Introduction
Breast cancer (BC) is one of the most common cancer
types and a leading cause of cancer-related mortality among women
worldwide, and it can be classified clinically based on the
expression of estrogen receptor (ER), progesterone receptor (PR)
and HER2 (1,2). In total, ~80% of patients with BC are
hormone receptor-positive and usually respond well to hormone
therapy (3). Patients who are
HER2+ can be treated more effectively with targeted
anti-HER2 treatment, such as trastuzumab. On the other hand,
patients with triple-negative BC (TNBC) respond poorly to hormone
or targeted therapy, and chemotherapy remains the primary treatment
option (4,5).
Molecularly, BCs are classified into four groups
that largely overlap with the clinical subtypes (6). Luminal A and B are characterized by
positive ER and/or PR expression, and generally have a more
favorable prognosis (6). Luminal A
usually has high ER/PR expression and is negative for HER2
expression, while luminal B has lower ER/PR expression and is
associated with a slightly worse prognosis than luminal A (6). HER2-enriched cancer types exhibit
amplification and upregulation of the HER2 gene (6). Basal cancer types are usually
triple-negative, with minimal expression of ER/PR and no HER2
amplification, and they are associated with aggressive pathology
and a poor prognosis (6).
Cancer stem cells (CSCs) are a small subpopulation
accounting for 1–5% of cell types within the tumor, which have the
potential to initiate clonal tumors to drive tumorigenesis and
maintain the heterogeneity of the tumor through maintaining their
self-renewal and differentiation abilities (7–9).
Moreover, these cells contribute to treatment resistance and
recurrence of tumors (7,10). Thus, identifying methods to
eradicate CSCs may be the key to curing cancer. In this regard,
CD44+/CD24−/low, aldehyde dehydrogenase 1
family member A1 (ALDH1) and BMI1 proto-oncogene, polycomb ring
finger (Bmi1) were shown to be common markers for breast (B)CSCs,
and signaling pathways including Hedgehog, Wnt and Notch were found
to be essential for their self-renewal (10).
Cyclin-dependent kinases (CDKs) are a group of
serine/threonine kinases that require the binding of various
cyclins to form different CDK/cyclin complexes to become activated
(11), and most of them are known
to play a role in cell cycle progression (12–15).
Furthermore, CDK1 and CDK2 are also involved in the regulation of
pluripotency and differentiation during embryonic development
(16–18). Intriguingly, other CDKs play
important roles in the self-renewal of SCs, neuronal functions and
transcriptional regulation (8,19–22).
Malignant cells are defined by their uncontrolled
proliferation, which often occurs as a result of malfunctioning
cell cycle checkpoints (23).
Accumulating evidence has indicated that a high expression of
various CDKs and cyclins is correlated with poor prognosis, therapy
resistance, tumor recurrence and CSC maintenance in BC (8,14).
Several CDK4/6 inhibitors, including palbociclib, ribociclib and
abemaciclib, have recently been approved by the US Food and Drug
Administration for combined use with letrozole or other aromatase
inhibitors in the first-line treatment of patients with hormone
receptor-positive advanced BC (24–26).
However, newly emerging evidence has suggested that ER-positive BC
could develop resistance to these CDK4/6 inhibitors (27–29).
Interestingly, drugs against other CDKs, such as dinaciclib (i.e.,
SCH727965), are being evaluated clinically for treating patients
with advanced or metastatic TNBC (30,31)
as CDK1 inhibition has been reported to trigger the apoptosis of
human basal-like TNBC cells (32).
Furthermore, CDK2 inhibition has been revealed to not only decrease
the CD44+/CD24−/low stem-like subpopulation,
but also restore the chemosensitivity of SUM149PT TNBC cells
(18).
The embryonic SC (ESC) markers Oct4, Nanog and Sox2
are transcription factors (TFs) that are not only highly expressed
in ESCs, but they also work together to regulate a set of target
genes that have important functions in ESC pluripotency (33). Interestingly, CDK2/cyclin E and
CDK6/cyclin D3 complexes have been shown to phosphorylate these ESC
markers, thereby promoting their interactions with peptidylprolyl
cis/trans isomerase, NIMA-interacting 1 (Pin1), which
protects them from polyubiquitination and proteasomal degradation
(34). Forkhead box M1 (FoxM1) is
another TF the activity of which has also been observed to be
stimulated by various CDK/cyclin complexes in either a
phosphorylation-dependent (35) or
-independent (36) manner. More
importantly, the aforementioned TFs have been shown to be involved
in the activation of the Hedgehog and Wnt signaling pathways that
are crucial for the maintenance of BCSCs (37–39).
Therefore, the present study aimed to investigate
the cytotoxic and the stemness-inhibitory effects of dinaciclib in
two human BC cell lines, MCF-7 (luminal A) and HCC-1806 (TNBC), as
well as to elucidate the mechanisms underlying its
stemness-suppressive effects.
Materials and methods
Cell culture
MCF-7 and HCC-1806 human BC cell lines were
purchased from the American Type Culture Collection (ATCC) and
maintained in DMEM and RPMI-1640 medium, respectively, which were
supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.),
100 U/ml penicillin, 100 µg/ml streptomycin and 25 µg/ml
amphotericin B (1% PSA; Biological Industries, USA) at 37°C with 5%
CO2. MDA-MB-231 cells purchased from the ATCC were
maintained in Leibovitz's L-15 medium supplemented with 10% FBS and
1% PSA at 37°C without CO2.
MTT assay
MCF-7 (2×103) and HCC-1806
(5×103) cells were seeded into each well of 96-well
plates the day before the indicated concentrations of dinaciclib
were added into the wells. After a 72-h incubation, 1 mg/ml MTT
reagent (Sigma-Aldrich; Merck KGaA) was added into each well for a
3-h incubation before the MTT medium was removed and replaced with
DMSO to solubilize the formazan crystals. The optical density at
570 nm (OD570) of each well was measured using an ELISA
reader (Bio-Rad Laboratories, Inc.). Cells treated with 0.1% DMSO
alone were considered as untreated cells and their viability was
designated as 100%. The half maximal inhibitory concentration
(IC50) values of drug in each cell line were calculated
using GraphPad Prism software version 4.0 (GraphPad Software,
Inc.).
Western blotting
For total lysate preparation, the cells were washed
with cold PBS and scraped into 1 ml cold PBS. After centrifugation,
the cells were lysed in RIPA buffer [50 mM Tris-HCl, 150 mM NaCl,
0.1% SDS and 1% Nonidet P-40 (NP-40); pH 7.4]. To prepare the
nuclear fraction, cells were first incubated in a low-salt buffer
(10 mM Tris-HCl, 10 mM NaCl, 3 mM MgCl2 and 0.5% NP-40;
pH 7.4) and the nucleus was then pelleted via centrifugation.
Nuclear proteins were subsequently extracted using a high-salt
buffer (20 mM HEPES, 25% glycerol, 0.4 M NaCl, 1.5 mM
MgCl2 and 0.2 mM EDTA; pH 7.9) on ice. Total lysates (20
µg) or nuclear proteins (40 µg) were separated via 10% SDS-PAGE and
processed for blocking using 5% skim milk in 1X TBS-Tween 20 (TBST)
for 1 h at room temperature. Next, immunoblotting was performed
using primary antibodies against β-actin (1:13,000; cat. no.
MAB1501; MilliporeSigma), FoxM1 (1:2,000; cat. no. GTX100276;
GeneTex, Inc.), phosphorylated (p-)FoxM1 (Ser35; 1:1,000; cat. no.
14170; Cell Signaling Technology, Inc.), CD44 (1:2,000; cat. no.
GTX102111; GeneTex, Inc.), ALDH1A1 (1:5,000; cat. no. GTX123973;
GeneTex, Inc.), Bmi1 (1:2,000; cat. no. GTX114008; GeneTex, Inc.),
Oct4 (1:1,000; cat. no. GTX101497; GeneTex, Inc.), Nanog (1:1,000;
cat. no. GTX100863; GeneTex, Inc.), Sox2 (1:1,000; cat. no.
GTX101506; GeneTex, Inc.), GLI family zinc finger 1 (GLI1; 1:1,000;
cat. no. sc-20687; Santa Cruz Biotechnology, Inc.) and Lamin A/C
(1:5,000; cat. no. E18-6056-2; EnoGene). After an overnight
incubation at 4°C, the blots were washed several times with 1X TBST
before being probed with the HRP-conjugated secondary antibodies,
including anti-mouse IgG or anti-rabbit IgG. Signals were detected
using an ECL reagent (T-Pro Biotechnology) under a Luminescence
Imaging system (FUJIFILM LAS-4000; FUJIFILM Wako Pure Chemical
Corp.) and their intensities were semi-quantified via densitometry
(ImageJ software version 1.50i; National Institutes of Health).
Sphere formation assay
MCF-7 (1×104) and HCC-1806
(2×104) cells were seeded on low attachment dishes
containing DMEM and RPMI-1640 medium, respectively, supplemented
with 2% B27 (cat. no. 17504044; Thermo Fisher Scientific, Inc.), 20
ng/ml EGF (PeproTech, Inc.), 10 ng/ml basic fibroblast growth
factor (PeproTech, Inc.), insulin (5 µg/ml), 0.4% BSA and 1% PSA
(referred to as defined media) in the absence or presence of
various doses of dinaciclib. MCF-7 cells were cultured for 14 days,
while HCC-1806 cells were cultured for 21 days before the spheres
formed, following which cells were stained with 1 mg/ml MTT reagent
at room temperature and counted using MetaMorph software version
7.8 (Molecular Devices LLC).
Sphere shrinkage assay
MCF-7 (1×104) and HCC-1806
(2×104) cells were seeded on low attachment dishes
containing defined media for 8 and 10 days, respectively, before
being treated without or with various doses of dinaciclib. After 10
days, the number of spheres was counted as described in the
‘Sphere formation assay’.
Soft agar colony formation assay
MCF-7 (5×102) and MDA-MB-231
(5×103) cells were resuspended in 2.7 ml DMEM and
Leibovitz's L-15 medium, respectively, without or with varying
doses of dinaciclib, before being mixed with 0.3 ml pre-warmed
(55°C) 3% agarose solution (in media). The cell suspension was then
seeded into three different wells of a 6-well plate (1 ml/well)
that were pre-coated with 2 ml 0.6% agarose in media. After 3 weeks
of culture, during which the media were replenished every 3 days,
the colonies were stained with MTT and counted using MetaMorph
software version 7.8 (Molecular Devices LLC).
Reverse transcription-quantitative
(RT-q)PCR
Total RNA was isolated from cells using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) and 5 µg RNA was reverse transcribed using a MMLV RT kit
(Thermo Fisher Scientific, Inc.). SYBR Green-based quantitative PCR
analysis was then carried using the CFX Connect™ Real-Time PCR
Detection system (Bio-Rad Laboratories, Inc.) with primer sets
designed to analyze the expression of specific genes including:
FOXM1 forward, 5′-CGTGGATTGAGGACCACTTT-3′ and reverse,
5′-TCTGCTGTGATTCCAAGTGC-3′; ACTB (β-actin) forward,
5′-TGGCATTGCCGACAGGAT-3′ and reverse, 5′-GCTCAGGAGGAGCAATGATCT-3′;
GLI1 forward, 5′-GTGCAAGTCAAGCCAGAACA-3′ and reverse,
5′-ATAGGGGCCTGACTGGAGAT-3′; PTCH1 forward,
5′-ACAAACTCCTGGTGCAAACC-3′ and reverse, 5′-CTTTGTCGTGGACCCATTCT-3′;
and Importin-7 (IPO7) forward, 5′-TCTGAAGGCATTTGCTGTTG-3′
and reverse, 5′-TGCCTTGTATATGGGGCTTC-3′. The reaction conditions
were: Initial denaturation at 95°C for 10 min, followed by 40
cycles at 95°C for 30 sec, 65°C for 30 sec and 72°C for 30 sec. The
relative quantity of target gene expression was calculated using
the comparative Cq method (ΔΔCq), which was normalized to
endogenous β-actin levels using CFX Manager version 3.1 (Bio-Rad
Laboratories, Inc.).
Examination of the involvement of the
proteasome in dinaciclib-induced decreases of the protein
expression levels of FoxM1 and three ESC markers in MCF-7
cells
Total lysates (20 µg) prepared from MCF-7 cells,
after they were treated without or with dinaciclib (10 nM) in the
absence or presence of 10 µM MG-132 (cat. no. BML-PI102-0005; Enzo
Life Sciences) for 24 h, were subjected to western blot analyses
using primary antibodies against FoxM1, p-FoxM1, Oct4, Nanog and
Sox2.
Establishment of the FoxM1-knockdown
stable clones from MCF-7 and MDA-MB-231 cells
Preparation of the lentivirus
The day before transfection, 8×105 293
cells were seeded into 6-cm dishes in DMEM. A DNA mixture
containing 0.25 µg pMDG, 2.25 µg pCMV-dR8.91 and 2.5 µg pLKO-FoxM1
short hairpin (sh)RNA (National RNAi Core Facility, Academia
Sinica, Taipei, Taiwan; sh1/shA, 5′-GCCAATCGTTCTCTGACAGAA-3′;
sh2/shB, 5′-AGGACCACTTTCCCTACTTTA-3′) was added into serum-free
DMEM to a final volume of 100 µl. Moreover, 7.5 µl PolyJet™ reagent
(Signagen Laboratories LLC) was mixed with serum-free DMEM to a
final volume of 100 µl. The diluted PolyJet™ reagent was then added
to the diluted DNA mixture, which was placed at room temperature
for 15 min to allow the formation of PolyJet/DNA complexes. Cells
were then treated with the aforementioned complexes at 37°C for 16
h before the medium was replaced with medium containing 1% BSA.
Viruses presenting in the medium were collected respectively at 24
and 48 h post-medium change and were stored at −80°C after being
passed through a 0.22-µm filter (MilliporeSigma).
Selection of stable clones
To establish the FoxM1-knockdown stable clones from
MCF-7 and MDA-MB-231 cell lines, cells were infected with the
aforementioned viruses expressing shRNA against FoxM1 for 48 h,
before being placed in a selection medium containing 1.5 µg/ml
puromycin. Single clones (designated as sh1 and sh2) were selected
and expanded, and the FoxM1 protein and mRNA expression levels of
them were examined via western blotting and RT-qPCR analyses,
respectively. The self-renewal abilities, as well as the protein
levels of various stemness markers and GLI1, in these clones were
analyzed as aforementioned.
Establishment of doxycycline-inducible
FoxM1-expressing HCC-1806 cells
The day before transfection, 6×105
HCC-1806 cells were seeded into 6-cm dishes containing RPMI-1640
medium. The plasmid pCW57.1-FOXM1b (Addgene, Inc.), which carries a
FOXM1 gene whose expression could be induced by doxycycline,
was added into serum-free RPMI-1640 medium to reach a final volume
of 100 µl. Furthermore, 7.5 µl PolyJet™ reagent was mixed with
serum-free RPMI-1640 medium to reach a final volume of 100 µl. Both
mixtures were left at room temperature for 15 min, before being
mixed and added to cells at 37°C for 16 h. The transfected cells
were selected using selection medium containing 1.5 µg/ml puromycin
and single clones were selected and expanded as aforementioned.
Doxycycline treatments
The aforementioned stable clones established from
HCC-1806 cells were then treated with varying concentrations of
doxycycline (0, 0.5, 1 and 2 µg/ml) at 37°C for 72 h before western
blotting was performed to determine what clones and concentration
of doxycycline should be used for the subsequent experiments. In
total, 1 µg/ml doxycycline was selected for inducing FoxM1
expression in HCC-1806 cells to assess the effects of FoxM1
upregulation on the cytotoxicity and sphere-forming inhibition of
dinaciclib in these cells.
Statistical analysis
Data are presented as the mean ± SD of three
independent experiments. Significant differences between untreated
or wild-type cells were determined by multiple comparison procedure
using one-way ANOVA with Tukey's post hoc test. P<0.05 was
considered to indicate a statistically significant difference.
Results
Examination of the cytotoxic effects
of dinaciclib on ER-positive MCF-7 and triple-negative HCC-1806
human BC cells
MTT assays were first performed to examine the
cytotoxic effects of dinaciclib on MCF-7 human luminal A
(ER+/PR+/HER2−) and HCC-1806 human
TNBC cells. As shown in Fig. S1,
the IC50 values of this drug on MCF-7 and HCC-1806 cells
were 8.3 and 29.0 nM, respectively.
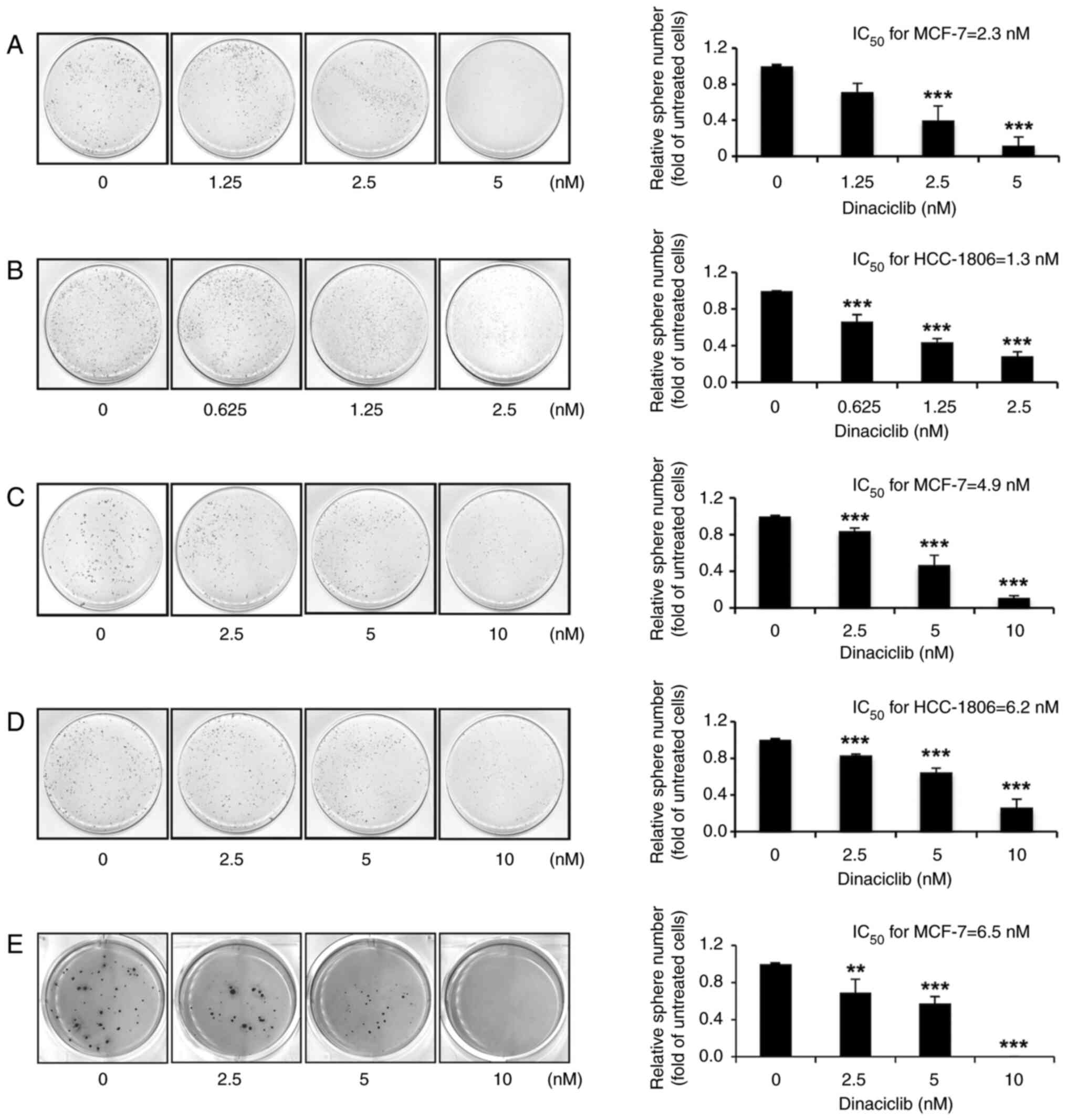
Dinaciclib is effective in suppressing
self-renewal and anchorage-independent growth abilities of MCF-7
and HCC-1806 cells
Having verified the cytotoxic effects of dinaciclib
on two human BC cell lines, it was next examined whether this drug
could reduce the self-renewal and anchorage-independent growth
abilities in these cells. By performing a sphere formation assay,
it was found that dinaciclib dose-dependently decreased the
sphere-forming abilities of both cells, and the respective
IC50 values of dinaciclib in suppressing the
self-renewal abilities of MCF-7 (Fig.
1A) and HCC-1806 (Fig. 1B)
cells were 2.3 and 1.3 nM. Subsequently, a sphere shrinkage assay
was used to analyze whether dinaciclib could induce shrinkage of
the pre-formed spheres. The results demonstrated that dinaciclib
also dose-dependently induced the shrinkage of the spheres formed
from both MCF-7 and HCC-1806 cells, with respective IC50
values of 4.9 (Fig. 1C) and 6.2 nM
(Fig. 1D).
Moreover, as the anchorage-independent growth
ability is considered as a hallmark of carcinogenesis, a soft agar
colony formation assay was conducted to evaluate the effects of
dinaciclib on this ability of MCF-7 cells, but not HCC-1806 cells,
as they failed to form colonies in soft agar (data not shown). As
expected, dinaciclib dose-dependently inhibited the
anchorage-independent growth of MCF-7 cells, with an
IC50 of 6.5 nM (Fig.
1E). These findings suggest that dinaciclib is capable of
inhibiting crucial stemness properties of human BC cells.
Dinaciclib decreases the protein
expression levels of various BCSC and ESC markers in MCF-7 and
HCC-1806 cells
Next, the protein expression levels of three
well-known BCSC markers (CD44, ALDH1 and Bmi1) and three ESC
markers (Oct4, Nanog and Sox2) were examined via western blotting
in MCF-7 and HCC-1806 cells treated without or with dinaciclib. In
agreement with the aforementioned observations, dinaciclib
decreased the expression levels of not only the three BCSC markers,
but also the three ESC markers in a dose-dependent manner in MCF-7
(Fig. 2A) and HCC-1806 (Fig. 2B) cells. Since these BCSC and ESC
markers are commonly used together for identifying BCSCs, the
current results further indicated the potential of dinaciclib in
diminishing the CSC populations in both BC lines.
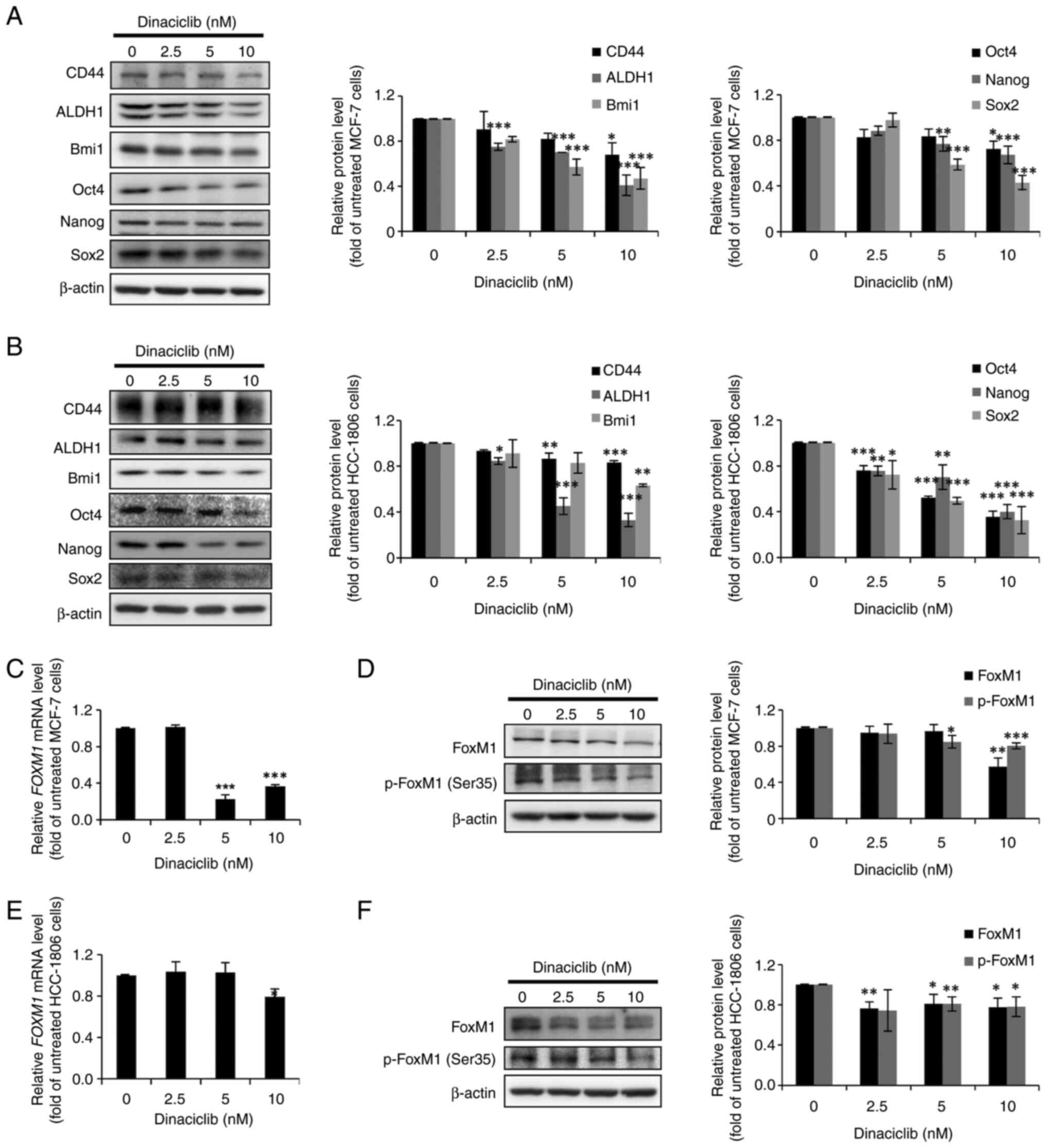 | Figure 2.Dinaciclib reduces the protein
expression levels of several breast cancer stem cell and embryonic
stem cell markers, as well as those of FoxM1 in MCF-7 and HCC-1806
cells. Total lysates (20 µg) prepared from (A and D) MCF-7 and (B
and F) HCC-1806 cells after being treated without or with indicated
doses of dinaciclib for 24 h were subjected to western blot
analysis using primary antibodies against CD44, ALDH1, Bmi1, Oct4,
Nanog, Sox2, total FoxM1 and p-FoxM1 (Ser35). β-actin was used as a
loading control. The semi-quantitative results were obtained via
densitometry. Total RNAs (5 µg) extracted from (C) MCF-7 and (E)
HCC-1806 cells after being treated without or with indicated doses
of dinaciclib for 24 h were subjected to reverse
transcription-quantiative PCR analysis to determine the mRNA
expression levels of FOXM1. Data are presented as the mean ±
SD of three independent experiments. *P<0.05, **P<0.01 and
***P<0.001 vs. untreated cells (using one-way ANOVA). ALDH1,
aldehyde dehydrogenase 1 family member A1; Bmi1, BMI1
proto-oncogene, polycomb ring finger; p-, phosphorylated; FoxM1,
forkhead box M1. |
FoxM1 expression and phosphorylation
are decreased by dinaciclib in MCF-7 and HCC-1806 cells
Given that certain CDKs can regulate the
transcriptional activity and stability of FoxM1 by phosphorylating
various serine and threonine residues, including Ser35 (14), and the fact that FoxM1 plays a
critical role in maintaining the stemness of a wide variety of
cancers including BC (15), we
hypothesized that some of the stemness-inhibitory effects of
dinaciclib in MCF-7 and HCC-1806 cells may be due to its effects on
the expression and/or stability of FoxM1. The RT-qPCR results
demonstrated that the mRNA expression levels of FoxM1 were
significantly decreased by dinaciclib treatment in both cell lines
(Fig. 2C and E). Furthermore, the
protein expression levels of total FoxM1 and p-FoxM1 (Ser35) in
both cell lines were also markedly decreased by dinaciclib
(Fig. 2D and F). Taken together,
these data demonstrated that dinaciclib could not only downregulate
the expression of FoxM1, but also decrease its phosphorylation at
Ser35 in MCF-7 and HCC-1806 cells.
Co-treatment with MG-132 can prevent
dinaciclib-induced reduction in the protein expression levels of
FoxM1, Nanog and Sox2 in MCF-7 cells
Since the phosphorylation of FoxM1 and several ESC
factors (e.g., Oct4, Nanog and Sox2) by various CDKs has been shown
to increase their respective stabilities by inhibiting
proteasome-mediated degradation (34), it was assessed whether the
decreased protein expression levels of FoxM1 and three ESC markers
in MCF-7 cells caused by dinaciclib treatment was due to
proteasomal degradation. As shown in Fig. 3, co-treatment with MG-132, a
proteasome inhibitor, completely prevented the decreases in the
protein expression levels of total FoxM1, p-FoxM1 (Ser35) and
Nanog, as well as partially hindered the reduction of Sox2
expression in cells induced by dinaciclib. However, no significant
recovery of Oct4 expression was observed after this co-treatment.
These findings suggested that dinaciclib decreased the stability of
FoxM1, Nanog and Sox2 proteins by inducing their proteasomal
degradation in MCF-7 cells.
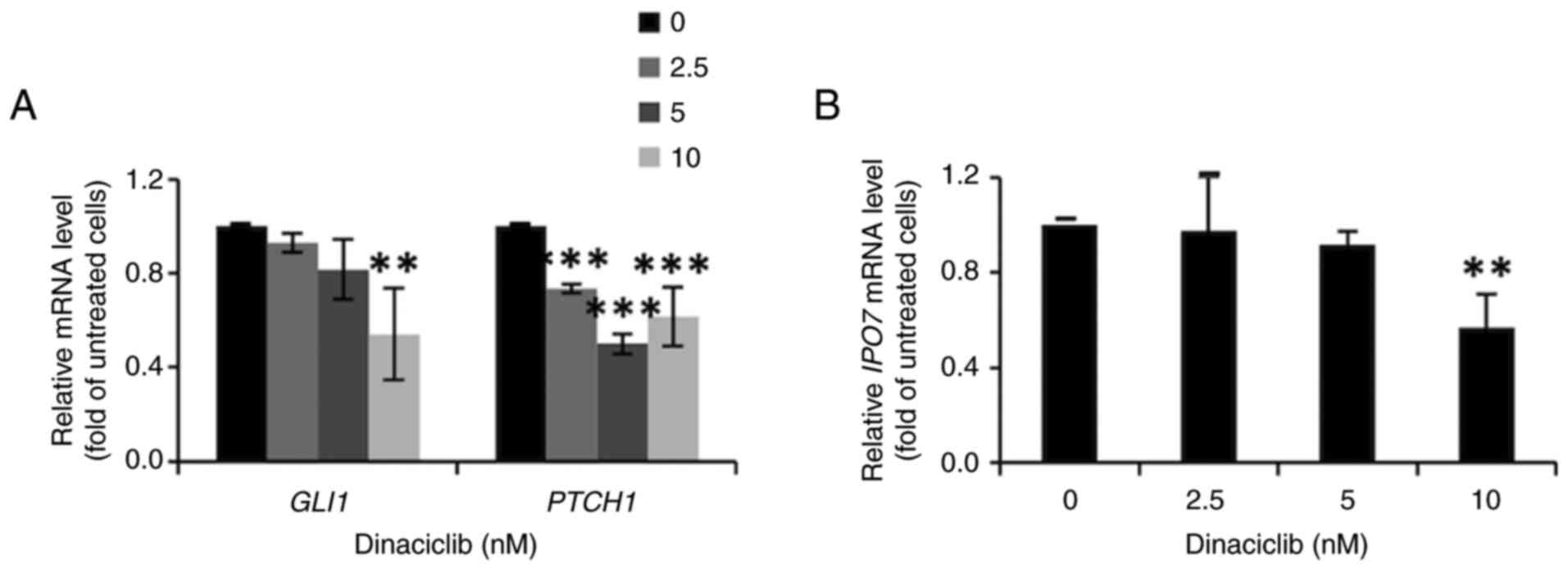
Dinaciclib inhibits the Hedgehog
pathway by decreasing GLI1 protein expression in MCF-7 cells
To determine the stemness-inhibitory mechanisms of
dinaciclib in MCF-7 cells, the effects of this drug on Wnt, Notch
and Hedgehog pathways, three of the main stemness-regulatory
signaling pathways, were examined in these cells. After excluding
the involvements of the first two pathways (data not shown), the
current study focused on the Hedgehog pathway by analyzing the
nuclear expression level of GLI1, the crucial mediator of this
pathway, as well as the mRNA expression levels of its downstream
target genes, including GLI1 and PTCH1. The western
blotting results showed dinaciclib dose-dependently decreased the
nuclear expression level of GLI1 in MCF-7 cells (Fig. S2). In accordance with these
findings, the mRNA expression levels of two downstream genes
GLI1 and PTCH1 were also markedly diminished by this
drug (Fig. 4A).
Since the nuclear translocation of GLI1 has been
shown to be enhanced by IPO7, whose promoter is able to be
activated by FoxM1 (37), it was
next examined whether dinaciclib affected the expression of
IPO7. Indeed, the RT-qPCR results indicated that the mRNA
expression levels of IPO7 in MCF-7 cells were significantly
downregulated after treatment with 10 nM dinaciclib (Fig. 4B). Collectively, these findings
suggest that the stemness-inhibitory effect of dinaciclib in MCF-7
cells was attributed, at least in part, to its suppression of the
Hedgehog pathway.
FoxM1 knockdown in MCF-7 cells
diminishes their stemness properties and inhibits the expression of
GLI1
Having demonstrated that dinaciclib could suppress
both the expression and phosphorylation of FoxM1 in both cell lines
(Fig. 2C-F), it was then assessed
whether their stemness was also regulated by this TF. Thus,
FoxM1-knockdown stable clones were established (designated as sh1
and sh2, or as shA and shB for MCF-7 and MDA-MB-231 cells,
respectively), whose FOXM1 mRNA (Fig. 5A and C) and protein (Fig. 5B and D) expression levels were
significantly lower than their parental counterparts.
Sphere formation assays were then conducted to
investigate the self-renewal abilities of FoxM1-knockdown MCF-7
clones. As expected, the sphere-forming abilities of the
FoxM1-knockdown MCF-7 clones were markedly reduced (Fig. 5E). Moreover, the
anchorage-independent growth abilities of the clones derived from
MDA-MB-231 cells were diminished (Fig.
5F). In addition, the western blotting results demonstrated
notable and significant decreases in the protein expression levels
of three BCSC (CD44, ALDH1 and Bmi1) and three ESC (Oct4, Nanog,
and Sox2) markers in the FoxM1-knockdown MCF-7 stable clones
(Fig. 6A). The protein expression
levels of CD44 and ALDH1 were also significantly decreased in the
FoxM1-knockdown MDA-MB-231 clones (Fig. 6B). It was also assessed whether
FoxM1 knockdown in MCF-7 cells could diminish total GLI1 expression
as was observed with dinaciclib treatment. As expected, the mRNA
and protein expression levels of GLI1 were significantly reduced in
these clones (Fig. 6C and D).
Collectively, these findings demonstrated an important role of
FoxM1 in maintaining the stemness of two types of BC cells, and
suggested that dinaciclib may inhibit their stemness properties by
targeting two well-known stemness-regulatory TFs, FoxM1 and
GLI1.
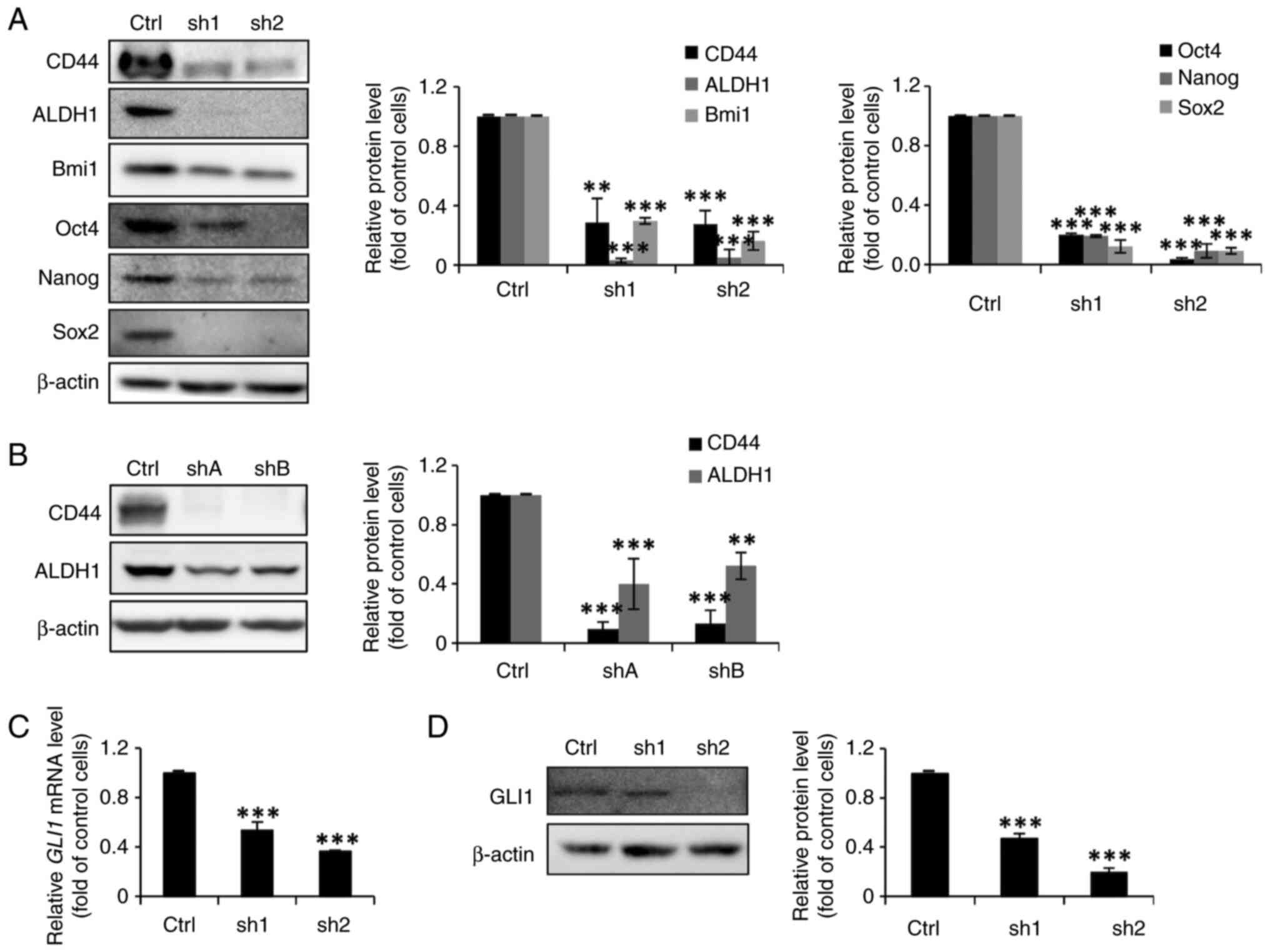 | Figure 6.FoxM1 knockdown in MCF-7 and
MDA-MB-231 cells decreases the protein expression levels of several
stem cell markers and GLI1. (A, B and D) Total lysates (20 µg)
prepared from the FoxM1-knockdown stable clones and their Ctrl
MCF-7 (sh1 and sh2 in panels A and D) and MDA-MB-231 (shA and shB
in panel B) cells were subjected to western blot analysis, using
primary antibodies against CD44, ALDH1, Bmi1, Oct4, Nanog, Sox2 and
GLI1. β-actin was used as a loading control. The semi-quantitative
results were obtained via densitometry. (C) Total RNAs (5 µg)
extracted from the MCF-7 FoxM1-knockdown stable clones were
subjected to reverse transcription-quantitative PCR analysis to
determine the mRNA expression levels of GLI1. Data are
presented as the mean ± SD of three independent experiments.
**P<0.01 and ***P<0.001 vs. Ctrl cells (using one-way ANOVA).
Ctrl, wild-type cells; sh, short hairpin RNA; FoxM1, forkhead box
M1; GLI1, GLI family zinc finger 1; ALDH1, aldehyde dehydrogenase 1
family member A1; Bmi1, BMI1 proto-oncogene, polycomb ring
finger. |
Upregulation of FoxM1 in HCC-1806
cells increases their resistance to the cytotoxic and
sphere-forming inhibitory effects of dinaciclib
To further investigate whether the upregulation of
FoxM1 in human BC cells could affect the effects of dinaciclib, a
doxycycline-inducible FoxM1-expressing clone was established from
HCC-1806 cells. The results demonstrated that the protein
expression levels of FoxM1, as well as CD44 and ALDH1, two stemness
markers, were dose-dependently elevated by doxycycline in this
clone (Fig. S3A). MTT assays were
then performed to examine whether the upregulation of FoxM1 in
these cells affected their sensitivity to dinaciclib. As shown in
Fig. S3B, FoxM1 upregulation
markedly increased the resistance of doxycycline-inducible
FoxM1-expressing HCC-1806 cells to the cytotoxic effects of
dinaciclib. Finally, the results from a sphere formation assay
further suggested that the self-renewal inhibitory effect of
dinaciclib was notably diminished by the upregulation of FoxM1 in
these clones (data not shown).
Discussion
Over the past few years, several cyclin-dependent
kinase (CDK)4/6 inhibitors, including palbociclib, abemaciclib and
ribociclib, have been approved by the US Food and Drug
Administration, for use in combination with the aromatase inhibitor
letrozole for treating patients with ER+HER2−
metastatic BC (24–26). However, whether palbociclib can
suppress the self-renewal of different types of breast cancer (BC)
remains controversial (24,28,29).
Dinaciclib is a novel CDK1/2/5/9 inhibitor, and its
therapeutic effects on several types of cancer, including BC,
leukemia and lymphoma, are being assessed in various clinical
trials (31,40–42).
Accumulating evidence suggests the involvements of CDK1/2/5/9 in
regulating the stemness of normal and cancer cells, as well as the
cancer stem cell (CSC)-suppressive effects of various selective
inhibitors against these CDKs. For example, overexpression of CDK1
enhances the spheroid-forming ability, tumorigenic potential and
tumor-initiating capacity of human melanoma cells by increasing the
phosphorylation, nuclear localization and transcriptional activity
of Sox2 (43). Moreover, the
aberrant activation of cyclin E/CDK2 oncogenic signaling is
essential for the maintenance and expansion of the breast cancer
stem cell (BCSC) subpopulation, which is more sensitive to SU9516,
a specific CDK2 inhibitor (18).
The Nestin/CDK5/dynamin-related protein 1 axis inhibits
mitochondrial oxidative phosphorylation, which is essential for the
maintenance of the stemness of neural stem/progenitor cells
(43). In addition, CDK9 signaling
plays an important role in brain tumor-initiating cell maintenance,
and dinaciclib treatment can attenuate tumor growth and prolong the
survival of tumor-bearing mice (44).
The present study aimed to identify whether
dinaciclib could inhibit the stemness of human BC cells. Estrogen
receptor-positive (ER+) MCF-7 and triple-negative breast
cancer (TNBC) HCC-1806 cell lines were selected as models, and it
was found that the former was more susceptible to dinaciclib.
Interestingly, although the protein expression levels of total and
p-FoxM1 were significantly reduced by dinaciclib in both MCF-7 and
HCC-1806 cells, the effects on the former could be accounted mainly
by the drug-induced transcriptional suppression, whereas those in
the latter may be attributed to the decreased stability of FoxM1
due to the inhibition of CDK1/2-mediated phosphorylation of this
transcription factor (TF) (15).
In terms of the regulation of FoxM1 protein, multiple CDK
phosphorylation sites on FoxM1 variants (FoxM1b and c) have been
identified, and the phosphorylation on some of them (e.g., Ser35)
can enhance both protein stability and transcriptional activity of
FoxM1 (15). As the protein level
of total FoxM1 should be composed of both the unphosphorylated and
phosphorylated forms of FoxM1, it was expected to see a more
dramatic decrease in total FoxM1 protein level. Thereby, it was not
too surprising to find that both total and phospho-(Ser35)-FoxM1
levels were increased by MG-132 co-treatment (Fig. 3).
In agreement with earlier observations made by other
investigators, the present study demonstrated that dinaciclib could
suppress the sphere formation and induce sphere shrinkage in MCF-7
and HCC-1806 cells, as well as inhibit the anchorage-independent
growth of the former, thereby confirming the stemness-suppressive
effects of this drug in human BC cells. Additionally, it was
discovered that dinaciclib significantly decreased the protein
expression levels of not only the three BCSC markers CD44, ALDH1
and Bmi1, but also of the three ESC markers Oct4, Nanog and
Sox2.
Regarding the inhibitory effects of this drug on the
protein expression levels of three ESC markers, previous studies
have shown that respective CDK-mediated phosphorylation at the
Ser/Thr-Pro motifs of Nanog and at the Ser12-Pro motif of Oct4
could promote their interactions with Pin1, leading to the
increases of their stability and transcriptional activity (45,46).
Moreover, the cyclin E/CDK2 and cyclin D3/CDK6 complexes have been
reported to phosphorylate Nanog, Oct4 and Sox2, thereby promoting
their interactions with Pin1 and protecting them from
polyubiquitylation and proteasomal degradation (34,43).
It has also been shown that the protein expression levels of Nanog,
Oct4 and Sox2 in murine ESCs, patient-derived glioblastoma
tumor-initiating cells or TNBC cells are notably decreased by
CVT-313 treatment, which is a highly selective inhibitor of CDK2
(34). CDK1 has also been reported
to bind to Sox2 and phosphorylate its Ser249, 250 and 251,
resulting in an increased nuclear translocation and transcriptional
activity of this factor, ultimately promoting the tumor-initiating
potential of human melanoma cells (34). In agreement with the aforementioned
observations, the present study identified that the decreased
protein expression levels of two ESC markers, as well as those of
total and p-FoxM1, triggered by dinaciclib were significantly
restored by the co-treatment with MG-132, a proteasome inhibitor.
These findings suggested that dinaciclib may inhibit the stemness
of human BC cells by suppressing the expression of FoxM1 and three
core pluripotency TFs in ESCs.
FoxM1 has been implicated in stimulating not only
BCSC (47), but also
TNBC/basal-like BC phenotypes (48). Although FoxM1 has been reported to
be a downstream component of the Wnt signaling pathway critical for
β-catenin transcriptional function in human glioma cells (38), the current study failed to detect
any effect of dinaciclib on the nuclear β-catenin expression in
MCF-7 cells (data not shown). Neither was Notch signaling, which
has not yet been associated with FoxM1, to the best of our
knowledge, affected by this drug (data not shown). Therefore, the
current study assessed the effects of dinaciclib on the
Hedgehog/Gli signaling pathway in MCF-7 cells, since Gli1 has been
shown to activate FoxM1 expression in murine cerebral neural SCs
(49), as well as in human basal
cell carcinomas (50) and
colorectal cancer cells (51). As
expected, dinaciclib reduced the nuclear protein expression level
of Gli1. Furthermore, this drug also suppressed the expression of
Gli1 and PTCH1, two well-known downstream target genes of Gli1 in
MCF-7 cells. The present study also detected a significant decrease
in IPO7 mRNA expression in these cells after they were treated with
10 nM dinaciclib, which may be due to the drug-induced reduction of
FoxM1, a known transcriptional activator of IPO7 (37). Additionally, as IPO7 has been shown
to facilitate the nuclear translocation of Gli1 in glioblastoma
multiforme cells (37), its
downregulation may also account in part for the decreased nuclear
Gli1 expression in drug-treated cells. Based on these findings, it
was suggested that FoxM1 may also act as an upstream regulator of
the Hedgehog signaling pathway. In line with this suggestion, it
was further demonstrated that the self-renewal ability and protein
expression levels of three embryonic SC (ESC) and three BCSC
markers, as well as those of Gli1, in MCF-7 cells were notably
suppressed by FoxM1 knockdown. Data of the time course experiments
of FoxM1 downregulation were not provided as previous results (data
not shown) suggested limited cytotoxic effects of FoxM1 knockdown
on cell growth which was in contrast to the findings in a number of
previous studies which showed a positive role of FoxM1 in
regulating the growth of various types of cancer cells (reviewed in
ref. 52) and the reasons to explain the discrepancy between our and
others' observations need further dissection. On the other hand,
transient upregulation of FoxM1 in HCC-1806 cells markedly
increased their resistance to both the cytotoxic and sphere-forming
inhibitory effects of dinaciclib, which indicated FoxM1 as the
major target of this novel CDK inhibitor.
In conclusion, the present study demonstrated that
dinaciclib, a CDK1/2/5/9 inhibitor, not only kills MCF-7
(ER-positive) and HCC-1806 (triple-negative) human BC cells, but
also effectively suppresses their stemness. In addition, the
stemness-inhibitory effects of this drug in the former cells are
likely achieved by diminishing the expression and phosphorylation
of FoxM1, resulting in decreases in the expression of three core
pluripotency TFs (Nanog, Oct4, and Sox2) as well as in the Hedgehog
signaling pathway. Thus, the present findings support the clinical
development of dinaciclib for BC treatment.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This work was supported by grants from the Taiwan Clinical
Oncology Research Foundation and the Ministry of Science and
Technology (grant nos. MOST 105-2320-B-010-030 and
106-2320-B-010-003).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
ANT was a major contributor who performed the
dinaciclib (and MG-132) treatment on both wild-type MCF-7 and
HCC-1806 cell lines to generate, analyze, and interpret the data
regarding sphere formation and shrinkage assays, soft agar colony
formation assays, western blot and RT-qPCR, and the MTT assay. ANT
established the FoxM1-knockdown stable clones from MCF-7 cell line,
performed the experiments regarding these clones, and generated,
analyzed, and interpreted the data. YSC established the
FoxM1-knockdown stable clones from MDA-MB-231 cell line and the
doxycycline-inducible FoxM1-expressing HCC-1806 cells, and used the
aforementioned cells to perform western blot analysis, RT-qPCR, and
soft agar colony formation assays. YCL used the
doxycycline-inducible FoxM1-expressing HCC-1806 cells established
by YC to perform the MTT assay. ANT analyzed and interpreted the
raw data generated by YCL and YSC. YS and TCC were the principal
investigators who supervised this study and provided significant
guidance and instructions for each result. All authors read and
approved the manuscript and agree to be accountable for all aspects
of the research in ensuring that the accuracy or integrity of any
part of the work, in particular the data, are appropriately
investigated and resolved.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declared that they have no competing
interests.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ghoncheh M, Pournamdar Z and Salehiniya H:
Incidence and mortality and epidemiology of breast cancer in the
world. Asian Pac J Cancer Prev. 17:43–46. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Nguyen PL, Taghian AG, Katz MS, Niemierko
A, Abi Raad RF, Boon WL, Bellon JR, Wong JS, Smith BL and Harris
JR: Breast cancer subtype approximated by estrogen receptor,
progesterone receptor, and HER-2 is associated with local and
distant recurrence after breast-conserving therapy. J Clin Oncol.
26:2373–2378. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sotiriou C and Pusztai L: Gene-expression
signatures in breast cancer. N Engl J Med. 360:790–800. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
André F and Zielinski CC: Optimal
strategies for the treatment of metastatic triple-negative breast
cancer with currently approved agents. Ann Oncol. 23 (Suppl
6):vi46–vi51. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Perou CM, Sørlie T, Eisen MB, van de Rijn
M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA,
et al: Molecular portraits of human breast tumours. Nature.
406:747–752. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Al-Hajj M, Wicha MS, Benito-Hernandez A,
Morrison SJ and Clarke MF: Prospective identification of
tumorigenic breast cancer cells. Proc Natl Acad Sci USA.
100:3983–3988. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Malumbres M and Barbacid M: Cell cycle,
Cdks and cancer: A changing paradigm. Nat Rev Cancer. 9:153–166.
2009. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Plaks V, Kong N and Werb Z: The cancer
stem cell niche: How essential is the niche in regulating stemness
of tumor cells? Cell Stem Cell. 16:225–238. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Pavlopoulou A, Oktay Y, Vougas K, Louka M,
Vorgias CE and Georgakilas AG: Determinants of resistance to
chemotherapy and ionizing radiation in breast cancer stem cells.
Cancer Lett. 380:485–493. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Evans T, Rosenthal ET, Youngblom J, Distel
D and Hunt T: Cyclin: A protein specified by maternal mRNA in sea
urchin eggs that is destroyed at each cleavage division. Cell.
33:389–396. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Suryadinata R, Sadowski M and Sarcevic B:
Control of cell cycle progression by phosphorylation of
cyclin-dependent kinase (CDK) substrates. Biosci Rep. 30:243–255.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Nurse P and Thuriaux P: Regulatory genes
controlling mitosis in the fission yeast Schizosaccharomyces pombe.
Genetics. 96:627–637. 1980. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lim S and Kaldis P: Cdks, cyclins and
CKIs: Roles beyond cell cycle regulation. Development.
140:3079–3093. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Song X, Kenston SSF, Zhao J, Yang D and Gu
Y: Roles of FoxM1 in cell regulation and breast cancer targeting
therapy. Med Oncol. 34:412017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lange C and Calegari F: Cdks and cyclins
link G1 length and differentiation of embryonic, neural and
hematopoietic stem cells. Cell Cycle. 9:1893–1900. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Neganova I, Tilgner K, Buskin A,
Paraskevopoulou I, Atkinson SP, Peberdy D, Passos JF and Lako M:
CDK1 plays an important role in the maintenance of pluripotency and
genomic stability in human pluripotent stem cells. Cell Death Dis.
5:e15082014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Opyrchal M, Salisbury JL, Iankov I, Goetz
MP, McCubrey J, Gambino MW, Malatino L, Puccia G, Ingle JN, Galanis
E and D'Assoro AB: Inhibition of Cdk2 kinase activity selectively
targets the CD44+/CD24−/low stem-like
subpopulation and restores chemosensitivity of SUM149PT
triple-negative breast cancer cells. Int J Oncol. 45:1193–1199.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sun LH, Ban T, Liu CD, Chen QX, Wang X,
Yan ML, Hu XL, Su XL, Bao YN, Sun LL, et al: Activation of Cdk5/p25
and tau phosphorylation following chronic brain hypoperfusion in
rats involves microRNA-195 down-regulation. J Neurochem.
134:1139–1151. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
De Falco G and Giordano A: CDK9: From
basal transcription to cancer and AIDS. Cancer Biol Ther.
1:342–347. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hatch VL, Marin-Barba M, Moxon S, Ford CT,
Ward NJ, Tomlinson ML, Desanlis I, Hendry AE, Hontelez S, van
Kruijsbergen I, et al: The positive transcriptional elongation
factor (P-TEFb) is required for neural crest specification. Dev
Biol. 416:361–372. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cortés N, Guzmán-Martínez L, Andrade V,
González A and Maccioni RB: CDK5: A unique CDK and its multiple
roles in the nervous system. J Alzheimers Dis. 68:843–855. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hanahan D and Weinberg RA: The hallmarks
of cancer. Cell. 1:57–70. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Turner NC, Slamon DJ, Ro J, Bondarenko I,
Im SA, Masuda N, Colleoni M, DeMichele A, Loi S, Verma S, et al:
Overall survival with palbociclib and fulvestrant in advanced
breast cancer. N Engl J Med. 379:1926–1936. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Im SA, Lu YS, Bardia A, Harbeck N,
Colleoni M, Franke F, Chow L, Sohn J, Lee KS, Campos-Gomez S, et
al: Overall survival with ribociclib plus endocrine therapy in
breast cancer. N Engl J Med. 381:307–316. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Goetz MP, Toi M, Campone M, Sohn J,
Paluch-Shimon S, Huober J, Park IH, Trédan O, Chen SC, Manso L, et
al: MONARCH 3: Abemaciclib as initial therapy for advanced breast
cancer. J Clin Oncol. 35:3638–3646. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Portman N, Alexandrou S, Carson E, Wang S,
Lim E and Caldon CE: Overcoming CDK4/6 inhibitor resistance in
ER-positive breast cancer. Endocr Relat Cancer. 26:R15–R30. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chen L, Yang G and Dong H: Everolimus
reverses palbociclib resistance in ER+ human breast cancer cells by
inhibiting phosphatidylinositol 3-kinase(PI3K)/Akt/mammalian target
of rapamycin (mTOR) pathway. Med Sci Monit. 25:77–86. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kettner NM, Vijayaraghavan S, Durak MG,
Bui T, Kohansal M, Ha MJ, Liu B, Rao X, Wang J, Yi M, et al:
Combined Inhibition of STAT3 and DNA repair in
palbociclib-resistant ER-positive breast cancer. Clin Cancer Res.
25:3996–4013. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chien AJ, Rahmaputri S, Dittrich HF,
Majure MC, Rugo HS, Melisko ME and Goga A: A phase Ib trial of the
cyclin-dependent kinase inhibitor dinaciclib (dina) in combination
with pembrolizumab (P) in patients with advanced triple-negative
breast cancer (TNBC). J Clin Oncol. 37 (Suppl 15):S10722019.
View Article : Google Scholar
|
|
31
|
Rajput S, Khera N, Guo Z, Hoog J, Li S and
Ma CX: Inhibition of cyclin dependent kinase 9 by dinaciclib
suppresses cyclin B1 expression and tumor growth in triple negative
breast cancer. Oncotarget. 7:56864–56875. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kang J, Sergio CM, Sutherland RL and
Musgrove EA: Targeting cyclin-dependent kinase 1 (CDK1) but not
CDK4/6 or CDK2 is selectively lethal to MYC-dependent human breast
cancer cells. BMC Cancer. 14:322014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tay Y, Zhang J, Thomson AM, Lim B and
Rigoutsos I: MicroRNAs to Nanog, Oct4 and Sox2 coding regions
modulate embryonic stem cell differentiation. Nature.
455:1124–1128. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Liu L, Michowski W, Inuzuka H, Shimizu K,
Nihira NT, Chick JM, Li N, Geng Y, Meng AY, Ordureau A, et al: G1
cyclins link proliferation, pluripotency and differentiation of
embryonic stem cells. Nat Cell Biol. 19:177–188. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Anders L, Ke N, Hydbring P, Choi YJ,
Widlund HR, Chick JM, Zhai H, Vidal M, Gygi SP, Braun P and
Sicinski P: A systematic screen for CDK4/6 substrates links FOXM1
phosphorylation to senescence suppression in cancer cells. Cancer
Cell. 20:620–634. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wierstra I: Cyclin D1/Cdk4 increases the
transcriptional activity of FOXM1c without phosphorylating FOXM1c.
Biochem Biophys Res Commun. 431:753–759. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Xue J, Zhou A, Tan C, Wu Y, Lee HT, Li W,
Xie K and Huang S: Forkhead box M1 is essential for nuclear
localization of glioma-associated oncogene homolog 1 in
glioblastoma multiforme cells by promoting importin-7 expression. J
Biol Chem. 290:18662–18670. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhang N, Wei P, Gong A, Chiu WT, Lee HT,
Colman H, Huang H, Xue J, Liu M, Wang Y, et al: FoxM1 promotes
β-catenin nuclear localization and controls Wnt target-gene
expression and glioma tumorigenesis. Cancer Cell. 20:427–442. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Elango R, Vishnubalaji R, Manikandan M,
Binhamdan SI, Siyal AA, Alshawakir YA, Alfayez M, Aldahmash A and
Alajez NM: Concurrent targeting of BMI1 and CDK4/6 abrogates tumor
growth in vitro and in vivo. Sci Rep. 9:136962019. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ghia P, Scarfò L, Perez S, Pathiraja K,
Derosier M, Small K, McCrary Sisk C and Patton N: Efficacy and
safety of dinaciclib vs ofatumumab in patients with
relapsed/refractory chronic lymphocytic leukemia. Blood.
129:1876–1878. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Gregory G, Walker P, Mahadevan D, Wang D,
Chang J, Hernandez-Ilizaliturri F, Klein A, Rybka W,
Wagner-Johnston N, Escobar C, et al: Antitumor activity of
pembrolizumab plus dinaciclib in patients with diffuse large b cell
lymphoma: The phase 1b keynote-155 study. Hematol Oncol.
37:328–329. 2019. View Article : Google Scholar
|
|
42
|
Ravindran Menon D, Luo Y, Arcaroli JJ, Liu
S, KrishnanKutty LN, Osborne DG, Li Y, Samson JM, Bagby S, Tan AC,
et al: CDK1 interacts with Sox2 and promotes tumor initiation in
human melanoma. Cancer Res. 78:6561–6574. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wang J, Huang Y, Cai J, Ke Q, Xiao J,
Huang W, Li H, Qiu Y, Wang Y, Zhang B, et al: A
nestin-cyclin-dependent kinase 5-dynamin-related protein 1 axis
regulates neural stem/progenitor cell stemness via a metabolic
shift. Stem Cells. 36:589–601. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Xie Q, Wu Q, Kim L, Miller TE, Liau BB,
Mack SC, Yang K, Factor DC, Fang X, Huang Z, et al: RBPJ maintains
brain tumor-initiating cells through CDK9-mediated transcriptional
elongation. J Clin Invest. 126:2757–2772. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Moretto-Zita M, Jin H, Shen Z, Zhao T,
Briggs SP and Xu Y: Phosphorylation stabilizes Nanog by promoting
its interaction with Pin1. Proc Natl Acad Sci USA. 107:13312–13317.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Nishi M, Akutsu H, Masui S, Kondo A,
Nagashima Y, Kimura H, Perrem K, Shigeri Y, Toyoda M, Okayama A, et
al: A distinct role for Pin1 in the induction and maintenance of
pluripotency. J Biol Chem. 286:11593–11603. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Yang N, Wang C, Wang Z, Zona S, Lin SX,
Wang X, Yan M, Zheng FM, Li SS, Xu B, et al: FOXM1 recruits nuclear
Aurora kinase A to participate in a positive feedback loop
essential for the self-renewal of breast cancer stem cells.
Oncogene. 36:3428–3440. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Ring A, Nguyen C, Smbatyan G, Tripathy D,
Yu M, Press M, Kahn M and Lang JE: CBP/β-catenin/FOXM1 is a novel
therapeutic target in triple negative breast cancer. Cancers
(Basel). 10:5252018. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Besharat ZM, Abballe L, Cicconardi F,
Bhutkar A, Grassi L, Le Pera L, Moretti M, Chinappi M, D'Andrea D,
Mastronuzzi A, et al: Foxm1 controls a pro-stemness microRNA
network in neural stem cells. Sci Rep. 8:35232018. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Teh MT, Wong ST, Neill GW, Ghali LR,
Philpott MP and Quinn AG: FOXM1 is a downstream target of Gli1 in
basal cell carcinomas. Cancer Res. 62:4773–4780. 2002.PubMed/NCBI
|
|
51
|
Wang D, Hu G, Du Y, Zhang C, Lu Q, Lv N
and Luo S: Aberrant activation of hedgehog signaling promotes cell
proliferation via the transcriptional activation of forkhead box M1
in colorectal cancer cells. J Exp Clin Cancer Res. 36:232017.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Kalathil D, John S and Nair AS: FOXM1 and
cancer: Faulty cellular signaling derails homeostasis. Front Oncol.
10:6268362021. View Article : Google Scholar : PubMed/NCBI
|