable.
Funding
This work was supported by the Science and
Technology Project of Henan Province (grant no. 212102310121) and
the Medical Science Research Project of Henan Province (grant no.
LHGJ20191039).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author upon reasonable
request.
Authors' contributions
FL and RL performed the majority of the experiments
in the study. RZ collected clinical specimens. YZ and MH
contributed to the analysis of experimental data. FL and YZ
contributed to the study design, manuscript writing and provided
experimental funding support. All authors have read and approved
the final manuscript. FL and YZ checked and confirmed the
authenticity of all the raw data.
Ethics approval and consent to
participate
All aspects of experimental design and protocols
were reviewed and approved by the Committee for the Protection of
Human Subjects at Zhengzhou Central Hospital (Zhengzhou,
China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD, Fedewa SA, Butterly
LF, Anderson JC, Cercek A, Smith RA and Jemal A: Colorectal cancer
statistics, 2017. CA Cancer J Clin. 67:177–193. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Dekker E, Tanis PJ, Vleugels J, Kasi PM
and Wallace MB: Colorectal cancer. Lancet. 394:1467–1480. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Meng S, Zhou H, Feng Z, Xu Z, Tang Y, Li P
and Wu M: circRNA: Functions and properties of a novel potential
biomarker for cancer. Mol Cancer. 16:942017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kristensen LS, Andersen MS, Stagsted L,
Ebbesen KK, Hansen TB and Kjems J: The biogenesis, biology and
characterization of circular RNAs. Nat Rev Genet. 20:675–691. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Soslau G: Circular RNA (circRNA) was an
important bridge in the switch from the RNA world to the DNA world.
J Theor Biol. 447:32–40. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ruan H, Xiang Y, Ko J, Li S, Jing Y, Zhu
X, Ye Y, Zhang Z, Mills T, Feng J, et al: Comprehensive
characterization of circular RNAs in ~1000 human cancer cell lines.
Genome Med. 11:552019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lei B, Tian Z, Fan W and Ni B: Circular
RNA: A novel biomarker and therapeutic target for human cancers.
Int J Med Sci. 16:292–301. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chen LY, Wang L, Ren YX, Pang Z, Liu Y,
Sun XD, Tu J, Zhi Z, Qin Y, Sun LN and Li JM: The circular RNA
circ-ERBIN promotes growth and metastasis of colorectal cancer by
miR-125a-5p and miR-138-5p/4EBP-1 mediated cap-independent
HIF-1alpha translation. Mol Cancer. 19:1642020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Shang A, Gu C, Wang W, Wang X, Sun J, Zeng
B, Chen C, Chang W, Ping Y, Ji P, et al: Exosomal circPACRGL
promotes progression of colorectal cancer via the
miR-142-3p/miR-506-3p- TGF-β1 axis. Mol Cancer. 19:1172020.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li C, He X, Zhang L, Li L and Zhao W: A
pair-wise meta-analysis highlights circular RNAs as potential
biomarkers for colorectal cancer. BMC Cancer. 19:9572019.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liang L and Li L: Down-regulation of
circNRIP1 promotes the apoptosis and inhibits the migration and
invasion of gastric cancer cells by miR-182/ROCK1 axis. Onco
Targets Ther. 13:6279–6288. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Fang D and Lu G: Expression and role of
nuclear receptor-interacting protein 1 (NRIP1) in stomach
adenocarcinoma. Ann Transl Med. 8:12932020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ni XF, Zhao LH, Li G, Hou M, Su M, Zou CL
and Deng X: MicroRNA-548-3p and microRNA-576-5p enhance the
migration and invasion of esophageal squamous cell carcinoma cells
via NRIP1 down-regulation. Neoplasma. 65:881–887. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liang ZZ, Guo C, Zou MM, Meng P and Zhang
TT: circRNA-miRNA-mRNA regulatory network in human lung cancer: An
update. Cancer Cell Int. 20:1732020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xiong DD, Dang YW, Lin P, Wen DY, He RQ,
Luo DZ, Feng ZB and Chen G: A circRNA-miRNA-mRNA network
identification for exploring underlying pathogenesis and therapy
strategy of hepatocellular carcinoma. J Transl Med. 16:2202018.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhong Y, Du Y, Yang X, Mo Y, Fan C, Xiong
F, Ren D, Ye X, Li C, Wang Y, et al: Circular RNAs function as
ceRNAs to regulate and control human cancer progression. Mol
Cancer. 17:792018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ji Q, Zhang C, Sun X and Li Q: Circular
RNAs function as competing endogenous RNAs in multiple types of
cancer. Oncol Lett. 15:23–30. 2018.PubMed/NCBI
|
|
19
|
Li Q, Pan X, Zhu D, Deng Z, Jiang R and
Wang X: Circular RNA MAT2B promotes glycolysis and malignancy of
hepatocellular carcinoma through the miR-338-3p/PKM2 axis under
hypoxic stress. Hepatology. 70:1298–1316. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen X, Chen RX, Wei WS, Li YH, Feng ZH,
Tan L, Chen JW, Yuan GJ, Chen SL and Guo SJ: PRMT5 circular RNA
promotes metastasis of urothelial carcinoma of the bladder through
sponging miR-30c to induce epithelial-mesenchymal transition. Clin
Cancer Res. 24:6319–6330. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Xu D, Wu Y, Wang X, Hu X, Qin W, Li Y,
Wang Y, Zhang Z, Lu S, Sun T, et al: Identification of functional
circRNA/miRNA/mRNA regulatory network for exploring prospective
therapy strategy of colorectal cancer. J Cell Biochem.
1:297032020.
|
|
22
|
Li X, Ma N, Zhang Y, Wei H, Zhang H, Pang
X, Li X, Wu D, Wang D, Yang Z and Zhang S: Circular RNA circNRIP1
promotes migration and invasion in cervical cancer by sponging
miR-629-3p and regulating the PTP4A1/ERK1/2 pathway. Cell Death
Dis. 11:3992020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
McQuade RM, Stojanovska V, Bornstein JC
and Nurgali K: Colorectal cancer chemotherapy: The evolution of
treatment and new approaches. Curr Med Chem. 24:1537–1557. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Blondy S, David V, Verdier M, Mathonnet M,
Perraud A and Christou N: 5-fluorouracil resistance mechanisms in
colorectal cancer: From classical pathways to promising processes.
Cancer Sci. 111:3142–3154. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yang W, Gu J, Wang X, Wang Y, Feng M, Zhou
D, Guo J and Zhou M: Inhibition of circular RNA CDR1as increases
chemosensitivity of 5-FU-resistant BC cells through up-regulating
miR-7. J Cell Mol Med. 23:3166–3177. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ren TJ, Liu C, Hou JF and Shan FX:
circDDX17 reduces 5-fluorouracil resistance and hinders
tumorigenesis in colorectal cancer by regulating miR-31-5p/KANK1
axis. Eur Rev Med Pharmacol Sci. 24:1743–1754. 2020.PubMed/NCBI
|
|
27
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Dong Z, Liu Y, Wang Q, Wang H, Ji J, Huang
T, Khanal A, Niu H and Cao Y: The circular RNA-NRIP1 plays
oncogenic roles by targeting microRNA-505 in the renal carcinoma
cell lines. J Cell Biochem. 121:2236–2246. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhang X, Wang S, Wang H, Cao J, Huang X,
Chen Z, Xu P, Sun G, Xu J, Lv J and Xu Z: Circular RNA circNRIP1
acts as a microRNA-149-5p sponge to promote gastric cancer
progression via the AKT1/mTOR pathway. Mol Cancer. 18:202019.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ouyang X, Yao L, Liu G, Liu S, Gong L and
Xiao Y: Loss of androgen receptor promotes HCC invasion and
metastasis via activating circ-LNPEP/miR-532-3p/RAB9A signal under
hypoxia. Biochem Biophys Res Commun. 557:26–32. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ye J, Liu J, Tang T, Xin L, Bao X and Yan
Y: LINC00963 affects the development of colorectal cancer via
miR-532-3p/HMGA2 axis. Cancer Cell Int. 21:872021. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Jin L, Huang S, Guan C and Chang S:
ETS1-activated SNHG10 exerts oncogenic functions in glioma via
targeting miR-532-3p/FBXL19 axis. Cancer Cell Int. 20:5892020.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Dai X, Liu J, Guo X, Cheng A, Deng X, Guo
L and Wang Z: Circular RNA circFGD4 suppresses gastric cancer
progression via modulating miR-532-3p/APC/beta-catenin signalling
pathway. Clin Sci (Lond). 134:1821–1839. 2020.PubMed/NCBI
|
|
34
|
Lin J, Qin H, Han Y, Li X, Zhao Y and Zhai
G: circNRIP1 Modulates the miR-515-5p/IL-25 axis to control 5-Fu
and cisplatin resistance in nasopharyngeal carcinoma. Drug Des
Devel Ther. 15:323–330. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Xu G, Li M, Wu J, Qin C, Tao Y and He H:
Circular RNA circNRIP1 sponges microRNA-138-5p to maintain
hypoxia-induced resistance to 5-fluorouracil through
HIF-1α-dependent glucose metabolism in gastric carcinoma. Cancer
Manag Res. 12:2789–2802. 2020. View Article : Google Scholar : PubMed/NCBI
|




















