Introduction
In recent years, the incidence of malignant melanoma
has been increasing rapidly worldwide, with an annual growth rate
of 3–5%, and has become one of the most rapidly growing tumors
(1). Malignant melanoma is
associated with a poor prognosis and a high mortality rate; there
are also limited treatment options available (2). At present, although research progress
has been made as regards the pathogenesis of malignant melanoma,
the exact molecular mechanisms of malignant melanoma remain to be
fully elucidated.
It has been recently reported that the occurrence of
melanoma may not only be dependent on genetic factors, but may be
also affected by epigenetic modifications, particularly DNA
methylation (3). CpG island
methylation of tumor suppressor gene promoters has been proven to
be related to the occurrence and progression of multiple human
malignant tumors, and can also be used as a biomarker to evaluate
the prognosis of melanoma (4). For
example, Tanemura et al (5)
analyzed the CpG island methylation status of tumor-related gene
promoter region and revealed that the hypermethylation of RASSF1A,
WIF1, SOCS1 and TFPI2 was increased with the progression of
malignant melanoma clinical stage, with MINT31 methylation in stage
III melanoma being related to disease prognosis. Therefore, the
further understanding of melanoma-related DNA methylation genes may
prove to be of utmost importance for elucidating the regulatory
mechanisms of gene expression in melanoma and melanoma occurrence
and development.
In addition, long non-coding RNAs (lncRNAs) play a
crucial regulatory role in cell physiological activities, and their
abnormal expression has been closely related to the occurrence and
development of tumors (6). Among
lncRNAs, growth arrest-specific transcript 5 (GAS5) is a lncRNA
with 651 nucleotides that is expressed at low levels in a variety
of tumors. The GAS5 expression level has been shown to be closely
related to the clinicopathological characteristics of tumors and
patient prognosis, and GAS5 overexpression has been reported to
inhibit the growth of metastasized tumors in vivo (7,8).
Studies have demonstrated that lncRNAs may be also affected by DNA
methylation (9,10). For example, Cheng et al
(10) previously revealed that
nine lncRNA methylated genes were significantly related to the
overall survival of patients with glioma, which could be used to
predict the risk of glioma patients. Although the expression of
GAS5 has been reported to be downregulated in melanoma (8), it has not been specified whether this
may be caused by methylation modification.
Gene silencing caused by DNA methylation has been
reported to be important for tumor occurrence and development, and
demethylation agents can restore the expression of silenced genes
and continue to exert their antitumor biological effects (11). 5-Aza-2′-deoxycytidine (5-Aza-dC) is
currently known as a markedly effective demethylating agent that
can block cell cycle, induce apoptosis, promote differentiation,
reduce invasion and metastasis, and inhibit tumor cell growth
(12). However, there are few
studies on 5-Aza-dC in melanoma (13,14).
Therefore, present study aimed to investigate GAS5 methylation
levels in melanoma and to confirm the role of 5-Aza-dC in melanoma,
in order to provide therapeutic targets and diagnostic methods for
the treatment of melanoma.
Materials and methods
Patient samples and bioinformatics
analysis
Fresh-frozen and paraffin-embedded tissues from
patients diagnosed with melanoma (n=8) at Yunnan Cancer Hospital
were analyzed. Fresh-frozen biopsy specimens from the nevus of
eight individuals were used as control samples. All skin biopsy
specimens contained at least 70% of the cells as normal or melanoma
cells. The present study was approved by the Ethics Committee of
Yunnan Cancer Hospital, the Third Affiliated Hospital of Kunming
Medical University (KY201939). Written informed consent was
obtained from each patient included in this study. The analysis of
GAS5 expression level was derived from The Cancer Genome Atlas
(TCGA; http://portal.gdc.cancer.gov/)
database, and the correlation between GAS5 methylation and
expression was downloaded from the MEXPRESS (https://mexpress.be/) database.
Cells and cell culture
Human melanoma cell line A375 (SCSP-533) was
purchased from The Cell Bank of Type Culture Collection of the
Chinese Academy of Sciences, and SK-MEL-110 was provided by the
authors' laboratory. The cells were cultured in Dulbecco's modified
Eagle medium (DMEM; Invitrogen; Thermo Fisher Scientific, Inc.)
supplemented with 10% fetal bovine serum (FBS; Invitrogen; Thermo
Fisher Scientific, Inc.), 100 U/ml penicillin (Gibco; Thermo Fisher
Scientific, Inc.) and 100 µg/ml streptomycin (Gibco; Thermo Fisher
Scientific, Inc.). All cells were maintained at 37°C in an
incubator with 5% (v/v) CO2. Cells were incubated with
or without 20 µM 5-Aza-dC (Sigma-Adrich; Merck KGaA) for 48 h as
previously described (15).
Transfection
GAS5 siRNA (5′-CUUGCCUGGACCAGCUUAATT-3′) and
negative control (NC) siRNA (5′-UUCUCCGAACGUGUCACGUTT-3′) were
synthesized by Guangzhou RiboBio Co., Ltd. A total quantity of 50
nM GAS5 siRNA or NC siRNA was transfected into melanoma cells at
37°C using Lipofectamine 3000® (Invitrogen; Thermo
Fisher Scientific, Inc.) according to the manufacturer's protocol.
siRNA and transfection reagent were mixed and incubated for 20 min
at room temperature, and then added to the cells. Following 4 h of
culture, the cells were replaced with fresh medium to continue
culture. Subsequently, the GAS5 expression level was analyzed at 24
h post-transfection.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted using TRIzol®
(Invitrogen; Thermo Fisher Scientific, Inc.) from melanoma cells or
tissues, and mRNA expression levels were analyzed using a One Step
TB Green PrimeScript RT-PCR kit (Takara Biotechnology Co., Ltd.)
according to the manufacturer's instructions. The reactions were
performed as followed: 5 min at 42°C, 10 at 95°C, and 30 cycles of
5 sec at 95°C, 30 sec at 60°C. The primer sequences used were as
follows: GAS5 forward, 5′-GCACACAGGCATTAGACAGA-3′ and reverse,
5′-AAGCCGACTCTCCATACCTT-3′; U6 (used as the internal control)
forward, 5′-CTCGCTTCGGCAGCACA-3′ and reverse,
5′-AACGCTTCACGAATTTGCGT-3′. The relative quantification value of
RNA was calculated by applying the 2−ΔΔCq method
(16).
Genomic DNA extraction
Melanoma cells were collected by centrifugation
(8,000 × g) at room temperature for 5 min, and genomic DNA was
extracted with the Wizard Genomic DNA Purification kit (Promega
Corporation). Cell precipitates were then lysed with 600 µl nuclei
lysis solution (included with the kit), and then incubated at 65°C
for 15 min. RNase A (3 µl) was added to the cell lysate and
incubated at 37°C for 15 min, and subsequently, 200 µl protein
precipitation solution (included with the kit) were added.
Following centrifugation at 14,000 × g for 4 min at room
temperature, the supernatant was pipetted into an EP tube and 600
µl isopropanol (Sigma-Adrich; Merck KGaA) was added. Following
centrifugation at 14,000 × g for 1 min at room temperature, 600 µl
of 70% ethanol were added to wash the precipitate. Finally,
following centrifugation at 14,000 × g for 2 min at room
temperature for ethanol removal, 30 µl of DNA rehydration solution
(included with the kit) were added, in order to dissolve DNA.
Bisulfite modification of DNA
DNA methylation levels were detected using the
EpiTect Bisulfite kit (Qiagen GmbH). A total DNA quantity of 500 ng
was added to the Bisulfite Mix and DNA Protect Buffer, and then
place into a PCR thermal cycler for bisulfite modification. The
conversion conditions were applied as follows: 95°C, 5 min; 60°C,
25 min; 95°C, 5 min; 60°C, 85 min; 95°C, 5 min; 60°C, 175 min; 20°C
storage. This modification resulted in the conversion of
unmethylated cytosine residues into uracil, while the methylated
cytosines remained unchanged. Following bisulfite conversion, 560
µl Buffer BL were added to the above reaction solution, and all
solutions were added to the EpiTect spin column. Subsequently, 500
µl washing Buffer BW was added following centrifugation at 14,000 ×
g for 1 min at room temperature, and then 500 µl of Buffer BD was
added to incubate for 15 min. Following centrifugation at 14,000 ×
g for 1 min at room temperature, the EpiTect spin column was washed
twice with Buffer BW and then placed into a new Eppendorf tube.
Buffer EB (20 µl) was added, to dissolve the converted DNA.
Following centrifugation at 14,000 × g for 2 min at room
temperature, the converted DNA was stored at −20°C for later
use.
Methylation-specific polymerase chain
reaction (MSP)
The methylation levels of the CpG-rich region in the
GAS5 promoter were analyzed using MSP. Firstly, an online program
MethPrimer (http://www.urogene.org/cgi-bin/methprimer/methprimer.cgi)
dedicated to methylation analysis was used to predict the CpG
islands of GAS5 promoter. When the sequence of GAS5 promoter region
was input, MethPrimer could automatically analyze the possible CpG
islands and design usable primers. The primer pair sequences used
were as follows: methylated-sense, 5′-GTTGGAATGTAGTGGTTCGATA-3′,
and methylated-antisense, 5′-GCCAACATAATAAAACCCCGT-3′;
unmethylated-sense, 5′-AGGTTGGAATGTAGTGGTTT-3′, and
unmethylated-antisense, 5′-ACCAACATAATAAAACCCCATCT-3′. The
methylated primers amplified methylation-specific PCR products (M),
when CpG sites were methylated, whereas unmethylation-specific PCR
product (U) was present when CpG sites were unmethylated. When the
sites were partially methylated, M and U bands were simultaneously
present. Ex Taq HS (Takara Biotechnology Co., Ltd.) was used for
PCR amplification and the reactions were performed as follows: 5
min at 95°C, and 30 cycles of 30 sec at 95°C, 30 sec at 60°C, 40
sec at 72°C.
Cell counting kit-8 (CCK-8) assay
The concentration of melanoma cells was adjusted to
1×105 cells/ml, and 100 µl cells per well were seeded
onto 96-well plates. Subsequently, 10 µl CCK-8 solution (Dojindo
Laboratories, Inc.) were added at various time points (0, 24, 48
and 72 h). Following incubation for 4 h at 37°C, data were read
using a microplate reader (multiscan MK3; Thermo Fisher Scientific,
Inc.) at a wavelength of 450 nm.
Apoptosis analysis
Culture supernatants of each group were collected
and placed on ice. Following trypsinization for 2 min, the cells
were resuspended in the culture supernatant collected above, and
the cell density was then adjusted to ~1×106 cells/ml.
Subsequently, 1.25 µl Annexin V-FITC (Nanjing KeyGen Biotech Co.,
Ltd.) were added into 0.5 ml of the above cell suspension, and the
reaction was kept away from light at room temperature (18-24°C) for
15 min. The supernatant was then removed by centrifugation at 1,000
× g for 5 min at room temperature. The cells were gently
resuspended with 0.5 ml of pre-cooled 1X binding buffer, and then
incubated with 10 µl propidium iodide (Nanjing KeyGen Biotech Co.,
Ltd.) in the dark at 4°C for 15 min. Finally, a flow cytometer BD
FACScalibur (BD Biosciences) was used to analyze cell apoptosis and
data analysis was performed using FlowJo V10 software.
Cell migration assay
A total quantity of 1×105 cells were
resuspended in 100 µl serum-free DMEM, and then added to the upper
Transwell chamber (BD Biosciences) of Transwell inserts (8-µm pore
size, BD Biosciences), while 600 µl complete DMEM were added to the
lower chamber. After the cells were cultured for 12–48 h, the cells
in the upper chamber were removed by using cotton swabs. The
migratory cells were fixed with 4% paraformaldehyde for 20 min,
washed with PBS, then stained with crystal violet (Sigma-Adrich;
Merck KGaA) for 10 min at room temperature, and photographed using
a Leica DM4000B microscope (Leica Microsystems GmbH), for
subsequent experimental result statistical analysis.
Cell invasion
For cell invasion assay, 40 µl Matrigel (BD
Biosciences) were added into the pre-cooled upper Transwell chamber
of Transwell inserts (8-µm pore size), and following coagulation,
serum-free DMEM was added to balance overnight. Afterwards, 100 µl
serum-free DMEM containing 1×105 cells were added to the
upper chamber, and 600 µl complete DMEM was added to the lower
chamber. The following experimental steps were carried out
according to the detection method previously described for cell
migration evaluation.
Nude mouse tumorigenicity assay and
immunohistochemistry
A total of 16 male BALB/c nude mice (SPF grade, 4–5
weeks old, weighing 18–20 g) were purchased from the Experimental
Animal Center of Guangzhou University of Chinese Medicine. All mice
were housed (21-25°C with 45–55% humidity) at a 12-h light-dark
cycle and provided with food and water ad libitum. A A375
cell suspension (0.1 ml/mouse, containing 5×106 cells)
was injected subcutaneously into nude mice. After 1 week, a
subcutaneous tumor mass appeared. Subsequently, 5-Aza-dC was
injected intraperitoneally at a dose of 3 mg/kg for 3 consecutive
weeks as previously described (17). Tumor size was measured twice a
week, and the tumor size and weights were calculated and recorded.
When the tumor volume reached ~500 mm3, the mice were
anesthetized using 3% sevoflurane, then euthanized by using
CO2 at flow rate of 30%/min for 7 min, followed by
cervical dislocation, and tumors were excised on day 31. Xenograft
tumors were fixed in formalin (10%) for 24 h at room temperature
and then immunohistochemical evaluation was performed, using an
antibody against Ki-67 (1:200; cat. no. ab15580, Abcam). Dense
tumor segments (~4 µm) were blocked with 3% bovine serum albumin
(BSA; Beyotime Institute of Biotechnology) for 30 min at room
temperature, and incubated with Ki-67 antibody (0.5 µg/ml)
overnight at 4°C. Subsequently, 4-µm-thick slices were incubated
with goat anti-mouse horseradish peroxidase (HRP)-conjugated
secondary antibody (1:100; cat. no. ab6789; Abcam) for 30 min at
37°C. All sections were photographed and observed under an Eclipse
TS100 microscope (Nikon Corporation). Caspase-3 activity was
detected using a Caspase-3 Assay kit (cat. no. ab39401, Abcam)
according to the manufacturer's instructions. The present study was
approved by the Ethics Committee of Animal Research Institute of
Yunnan Cancer Hospital.
Statistical analysis
The data are presented as the mean ± standard
deviation (SD), and all the experiments were repeated at least
three times. Statistical analysis was performed using SPSS 20.0
software (IBM Corp.) by applying an unpaired Student's t-test for
comparisons between 2 groups, a two-way analysis of variance
(ANOVA) to compare between groups or one-way ANOVA followed by
Tukey's post-hoc test for mean separation including correction for
multiple comparisons. P<0.05 was considered to indicate a
statistically significant difference.
Results
GAS5 is hypermethylated in melanoma
tissues and cells
GAS5 has been reported to be an important tumor
suppressor gene in a variety of tumors (7); however, the association between GAS5
expression and methylation levels in melanoma has not been
reported, at least to the best of our knowledge. In the present
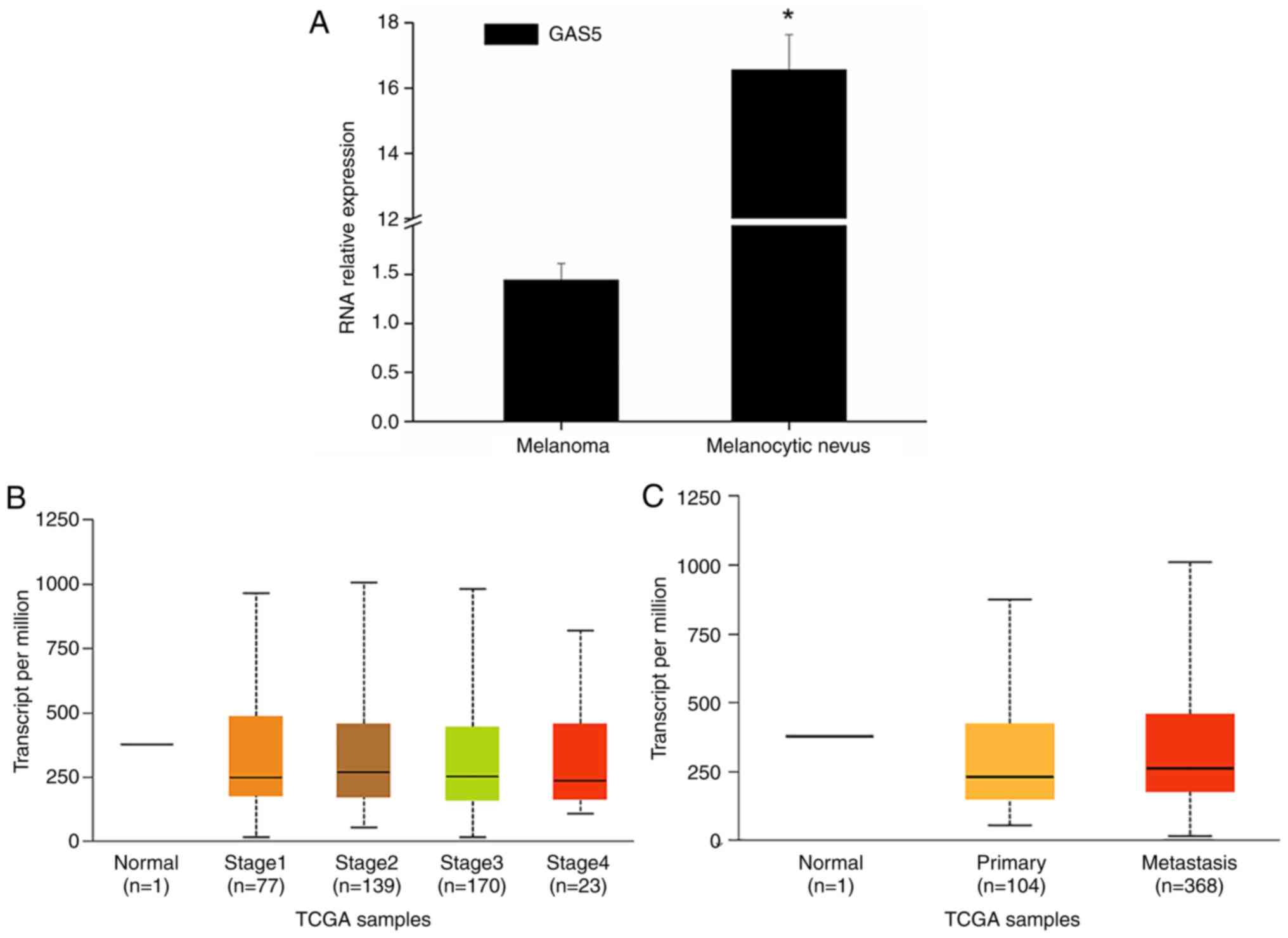
study, it was revealed that GAS5 expression in melanoma tissues was
downregulated in general (Fig.
1A). TCGA data analysis also revealed that the expression of
GAS5 was significantly downregulated in melanoma (Fig. 1B and C). At the same time, MEXPRESS
data evaluation revealed that GAS5 expression and methylation
levels in melanoma had a certain negative correlation
(r=−0.094~-0.308; Fig. 2),
indicating that the downregulation of GAS5 expression in melanoma
may be related to the modification of methylation. The GAS5
promoter region exhibited evident CpG islands (Fig. 3A). The methylation level of GAS5
promoter detected using MSP revealed that GAS5 did have methylation
modifications in melanoma tissues, since the methylated primers
amplified methylation-specific PCR products (M); however, no GAS5
methylation was detected in normal tissues, as the band of
unmethylation-specific PCR product (U) was present (Fig. 3B). The simultaneous detection of
GAS5 methylation level in A375 and SK-MEL-110 melanoma cells also
revealed that GAS5 was methylated, and the methylation of GAS5 was
significantly reduced after 5-Aza-dC treatment (Fig. 3C). At the same time, 5-Aza-dC
significantly upregulated the expression of GAS5, while GAS5 siRNA
significantly inhibited the expression of GAS5 and blocked the
expression-promoting effect of 5-Aza-dC (Fig. 4A and B). Therefore, the
downregulation of GAS5 expression in melanoma may be related to its
methylation modification, and 5-Aza-dC may be able to reverse GAS5
methylation modification.
5-Aza-dC inhibits melanoma cell
proliferation and promotes apoptosis
The evaluation of the effects of 5-Aza-dC on the
proliferation of melanoma cells demonstrated that 5-Aza-dC
inhibited the proliferation of A375 cells in a time-dependent
manner (Fig. 4C) and similar
results were also observed in another melanoma cell line,
SK-MEL-110 (Fig. 4D). However,
when the expression of GAS5 was silenced, the effect of 5-Aza-dC on
cell proliferation was not significant compared to the control
(Fig. 4C and D). The continued
detection of cell apoptosis demonstrated that 5-Aza-dC
significantly increased the apoptotic level of A375 cells (Fig. 4E and F), and also significantly
enhanced the apoptosis of SK-MEL-110 cells (Fig. 4G and H). However, GAS5 siRNA
attenuated the pro-apoptotic effects of 5-Aza-dC (Fig. 4G and H). Therefore, 5-Aza-dC
significantly inhibited the proliferation of melanoma cells and
promoted apoptosis by upregulating the expression of GAS5,
indicating that 5-Aza-dC may present with an evident tumor
suppressor function.
5-Aza-dC inhibits the migration and
invasion of melanoma cells
Migration and invasion are the main biological
characteristics of tumor cells. The evaluation of the effects of
5-Aza-dC on melanoma cell migration revealed that 5-Aza-dC
significantly inhibited A375 cell migration (Fig. 5A and B), and similar results were
also observed in the SK-MEL-110 cells (Fig. 5A and B). Following the addition of
5-Aza-dC, the migration of the cells was markedly inhibited
(Fig. 5A and B). At the same time,
the evaluation of cell invasion level revealed that 5-Aza-dC
significantly inhibited A375 and SK-MEL-110 cell invasion levels
(Fig. 5C and D). However, when the
expression of GAS5 was silenced, the inhibitory effects of 5-Aza-dC
on cell migration and invasion were significantly impeded (Fig. 5). Therefore, 5-Aza-dC may inhibit
the migration and invasion of melanoma cells by upregulating the
expression of GAS5.
5-Aza-dC inhibits the growth of
melanoma xenografts
Although there have been several studies on the role
of 5-Aza-dC in tumors (12–15),
there are few in vivo studies on 5-Aza-dC and melanoma.
Thus, the role of 5-Aza-dC in vivo remains to be elucidated.
In the present study, the subcutaneous injection of A375 cells in
nude mice successfully led to the formation of xenograft tumors.
Following the intraperitoneal injection of 5-Aza-dC, although
5-Aza-dC had a certain effect on the body weight of nude mice
(Fig. 6A), it significantly
inhibited the growth of tumors, demonstrating that the volume and
weight of xenograft tumors were significantly reduced (Fig. 6B-D). In addition, the expression of
Ki-67 was significantly decreased (Fig. 6E and F), while caspase-3 activity
and GAS5 expression were significantly increased (Fig. 6G and H) in the mice treated with
5-Aza-dC. However, when the expression of GAS5 was silenced, the
antitumor effect of 5-Aza-dC was markedly inhibited (Fig. 6B-D) and the expression level of
Ki-67 and caspase-3 activity were markedly restored to the control
levels (Fig. 6E-G). Therefore,
5-Aza-dC may possibly inhibit the growth of melanoma xenografts
in vivo, and its effect is closely related to GAS5
levels.
Discussion
Malignant melanoma is highly invasive and highly
metastatic, and its morbidity and mortality have been continuously
increasing (2). Therefore, the
further exploration of malignant melanoma pathogenesis and the
designation of novel therapeutic targets for the treatment of
malignant melanoma is of utmost urgency.
GAS5 is an important tumor suppressor gene, playing
a crucial role by regulating a variety of intracellular biological
processes, including growth arrest, apoptosis, proliferation,
metastasis, and DNA damage repair (7). For example, Chen et al
(18) revealed that the
downregulation of GAS5 may increase cyclin D1, CDK4, p27, NADPH
oxidase 2 (NOX4), glucose-6-phosphate dehydrogenase (G6PD) and
Bcl-2 expression, thereby inducing G1/S cell cycle progression and
redox imbalance, inhibiting apoptosis and then promoting the growth
of melanoma cells. More and more studies have shown that GAS5 is
low expressed in a variety of tumors (7). The analysis of clinical samples of
patients with malignant melanoma has also revealed that the
expression level of GAS5 in patients with advanced tumors is
significantly lower than that of adjacent non-cancerous tissues and
is also significantly associated with the TNM stage and distal
metastasis of melanoma (18–21).
Therefore, increasing the expression of GAS5 in tumor tissues may
be a possible tumor treatment strategy.
DNA methylation, a more widely studied and a main
modification process in epigenetics, is crucial for the study of
tumor pathogenesis. A large amount of research data have confirmed
that DNA methylation is significantly associated with
tumorigenesis, particularly the abnormal hypermethylation of CpG
islands in the promoter region of tumor suppressor genes, which
often leads to the inactivation and silencing of the transcription
level of relevant tumor suppressor genes, resulting in unrestricted
cancer cell growth, thus promoting cancer development (22,23).
In the present study, the analysis of GAS5 revealed that the
promoter region of GAS5 presented with apparent CpG islands, and
also presented with methylation modification in melanoma tissues
and cells. However, no GAS5 promoter methylation modification was
found in normal tissues. The data in the MEXPRESS database
demonstrated that the GAS5 expression levels in melanoma had a
certain negative correlation with the level of methylation
modification. Therefore, the downregulation of GAS5 expression in
melanoma may be likely attributed to methylation modification.
Studies on triple-negative breast cancer and cervical cancer have
also revealed that GAS5 methylation levels in cancer tissues are
significantly increased, culminating in the downregulation of GAS5
expression (24,25). However, Wang et al (26) demonstrated that although GAS5 was
downregulated in gliomas, its methylation levels was not
significantly altered. Another study [Wang et al (27)] indicated that a decreased GAS5
expression in esophageal squamous cell carcinoma may be related to
the increased expression of miR-196a, and may possibly not be
related to DNA methylation. Therefore, the downregulation of GAS5
expression in tumor cells may be related to a variety of regulatory
mechanisms, and methylation modification may be one of the
important regulatory mechanisms.
Demethylation drugs have been reported to inhibit
tumor suppressor gene methylation levels in most tumor cells,
thereby restoring gene expression and inhibiting tumor cell growth
(11). Following the treatment of
melanoma cells with 5-Aza-dC, the methylation of GAS5 was
significantly reversed in the present study, and 5-Aza-dC
significantly inhibited cell proliferation, migration and invasion,
also inducing cell apoptosis through the upregulation of GAS5
expression, also indicating that 5-Aza-dC possibly also exerts a
tumor-suppressive effect in melanoma cells. Rajaii et al
(13) also reported that treating
melanoma cells with 5-Aza-dC significantly inhibited cell invasion,
growth and colony formation, and reduced tumor metastasis in mouse
skin melanoma xenograft models. In the present study, further in
vivo experiments once again demonstrated that the
intraperitoneal injection of 5-Aza-dC significantly reduced the
volume and weight of xenograft tumors, also inhibiting tumor
growth. After implanting 5-Aza-dC-treated A375 melanoma cells into
mice, it has been also revealed that 5-Aza-dC might significantly
reduce tumor volumes, and may play a role by reducing the
methylation level of Thrombospondin-1 (TSP1) and then restoring the
expression level of TSP1 (14). In
another study, decitabine (5-Aza-dC), followed by carboplatin
treatment, significantly attenuated cell proliferation, resulting
in a greater apoptotic response in melanoma cells, as compared to
carboplatin alone (28). Moreover,
clinical trials have demonstrated that 5-Aza-dC exerts a positive
antitumor effect and is safe for use (29,30).
The aforementioned studies have demonstrated that
the important tumor suppressor gene GAS5 is significantly
methylated in melanoma, leading to the downregulation of GAS5;
5-Aza-dC may effectively inhibit the methylation of GAS5, decrease
cell proliferation, migration and invasion, promote apoptosis and
reduce xenograft tumor growth. This finding may provide suitable
therapeutic targets and approaches for the diagnosis and treatment
of melanoma.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Natural Science
Foundation of China (grant no. 81960496), the China Postdoctoral
Science Foundation (grant no. 2019M653501), the Yunnan Fundamental
Research Projects (grant nos. 202101AT070050, 202001AY070001-247
and 202101AY070001-157), the Scientific Research Fund of Yunnan
Education Department (grant no. 2019J1286), XingDianYingCai support
plan, and the Reserve talents of Medical Science of Yunnan Province
(H-2018055).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
HΤN and LC made substantial contributions to the
conception and design of the study. YJZ, RX and JJ conducted most
of the experiments, confirmed the authenticity of all the raw data
and drafted the manuscript. LZ, CHY and JZ made substantial
contributions to data analysis and data interpretation. XW and DXC
were involved in performing some of the experiments and data
analysis. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethical
Committee of Yunnan Cancer Hospital of the Third Affiliated
Hospital of Kunming Medical University (KY201939). Informed consent
was obtained from each patient included in this study. The use of
animals was approved by the Ethics Committee of Animal Research
Institute of Yunnan Cancer Hospital.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Elder DE: Melanoma progression. Pathology.
48:147–154. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Krathen M: Malignant melanoma: Advances in
diagnosis, prognosis, and treatment. Semin Cutan Med Surg.
31:45–49. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Micevic G, Theodosakis N and Bosenberg M:
Aberrant DNA methylation in melanoma: Biomarker and therapeutic
opportunities. Clin Epigenetics. 9:342017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Guo W, Zhu L, Zhu R, Chen Q, Wang Q and
Chen JQ: A four-DNA methylation biomarker is a superior predictor
of survival of patients with cutaneous melanoma. Elife.
8:e443102019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tanemura A, Terando AM, Sim MS, van Hoesel
AQ, de Maat MF, Morton DL and Hoon DS: CpG island methylator
phenotype predicts progression of malignant melanoma. Clin Cancer
Res. 15:1801–1807. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bhan A, Soleimani M and Mandal SS: Long
noncoding RNA and cancer: A new paradigm. Cancer Res. 77:3965–3981.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yang X, Xie Z, Lei X and Gan R: Long
non-coding RNA GAS5 in human cancer. Oncol Lett. 20:2587–2594.
2020. View Article : Google Scholar
|
|
8
|
Chen L, Yang H, Xiao Y, Tang X, Li Y, Han
Q, Fu J, Yang Y and Zhu Y: LncRNA GAS5 is a critical regulator of
metastasis phenotype of melanoma cells and inhibits tumor growth in
vivo. Onco Targets Ther. 9:4075–4087. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ma X, Yu L, Wang P and Yang X: Discovering
DNA methylation patterns for long non-coding RNAs associated with
cancer subtypes. Comput Biol Chem. 69:164–170. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cheng M, Sun L, Huang K, Yue X, Chen J,
Zhang Z, Zhao B and Bian E: A signature of nine lncRNA methylated
genes predicts survival in patients with glioma. Front Oncol.
11:6464092021. View Article : Google Scholar
|
|
11
|
Linnekamp JF, Butter R, Spijker R, Medema
JP and van Laarhoven HWM: Clinical and biological effects of
demethylating agents on solid tumours-a systematic review. Cancer
Treat Rev. 54:10–23. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Momparler RL: Epigenetic therapy of cancer
with 5-aza-2′-deoxycytidine (decitabine). Semin Oncol. 32:443–451.
2005. View Article : Google Scholar
|
|
13
|
Rajaii F, Asnaghi L, Enke R, Merbs SL,
Handa JT and Eberhart CG: The demethylating agent 5-Aza reduces the
growth, invasiveness, and clonogenicity of uveal and cutaneous
melanoma. Invest Ophthalmol Vis Sci. 55:6178–6186. 2014. View Article : Google Scholar
|
|
14
|
Lindner DJ, Wu Y, Haney R, Jacobs BS,
Fruehauf JP, Tuthill R and Borden EC: Thrombospondin-1 expression
in melanoma is blocked by methylation and targeted reversal by
5-Aza-deoxycytidine suppresses angiogenesis. Matrix Biol.
32:123–132. 2013. View Article : Google Scholar
|
|
15
|
Wang W, Wang J, Li M, Ying J and Jing H:
5-Azacitidine induces demethylation of PTPL1 and inhibits growth in
non-Hodgkin lymphoma. Int J Mol Med. 36:698–704. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Numoto K, Yoshida A, Sugihara S, Kunisada
T, Morimoto Y, Yoneda Y, Fujita Y, Nishida K, Ouchida M and Ozaki
T: Frequent methylation of RASSF1A in synovial sarcoma and the
anti-tumor effects of 5-aza-2′-deoxycytidine against synovial
sarcoma cell lines. J Cancer Res Clin Oncol. 136:17–25. 2010.
View Article : Google Scholar
|
|
18
|
Chen L, Yang H, Yi Z, Jiang L, Li Y, Han
Q, Yang Y, Zhang Q, Yang Z, Kuang Y and Zhu Y: LncRNA GAS5
regulates redox balance and dysregulates the cell cycle and
apoptosis in malignant melanoma cells. J Cancer Res Clin Oncol.
145:637–652. 2019. View Article : Google Scholar
|
|
19
|
Bian D, Shi W, Shao Y, Li P and Song G:
Long non-coding RNA GAS5 inhibits tumorigenesis via miR-137 in
melanoma. Am J Transl Res. 9:1509–1520. 2017.PubMed/NCBI
|
|
20
|
Qi Y, Cui Q, Zhang W, Yao R, Xu D and
Zhang F: Long non-coding RNA GAS5 targeting microRNA-21 to suppress
the invasion and epithelial-mesenchymal transition of uveal
melanoma. Cancer Manag Res. 12:12259–12267. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Xu W, Yan Z, Hu F, Wei W, Yang C and Sun
Z: Long non-coding RNA GAS5 accelerates oxidative stress in
melanoma cells by rescuing EZH2-mediated CDKN1C downregulation.
Cancer Cell Int. 20:1162020. View Article : Google Scholar
|
|
22
|
Moore LD, Le T and Fan G: DNA methylation
and its basic function. Neuropsychopharmacology. 38:23–38. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kulis M and Esteller M: DNA methylation
and cancer. Adv Genet. 70:27–56. 2010. View Article : Google Scholar
|
|
24
|
Li J, Li L, Yuan H, Huang XW, Xiang T and
Dai S: Up-regulated lncRNA GAS5 promotes chemosensitivity and
apoptosis of triple-negative breast cancer cells. Cell Cycle.
18:1965–1975. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yang W, Xu X, Hong L, Wang Q, Huang J and
Jiang L: Upregulation of lncRNA GAS5 inhibits the growth and
metastasis of cervical cancer cells. J Cell Physiol.
234:23571–23580. 2019. View Article : Google Scholar
|
|
26
|
Wang Y, Xin S, Zhang K, Shi R and Bao X:
Low GAS5 levels as a predictor of poor survival in patients with
lower-grade gliomas. J Oncol. 2019:17850422019. View Article : Google Scholar
|
|
27
|
Wang K, Li J, Xiong G, He G, Guan X, Yang
K and Bai Y: Negative regulation of lncRNA GAS5 by miR-196a
inhibits esophageal squamous cell carcinoma growth. Biochem Biophys
Res Commun. 495:1151–1157. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Budden T, van der Westhuizen A and Bowden
NA: Sequential decitabine and carboplatin treatment increases the
DNA repair protein XPC, increases apoptosis and decreases
proliferation in melanoma. BMC Cancer. 18:1002018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tawbi HA, Beumer JH, Tarhini AA, Moschos
S, Buch SC, Egorin MJ, Lin Y, Christner S and Kirkwood JM: Safety
and efficacy of decitabine in combination with temozolomide in
metastatic melanoma: A phase I/II study and pharmacokinetic
analysis. Ann Oncol. 24:1112–1119. 2013. View Article : Google Scholar
|
|
30
|
van der Westhuizen A, Knoblauch N, Graves
MC, Levy R, Vilain RE and Bowden NA: Pilot early phase II study of
decitabine and carboplatin in patients with advanced melanoma.
Medicine (Baltimore). 99:e207052020. View Article : Google Scholar : PubMed/NCBI
|