Introduction
The ubiquitin-proteasome system (UPS) is a
remarkable protein degradation system having diverse known and
unknown functions, including degradation of most proteins in
eukaryotic cells (1). This system
encompasses several mechanisms including ubiquitination, which is
mediated by the successive activities of E1, E2, and E3 enzymes. In
addition, ubiquitination not only regulates proteasomal
degradation, but also cellular functions through single or multiple
polyubiquitinations (2).
Specifically, ubiquitination is a post-translational modification
(PTM) where the ubiquitin binds to a target protein and regulates
its proteasomal degradation or cellular functions (3). Reportedly, ubiquitin contains seven
lysine residues (K6, K11, K27, K29, K33, K48, and K63) which form a
polyubiquitin chain; of these, K48 and K63 are the most well-known
polyubiquitination sites (4,5).
Notably, numerous potential unknown polyubiquitination sites are
being reported currently, and a great amount of research is being
focused on identifying them.
Deubiquitination is a reversal process of
ubiquitination using enzymes called deubiquitinating enzymes
(DUBs), which play an essential role in protein stabilization by
removing ubiquitins from the target proteins (6,7).
Approximately 100 DUBs have been identified to date, and they are
reportedly involved in diverse cellular functions through their
special abilities to stabilize or alter the functions of target
proteins (8). According to
previous research, the DUB family is composed of at least nine
classes. In particular, the cysteine protease class contains
ubiquitin-specific protease (USP), ovarian tumor protease (OTU),
ubiquitin C-terminal hydrolase (UCH), permutated papain fold
peptidases of dsDNA viruses and eukaryote (PPPDE), Machado-Joseph
disease protease (MJD), motif interacting with Ub-containing novel
DUB (MINDY), monocyte chemotactic protein-induced protease (MCPIP),
and zinc finger with UFM1-specific peptidase domain protein (ZUFSP)
family (9,10). The metalloprotease class belongs to
the Jab1/Pab1/MPN metalloenzyme motif protease (JAMM) family
(11).
Ubiquitin-specific protease 7 [USP7; also known as
herpes virus-associated ubiquitin-specific protease (HAUSP)] is
known to play an essential role in various diseases, including
cancers. Substrates modulated by USP7 are broadly involved in
cellular processes including the cell cycle, DNA repair, chromatin
remodeling, and epigenetic regulation (12). The important role of USP7 is to
release ubiquitin on substrates that enhance tumorigenesis
(12,13). USP7 acts as a negative or positive
regulator that modulates the activity of the oncogene or tumor
suppressor in numerous cancers (14). Therefore, potential substrates of
USP7 could be essential regulators involved in various cellular
mechanisms in cancer progression (15,16).
Thus, in order to understand the cellular pathway, it is imperative
to discover the binding partners of USP7, and explore the cellular
and molecular mechanisms of USP7.
In a previous study, substrates related to USP7 were
analyzed by two-dimension electrophoresis (2-DE) and
matrix-assisted laser desorption ionization time-of-flight mass
spectrometry (MALDI-TOF/MS) analysis, and the analysis confirmed
that protein phosphatase 2A (PP2A) is a putative substrate of USP7
(17). PP2A is one of the most
abundant cellular enzymes, acting on numerous substrates (18). Furthermore, PP2A regulates diverse
cellular functions including cell growth or division (19). Due to multiple cellular processes
regulated by PP2A, several different mechanisms are capable of
regulating the phosphatase activity, such as association with
specific regulatory subunits, or post-translational modifications
(PTMs) of PP2A (for example, phosphorylation, carboxymethylation,
and ubiquitination) (20).
Moreover, PP2A has also been identified as a tumor suppressor
(21), wherein it reverses the
phosphorylation of oncoproteins such as c-MYC, MEK, and ERK
(22,23). The family components of PP2A are
reported to be uncommonly mutated or deficient in various cancers
(24). Considering all the
aforementioned outcomes, PP2A has been determined to play an
important role in cancers and is regarded as a significant
therapeutic target. Therefore, the intercellular association
between USP7 and PP2A was investigated. In addition, confirmation
of the role of USP7 as a DUB for PP2A, and elucidation of the
regulatory mechanism of PP2A was undertaken.
In the present study, protein-protein interaction
between USP7 and PP2A was identified. It is well documented that
the UPS regulates PP2A and cellular functions of PP2A are also
related to DUBs or E3 ligases. Recent experimental studies have
shown that PP2A is a key regulator of progression and metastasis of
cancer cells (25). Thus,
considering the known functions of USP7 and PP2A, it is suggested
that this association between the two proteins is an important
feature in cancer progression. Therefore, the association between
the two proteins was analyzed by confirming the potential of USP7
as a DUB for PP2A.
Materials and methods
Cell culture and transfection
HeLa (cat. no. CRM-CCL2), MCF-7 (cat. no. HTB-22),
and 293T (cat. no. CRL-3216; all from ATCC) cell lines were
maintained in Dulbecco's modified Eagle's medium (DMEM) containing
10% fetal bovine serum (FBS), 1% antibiotic-antimycotic reagent
(containing penicillin, streptomycin, and amphotericin B; cat. no.
15240-062; Gibco: Thermo Fisher Scientific, Inc.) at 37°C in a 5%
CO2 incubator. A549 (cat. no. CRM-CL-185; ATCC) cells
were incubated in Roswell Park Memorial Institute 1640 (RPMI-1640;
Gibco; Thermo Fisher Scientific, Inc.) medium containing 10% FBS,
1% antibiotic-antimycotic reagent (as previously aforementioned) at
37°C in a 5% CO2 incubator. Transfection was performed
using 150 mM NaCl and 10 mM linear polyethyleneimine (PEI;
Polysciences, Inc.) and harvested after 48 h.
Plasmid DNA and antibodies
PP2A gene was subcloned into the pCS4-Flag
vector and pcDNA3.1-Myc vector. USP7 and USP7 (C223S,
a catalytically inactive mutant) genes were subcloned into the
pcDNA3.1-Myc vector (12) or
pCS4-Flag vector. USP7 gene was subcloned into the pGEX-4T-1
vector for GST pull-down assay (GST-USP7). The HA-tagged
ubiquitin (K48 and K63) was produced using site-directed
point mutagenesis (26). HA-K48 is
an HA-tagged ubiquitin in which other lysines (K6, K11, K27, K29,
K33, and K63) are mutated to an arginine except for K48. While
HA-K63 is an HA-tagged ubiquitin in which other lysines (K6, K11,
K27, K29, K33, and K48) are substituted with an arginine except for
K63. The purpose of HA-K48 and HA-63 is to specifically identify an
HA-K48-linked- or HA-K63-linked ubiquitin chain, respectively.
Anti-β-actin (cat. no. sc-4778; Santa Cruz Biotechnology, Inc.),
anti-HA (cat. no. 12CA5; product code 11583816001; Roche
Diagnostics), anti-Flag (cat. no. M185-3L; MBL International
Corporation), anti-Myc (cat. no. sc-40; Santa Cruz Biotechnology,
Inc.), anti-USP7 (cat. no. CSB-PA849973LA01HU; Cusabio Technology
LLC), anti-USP7 (cat. no. sc-137008; Santa Cruz Biotechnology,
Inc.), and anti-PP2A (cat. no. sc-6110; Santa Cruz Biotechnology,
Inc.) antibodies were used.
Western blotting and
immunoprecipitation (IP)
Harvested cells were lysed using a lysis buffer (50
mM Tris-HCL, 300 mM NaCl, 1 mM EDTA, 10% Glycerol, 1% Triton X-100)
and were incubated for 20 min on ice. Cell lysates were centrifuged
at 16,200 × g at 4°C for 20 min, and the supernatant was boiled
with 2X SDS buffer at 100°C. The protein amounts (30 µg) were
determined by Bradford assay using Bio-Rad protein assay dye
reagent (cat. no. 5000006; Bio-Rad Laboratories, Inc.). The samples
were run in an 8% SDS-page gel and transferred onto microporous
polyvinylidene fluoride membranes (cat. no. IPVH00010; EMD
Millipore; Merck KGaA). The membranes were then blocked with 5%
skim milk in TBS buffer, including 0.05% Tween-20 for 30 min at
room temperature, and incubated at 4°C overnight with a primary
antibody [β-actin (1:3,000), HA (1:1,000), Flag (1:5,000), Myc
(1:100), USP7 (1:500; cat. no. sc-137008; Santa Cruz Biotechnology,
Inc.), and PP2A (1:5,000)] in 2% skim milk with TBS buffer,
including 0.05% Tween-20. The probed membranes were incubated in 1%
skim milk with TBS buffer including 0.05% Tween-20 with KPL
peroxidase-labeled antibody to mouse IgG (H+L) (cat. no. 074-1806;
1:30,000; LGC SeraCare) and mouse anti-goat IgG-HRP (cat. no.
sc-2354; 1:30,000; Santa Cruz Biotechnology, Inc.) at room
temperature for 1 h. The proteins were detected using an ECL
reagent solution (Young In Frontier). For immunoprecipitation, 2 mg
of cell lysates was incubated with antibodies [Flag, Myc, USP7, and
PP2A (1 µg per 500 µg of total proteins)] at 4°C on a rotator
overnight. A resuspended volume (30 µl) of Protein A/G PLUS-agarose
beads (cat. no. sc-2003; Santa Cruz Biotechnology, Inc.) was then
added to the cell lysates and incubated at 4°C on a rotator for 2
h. The samples were then washed with washing buffer [50 mM
Tris-HCl, 300 mM NaCl, 1 mM EDTA, 10% glycerol, 1% Triton X-100, a
protease inhibitor cocktail (PIC; cat. no. 11697498001; Roche
Diagnostics), and phenylmethanesulfonyl fluoride (PMSF; cat. no.
P7626; Sigma-Aldrich; Merck KGaA)]. Subsequently, the samples were
centrifuged at 3,200 × g at 4°C for 5 min, and then boiled in 35 µl
of 2X SDS buffer at 100°C for 7 min. The supernatant (25 µl) was
then loaded onto 8% SDS-PAGE gel. The remaining steps were the same
as those described for the western blotting.
Ubiquitination and deubiquitination
assays
For ubiquitination assay, Flag-PP2A and
HA-ubiquitin (HA-Ub) (K48 or K63) were
transfected into 293T cells. The HA-tagged lysine mutants have been
generated by substituting each lysine site with an arginine except
one corresponding lysine site. After 48 h, cells were harvested and
lysed using a lysis buffer (50 mM Tris-HCL, 300 mM NaCl, 1 mM EDTA,
10% Glycerol, 1% Triton X-100). Cell lysates were used for
immunoprecipitation with anti-Flag antibody (1 µg per 500 µg of
total proteins). The ubiquitination level of Flag-PP2A was detected
through western blotting. For the deubiquitination assay,
Flag-PP2A, HA-Ub (K48 or K63)and Myc-USP7 were
transfected into 293T cells. To confirm whether
proteasome-dependent degradation of substrates, cells were treated
with 10 mM concentration of MG132 [a proteasome inhibitor, cat. no.
F1100; Ubiquitin Proteasome Biotechnologies, LLC (UBPbio)] for 4 h
before harvest and cultured at 37°C in a 5% CO2
incubator. The cells were harvested after 48 h and
immunoprecipitation was performed with anti-Flag antibody. Both
assays were performed using an ubiquitination assay kit according
to the manufacturer's manual (cat. no. UBAK-100; D&P Biotech
Inc.; http://www.dnpbiotech.com).
GST pull-down assay
GST-USP7 was transformed into the BL21
strain. After IPTG treatment, GST-USP7-transformed cells
were cultured in an 18°C shaking incubator. The cells were then
precipitated and released with phosphate-buffered saline (PBS)
containing Triton X-100, PIC, and PMSF and then the cells were
lysed using a sonicator. The Flag-PP2A-transfected 293T cell
lysates (2 mg) were incubated with GST-USP7-fastened Glutathione
Sepharose beads (300 µg per 2 mg of total protein; cat. no.
GE17-0756-01; GE Healthcare; Cytiva). The beads were then washed
with a lysis buffer (20 mM Tris-HCl, 1 mM EDTA, 1 mM
dithiothreitol, 150 mM NaCl, 1% Triton X-100). After removing the
lysis buffer, 35 µl of 2X SDS buffer was added and boiled at 100°C
for 7 min. The supernatants (25 µl) were then loaded onto 8%
SDS-PAGE gel. The remaining steps were the same as those described
for the western blotting. A portion of the sample was confirmed for
protein expression through western blotting, and the expression of
GST-USP7 using the remaining samples was confirmed through
Coomassie Brilliant Blue (CBB) staining. The gels (1.5 mm
thickness) were stained with 0.1% Coomassie Brilliant blue G 250
(cat. no. 1.15444; Sigma-Aldrich; Merck KGaA) at room temperature
for 15 min. Subsequently, the gels were washed by destaining buffer
(40% methanol and 10% acetic acid). Gel images were captured using
a DUALED Blue/White Transilluminator (cat. no. A6020; Bioneer
Corporation). Western blotting was performed to validate the
results.
Immunocytochemistry
HeLa, MCF-7, and A549 cells were plated on a sterile
glass and were washed with PBS. The cells (80% confluence) were
treated with 4% paraformaldehyde at room temperature for 10 min and
were then treated with 0.2% Triton X-100 in PBS at room temperature
for 1 min. Cells were washed with PBS and were blocked in 1% bovine
serum albumin (BSA) in PBS for 1 h. The samples were incubated with
primary antibodies, PP2A (1:200) and USP7 (1:200; cat. no.
CSB-PA849973LA01HU), at 4°C overnight. Cells were washed with PBS
including 0.5% Tween-20 at room temperature for 10 min and were
incubated with Alexa-Fluor-488-conjugated goat anti-mouse (cat. no.
A32723; 1:500) and with Alexa-Fluor-568-conjugated goat anti-rabbit
(cat. no. A11011; 1:500; Invitrogen; Thermo Fisher Scientific,
Inc.) at room temperature for 1 h. Following incubation, nuclei
were stained using DAPI (Invitrogen; Thermo Fisher Scientific,
Inc.) at room temperature for 10 min. Cell images were obtained by
a confocal microscope (Zeiss LSM880; Carl Zeiss Microscopy
GmbH).
Protein stability assay
The sequences of small interfering RNA negative
control (siNC) and siUSP7 were
5′-UUCUCCGAACGUGUCACGUTT-3′ and 5′-CAUGCACAAGCAGUGCUGAAGAUAA-3′,
respectively. siNC or siUSP7 was transfected into
HeLa cells, and then cells were cultured in a 60-mm dish at 37°C in
a 5% CO2 incubator for 24 h. Transfection with
siUSP7 was performed as previously described (27). After 24 h, cycloheximide (CHX)
(cat. no. 01810; Sigma-Aldrich; Merck KGaA) was added to a final
concentration of 100 µg/ml in siUSP7-transfected cells.
Next, the cells were harvested in a time-dependent manner (0, 12
and 24 h) and cell lysates were used for western blotting.
Statistical analysis
For all measured data, the values for all samples
obtained from at least three independent experiments were averaged,
and the standard deviation or standard error was subsequently
calculated. Statistical analysis was performed using the unpaired
t-test and one-way analysis of variance followed by Tukey's
multiple comparisons post hoc tests using GraphPad Prism version 5
(GraphPad Software, Inc.). Densitometric analysis was conducted
using ImageJ software (version 1.4.3; National Institutes of
Health).
Results
USP7 interacts with PP2A
In a previous study, 2-DE and MALDI-TOF/MS analysis
determined that PP2A is a putative substrate of USP7 (17), thereby suggesting the possibility
that PP2A could physically bind to USP7. To confirm the interaction
between PP2A and USP7, a binding assay using 293T cells was
performed. The results revealed binding between PP2A and USP7
(Fig. 1A), indicating the
formation of an interaction between endogenous or exogenous PP2A
and USP7. Additionally, in order to investigate the binding between
these two proteins, the binding of PP2A was investigated with the
expression of the differential amount of Myc-USP7. As a result, it
was determined that as the concentration of Myc-USP7 increased, the
amount of Myc-USP7 binding to PP2A also increased (Fig. 1B). Thereafter, the GST pull-down
assay was performed to identify the occurrence of direct binding
between the two proteins. GST-USP7 was incubated with overexpressed
Flag-PP2A in 293T cell lysates, and subsequently subjected to
Western blotting, which revealed that the two proteins bind
directly each other (Fig. 1C).
Furthermore, immunocytochemical analysis revealed that USP7 and
PP2A are co-localized in the cytoplasm and the nucleus in HeLa
cells, MCF-7 cells, and A549 cells. (Fig. 1D). Collectively, these results
demonstrated the direct and indirect interplay between USP7 and
PP2A in the cytoplasm and the nucleus.
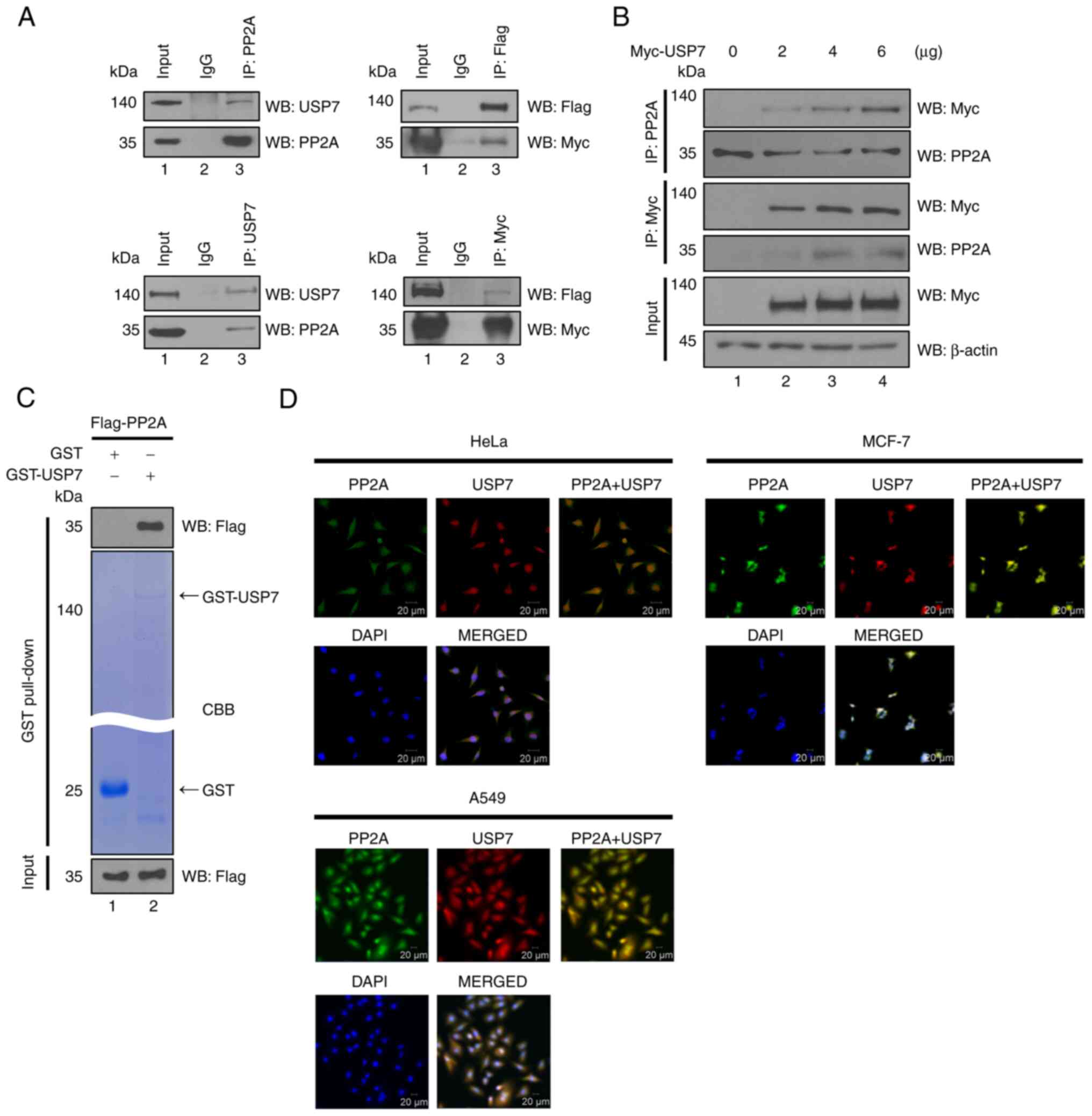 | Figure 1.USP7 interacts with PP2A. (A)
Endogenous binding assays were performed to confirm the endogenous
binding between PP2A and USP7 (left). Exogenous binding assays were
also performed to confirm the binding of Flag-PP2A with Myc-USP7,
using respective antibodies (right). (B) Myc-USP7 was
transfected into 293T cells in a dose-dependent manner. The
immunoprecipitation assay was performed using an anti-Myc or an
anti-PP2A antibody. (C) Flag-PP2A was transfected into 293T
cells for the GST pull-down assay with GST-USP7. (D)
Co-localization of USP7 and PP2A in HeLa, MCF-7, and A549 cells, as
determined by immunocytochemical analysis (green, PP2A; red, USP7;
and blue, DAPI). Scale bar, 20 µm. USP7, ubiquitin-specific
protease 7; PP2A, protein phosphatase 2A. |
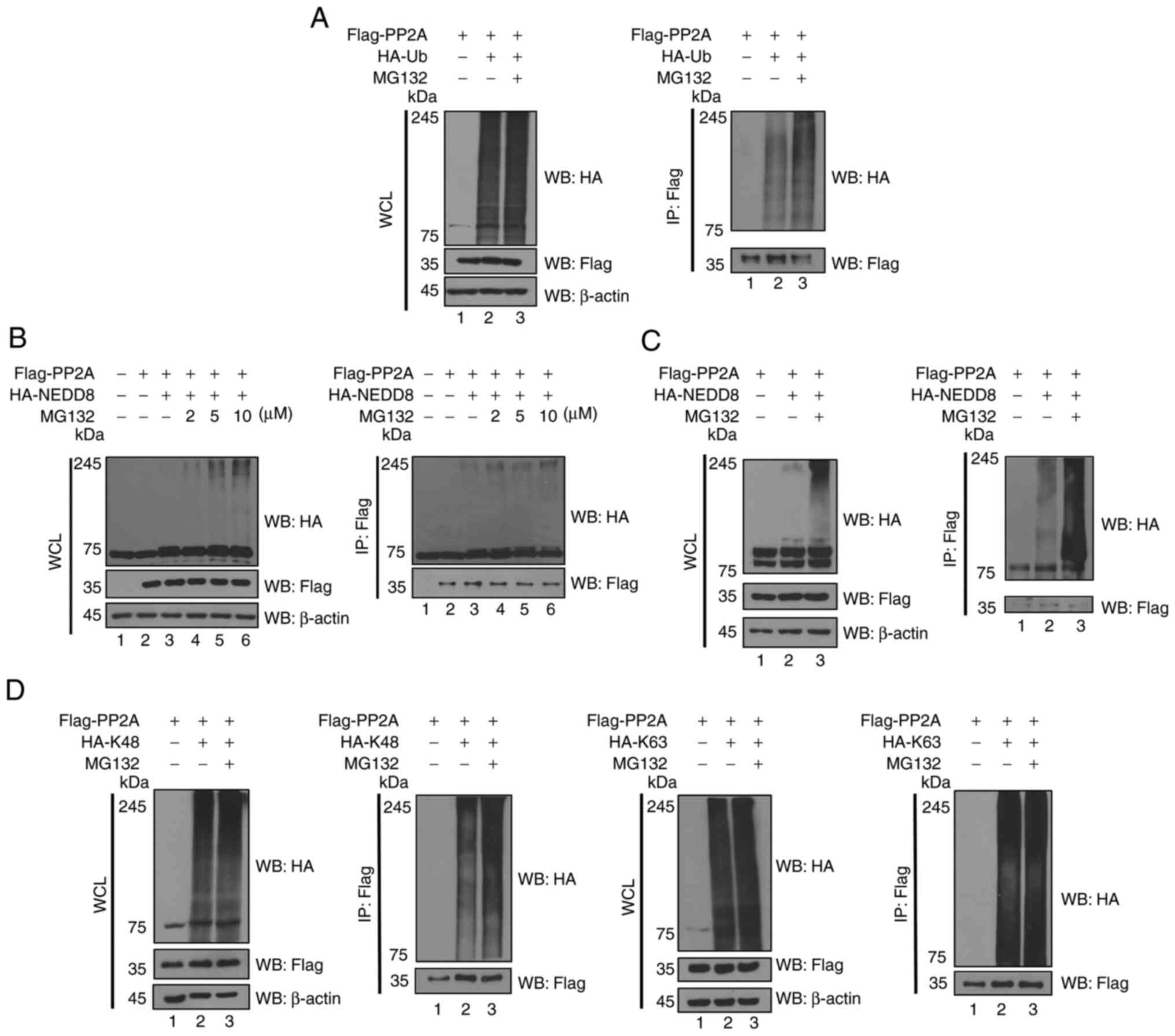
PP2A is regulated by ubiquitin and
ubiquitin-like molecules
The ubiquitination assay was performed to
investigate whether PP2A is regulated by the UPS (Fig. 2A). Results obtained indicated that
PP2A is degraded by the ubiquitin-mediated proteasomal degradation.
The PTMs involve neddylation, SUMOylation, and ISGylation as well
as ubiquitination (28). Moreover,
numerous proteins undergo a proteasome-dependent degradation
mechanism through ubiquitination, neddylation, SUMOylation, and
ISGlyation (29). First, when
MG132 was treated with the differential concentration, neddylation
of Flag-PP2A was not increased at 2 and 5 µM of MG132 (Fig. 2B); a minimum 10-µM concentration
was required to confirm the efficacy of MG132 on neddylation of
PP2A. To assume that PP2A would also be modulated by conjugation
with ubiquitin as well as ubiquitin-like molecules, IP was
performed using co-transfected HA-neuronal precursor cell-expressed
developmentally down-regulated protein 8 (HA-NEDD8) and
Flag-PP2A in 293T cells with a proteasome inhibitor, MG132
(Fig. 2C), and it was determined
that Flag-PP2A formed neddylation. However, SUMOylation and
ISGylation on PP2A were not formed (data not shown). These results
suggested that PP2A could also degrade via neddylation as well as
ubiquitination due to increased expression level of neddylation on
PP2A with the treatment of MG132. Therefore, PP2A could be
regulated by PTMs including ubiquitination and neddylation.
Increased levels of ubiquitinated or neddylated PP2A were observed
after exposure to MG132, indicating that PP2A is degraded in a
ubiquitination- or neddylation-mediated proteasome dependent
manner. In addition, the ubiquitination assay of PP2A was performed
with a K48-linked or K63-linked polyubiquitin chain (Fig. 2D). It was demonstrated that PP2A
formed the K48-linked and K63-linked polyubiquitin chains. This
suggested that PP2A may be affected by proteasome-dependent
degradation associated with K48-linked polyubiquitin chain and
intracellular effect by K63-linked ubiquitination (4). K63-linked polyubiquitin chains are
known to regulate the proteasome-independent pathways such as
signal transduction and endocytosis (5). By contrast, K48-linked polyubiquitin
chains are known to regulate the proteasome-dependent pathway
(5) Since PP2A formed a K63-linked
polyubiquitin chain as shown in Fig.
2D, it is possible that the K63-linked polyubiquitination of
PP2A is not related to proteasomal degradation. Furthermore, the
reciprocal co-immunoprecipitation assays were performed as
ubiquitination assays, as shown in Fig. S1. These results indicated that
Flag-PP2A is connected to the HA-tagged polyubiquitin chain or
polyneddylated chain.
PP2A is deubiquitinated by USP7
To identify whether USP7 acts as a DUB for PP2A, a
deubiquitination assay was performed subsequent to the expression
of Myc-USP7, Flag-PP2A, and HA-Ub in 293T cells. USP7 decreased the
ubiquitination level of PP2A, and inhibition of the enzymatic
activity of USP7 exerted no change in the ubiquitination level of
PP2A (Fig. 3A). These results
indicated that the catalytic activity of USP7 induced the
deubiquitination of PP2A. In addition, the effect of USP7 on the
K48-linked or K63-linked polyubiquitin chain on PP2A was
investigated (Fig. 3B). It was
determined that USP7 deubiquitinated the K48-linked polyubiquitin
chain on PP2A, with no associated difference in the K63-linked
ubiquitination level on PP2A. This result indicated that USP7
deubiquitinated the K48-linked polyubiquitin chain on PP2A, thereby
establishing that modulation of proteolysis is
proteasome-dependent.
USP7 affects the stability of
PP2A
Since USP7 affects the protein stability by
deubiquitinating the K48-linked polyubiquitin chain on PP2A, the
protein levels of PP2A were investigated to determine whether USP7
controls the PP2A stability in a dose-dependent manner of
siUSP7. First, the siUSP7 was used as previously
determined (Fig. 4A) (27); the experimental protocol to confirm
a decrease in the expression of USP7 was performed as previously
described (Fig. 4B) (27). The expression of PP2A was
dose-dependently reduced after exposure to varying concentrations
of siUSP7 (Fig. 4C). Since
the expression of PP2A was decreased subsequent to inhibition of
USP7, it was further investigated whether siUSP7 also
affects the ubiquitination of PP2A (Fig. 4D). The results confirmed that a
dose-dependent increase in the ubiquitination level of PP2A
subsequent to siUSP7 exposure indicated that the absence of
USP7 may fail to induce deubiquitination of PP2A. Collectively,
these experimental results revealed that the deubiquitination of
PP2A was suppressed following inhibition of USP7, and PP2A
stability was reduced by UPS. Conversely, overexpression of USP7
had no effect on the expression of PP2A (Fig. 4E); if the protein expression is too
strong, the band is affected by saturation (30,31).
Therefore, it appears that saturation of the PP2A protein signal in
the western blotting may hinder the ability to observe an increase
in the expression level of PP2A induced by USP7. Furthermore, the
half-life of PP2A tended to be modulated by siUSP7 in a
time-dependent manner of CHX exposure (Fig. 4F). This indicated that inhibition
of USP7 reduced the protein stability of PP2A.
 | Figure 4.USP7 regulates the stability of PP2A.
(A) A schematic diagram of the siRNA of USP7. (B) MCF-7
cells were transfected with siUSP7 or siNC.
Percentage of PP2A/β-actin from separate experiments (n=3;
**P<0.01). (C) siUSP7 was transfected into MCF-7 cells at
varying concentrations (0, 2, 4, and 8 mM). The expression levels
of PP2A/β-actin were analyzed by at least three independent
experiments (n=3; **P<0.01 and ***P<0.001). (D) HA-Ub
and Flag-PP2A were co-transfected into 293T cells, followed
by transfection with different concentrations of siUSP7 (0,
2, 4, and 8 mM). (E) Myc-USP7 was transfected into HeLa
cells in a dose-dependent manner (0, 2, 4, 6 µg). Percentage of
PP2A/β-actin from three independent experiments (n=3; ns). (F) The
half-life of PP2A was investigated with siUSP7, with
time-dependent exposure to CHX. USP7, ubiquitin-specific protease
7; PP2A, protein phosphatase 2A; siRNA, small interfering RNA;
siNC, siRNA negative control; HA-Ub, HA-ubiquitin;
ns, not significant; CHX, cycloheximide. |
Discussion
USP7 is one of the most prominent DUBs that has been
studied in various cancers (32).
USP7 has an N-terminal domain, a tumor necrosis factor
receptor-associated factor (TRAF) domain, a catalytic domain (CD),
and a C-terminal domain including five ubiquitin-like (UBL) domains
(33). The USP7 CD domain contains
the Ub hydrolase activity (34).
UBL domains are essential for the full activity of USP7 (35). The TRAF domain plays an important
role in recognition of target proteins (16). This domain recognizes target
proteins through the P/A/E-x-x-S motif. P/A/E-x-x-S motifs allow
numerous different interactors capable of binding with USP7, and
are probably responsible for the promiscuity of USP7 with other
binding partners (36). The
secondary binding domain in USP7 is located in UBL-2. UBL-2
interacts with a partner with K-x-x-x-K motifs (37). Interestingly, since PP2A has the
P/A/E-x-x-S motif (that binds to the TRAF domain) but no K-x-x-x-K
motifs, the possibility of interacting with the TRAF domain in USP7
was analyzed in a previous study (15). This suggests the possibility that
PP2A can interact with the TRAF domain in USP7. Moreover, the TRAF
domain is known to regulate intracellular signaling such as
ubiquitin-dependent degradation (38). Since ubiquitination in substrates
occurs in various ubiquitination motifs, further studies are
required to determine whether these motifs, in which USP7 and PP2A
bind to each other are related to protein activity and stability
exerted via ubiquitination. Furthermore, USP7 plays an important
role in tumor suppressor function by deubiquitinating both p53 and
MDM2, one of the E3 ligases for p53 (39). Conversely, USP7 is also considered
as an oncogene, enhancing the stability of c-MYC which contributes
to cancer progression (40). Thus,
USP7 paradoxically acts either as an oncogene or a tumor
suppressor. The substrates targeted by USP7 are therefore
considered to affect cancers through diverse functions, making
research focusing on substrates regulated by USP7 essential. In a
previous study by the authors, PP2A, a substrate to which USP7
binds, was identified through 2-DE and mass spectrometric analysis
(17).
PP2A controls various cell functions, including
protein synthesis, cell cycle regulation, and cell death through
signaling pathways. Therefore, alterations in the levels of PP2A
are considered an interesting topic in cancer research (41). PP2A enhances tumor immunity by
inhibiting the MAPK and PI3K pathways (including AKT, MEK, and
ERK), as well as inhibition of MYC through dephosphorylation
(23). In addition, PP2A contains
numerous subunits, including subunit A, B, and C, and mutations of
PP2A subunits have also been reported in various cancer types
(42). PP2A is known as a critical
tumor suppressor, since it functions as a negative regulator of
oncoproteins, including c-MYC, BCL2, ERK, and AKT (43). Thus, the PP2A subunits exert
numerous roles in cancer cells. Moreover, depending on the cancer
cell type, PP2A acts as a tumor suppressor or an oncogene. Thus,
there is increasing interest in the role of PP2A in cancer
development (44). It has been
confirmed that treatment with a PP2A inhibitor affects drug
resistance and cancer cell survival in various cancer cell types
(45–47). Thus, treatments targeting PP2A are
being considered as potential cancer treatments (48). PP2A has long been regarded as a
tumor suppressor, but recent arguments have contradicted the
original hypothesis that inhibition of PP2A induces cell apoptosis
(49). Therefore, treatment with
PP2A needs to be considered for both aspects of activity and
inhibition, depending on the cell type or type of drugs. As such,
PP2A is considered an important target for anticancer therapy
(50), and research on anticancer
treatment targeting PP2A is in progress. It is expected that
understanding of the enzyme targeting PP2A will also be required
for complete elucidation of the process. Thus, both USP7 and PP2A
play important roles in cancer therapy. The development of small
molecules that inhibit the interplay of USP7 and PP2A could be
considered as a new anticancer treatment.
The present study aimed to provide an understanding
of a new molecular mechanism by elucidating the association between
USP7 and PP2A, which are considered important modulators in cancer
progression. The direct binding of USP7 and PP2A was determined
(Fig. 1C). It was of interest
whether PP2A is neddylated, SUMOylated, and ISGylated as well. The
experimental results indicated that PP2A can be regulated not only
through ubiquitination but also through other processes including
neddylation, and further research on the enzymes modulating these
processes may be required to understand the regulatory mechanism of
PP2A. Results of the deubiquitination assay revealed the
deubiquitination of PP2A through USP7 (Fig. 3B), predicting that the PP2A
activity and stability could be modulated by the removal of
ubiquitins on PP2A by USP7. Furthermore, regulation of the
expression level of PP2A by USP7 was also established (Fig. 4A, B, E and F). These results
indicated that USP7 is a modulator of PP2A stability via the
K48-linked polyubiquitin chain. Since USP7 and PP2A are considered
to be tumor suppressors or oncoproteins, the results indicating
that both proteins are co-localized in various tumor cell lines,
and that USP7 regulates the PP2A stability, suggest that USP7 and
PP2A are intricately associated in various tumors. Further studies
validating the effect of USP7-mediated PP2A stability changes are
required for determining the oncogenic roles of USP7 as a positive
or a negative regulator to PP2A. Moreover, the expression levels of
PP2A and USP7 in cancer tissues need to be confirmed and verified
in vivo, thereby validating the potential of USP7 as a
therapeutic target for cancer in tumor malignant phenotypes. A
previous study demonstrated that USP7 interacts with PP2A and
controls the location of PP2A in the cytoplasm (51). In the present study, the direct
interaction between USP7 and PP2A was confirmed, and the role of
USP7 as a deubiquitinating enzyme was investigated by modulating
the K48-linked ubiquitin chain of PP2A. Additionally, it was
confirmed that PP2A not only forms polyubiquitin chains but also
proteasome-dependent degradation related to polyneddylation.
Collectively, the interplay between USP7 and PP2A provides an
essential understanding that should be considered for the
development of anticancer therapeutics.
Supplementary Material
Supporting Data
Acknowledgements
We would like to thank Ms. Hae-Seul Choi, Ms. So-Hee
Kim, Mr. Hong-Beom Park, and Mr. Sang-Soo Park (all from the
Department of Biomedical Science, CHA University) for proofreading
the manuscript.
Funding
The present study was supported by the Basic Science Research
Program through the National Research Foundation of Korea (NRF)
funded by the Ministry of Education (grant no.
2020R1I1A207500311).
Availability of data and materials
All data generated and/or analyzed during the
present study are included in this published article.
Authors' contributions
HAD and KHB designed the study and confirm the
authenticity of all the raw data. HAD performed most of the
experiments and wrote the manuscript. Both authors have read and
agreed to the published version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Tsuchiya H, Endo A and Saeki Y: Multi-step
ubiquitin decoding mechanism for proteasomal degradation.
Pharmaceuticals (Basel). 13:1282020. View Article : Google Scholar
|
|
2
|
Kim RQ and Sixma TK: Regulation of USP7: A
high incidence of E3 complexes. J Mol Biol. 429:3395–3408. 2017.
View Article : Google Scholar
|
|
3
|
Deng L, Meng T, Chen L, Wei W and Wang P:
The role of ubiquitination in tumorigenesis and targeted drug
discovery. Signal Transduct Target Ther. 5:112020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
French ME, Koehler CF and Hunter T:
Emerging functions of branched ubiquitin chains. Cell Discov.
7:62021. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tracz M and Bialek W: Beyond K48 and K63:
Non-canonical protein ubiquitination. Cell Mol Biol Lett. 26:12021.
View Article : Google Scholar
|
|
6
|
He M, Zhou Z, Shah AA, Zou H, Tao J, Chen
Q and Wan Y: The emerging role of deubiquitinating enzymes in
genomic integrity, diseases, and therapeutics. Cell Biosci.
6:622016. View Article : Google Scholar
|
|
7
|
Zhou X and Sun SC: Targeting ubiquitin
signaling for cancer immunotherapy. Signal Transduct Target Ther.
6:162021. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jacomin AC, Taillebourg E and Fauvarque
MO: Deubiquitinating enzymes related to autophagy: New therapeutic
opportunities? Cells. 7:1122018. View Article : Google Scholar
|
|
9
|
Kim SH and Baek KH: Regulation of cancer
metabolism by deubiquitinating enzymes: The Warburg effect. Int J
Mol Sci. 22:61732021. View Article : Google Scholar
|
|
10
|
Ajadi MB, Soremekun OS, Elrashedy AA,
Olotu FA, Kumalo HM and Soliman MES: Probing protein-protein
interactions and druggable site identification: Mechanistic binding
events between ubiquitin and zinc finger with UFM1-specific
peptidase domain protein (ZUFSP). Comb Chem High Throughput Screen.
25:831–837. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kim SY and Baek KH: TGF-β signaling
pathway mediated by deubiquitinating enzymes. Cell Mol Life Sci.
76:653–665. 2019. View Article : Google Scholar
|
|
12
|
Wang Z, Kang W, You Y, Pang J, Ren H, Suo
Z, Liu H and Zheng Y: USP7: Novel drug target in cancer therapy.
Front Pharmacol. 10:4272019. View Article : Google Scholar
|
|
13
|
Lu J, Zhao H, Yu C, Kang Y and Yang X:
Targeting ubiquitin-specific protease 7 (USP7) in cancer: A new
insight to overcome drug resistance. Front Pharmacol.
12:6484912021. View Article : Google Scholar
|
|
14
|
Veggiani G, Gerpe MCR, Sidhu SS and Zhang
W: Emerging drug development technologies targeting ubiquitination
for cancer therapeutics. Pharmacol Ther. 199:139–154. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Nininahazwe L, Liu B, He C, Zhang H and
Chen ZS: The emerging nature of ubiquitin-specific protease 7
(USP7): A new target in cancer therapy. Drug Discov Today.
26:490–502. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Valles GJ, Bezsonova I, Woodgate R and
Ashton NW: USP7 is a master regulator of genome stability. Front
Cell Dev Biol. 8:7172020. View Article : Google Scholar
|
|
17
|
Park JJ, Lim KH and Baek KH: Annexin-1
regulated by HAUSP is essential for UV-induced damage response.
Cell Death Dis. 6:e16542015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Seshacharyulu P, Pandey P, Datta K and
Batra SK: Phosphatase: PP2A structural importance, regulation and
its aberrant expression in cancer. Cancer Lett. 335:9–18. 2013.
View Article : Google Scholar
|
|
19
|
Wlodarchak N and Xing Y: PP2A as a master
regulator of the cell cycle. Crit Rev Biochem Mol Biol. 51:162–184.
2016. View Article : Google Scholar
|
|
20
|
Goguet-Rubio P, Amin P, Awal S, Vigneron
S, Charrasse S, Mechali F, Labbé JC, Lorca T and Castro A: PP2A-B55
holoenzyme regulation and cancer. Biomolecules. 10:15862020.
View Article : Google Scholar
|
|
21
|
Brautigan DL, Farrington C and Narla G:
Targeting protein phosphatase PP2A for cancer therapy: Development
of allosteric pharmaceutical agents. Clin Sci (Lond).
135:1545–1556. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sangodkar J, Farrington CC, McClinch K,
Galsky MD, Kastrinsky DB and Narla G: All roads lead to PP2A:
Exploiting the therapeutic potential of this phosphatase. FEBS J.
283:1004–1024. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Mazhar S, Taylor SE, Sangodkar J and Narla
G: Targeting PP2A in cancer: Combination therapies. Biochim Biophys
Acta Mol Cell Res. 1866:51–63. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Merisaari J, Denisova OV, Doroszko M, Le
Joncour V, Johansson P, Leenders WPJ, Kastrinsky DB, Zaware N,
Narla G, Laakkonen P, et al: Monotherapy efficacy of blood-brain
barrier permeable small molecule reactivators of protein
phosphatase 2A in glioblastoma. Brain Commun. 2:fcaa0022020.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Pandey P, Seshacharyulu P, Das S,
Rachagani S, Ponnusamy MP, Yan Y, Johansson SL, Datta K, Fong Lin M
and Batra SK: Impaired expression of protein phosphatase 2A
subunits enhances metastatic potential of human prostate cancer
cells through activation of AKT pathway. Br J Cancer.
108:2590–2600. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Park JH, Kim SY, Cho HJ, Lee SY and Baek
KH: YOD1 deubiquitinates NEDD4 involved in the hippo signaling
pathway. Cell Physiol Biochem. 54:1–14. 2020.PubMed/NCBI
|
|
27
|
Choi HS, Pei CZ, Park JH, Kim SY, Song SY,
Shin GJ and Baek KH: Protein stability of pyruvate kinase isozyme
M2 is mediated by HAUSP. Cancers (Basel). 12:15482020. View Article : Google Scholar
|
|
28
|
Song L and Luo ZQ: Post-translational
regulation of ubiquitin signaling. J Cell Biol. 218:1776–1786.
2019. View Article : Google Scholar
|
|
29
|
Lee HJ, Kim MS, Kim YK, Oh YK and Baek KH:
HAUSP, a deubiquitinating enzyme for p53, is polyubiquitinated,
polyneddylated, and dimerized. FEBS Lett. 579:4867–4872. 2005.
View Article : Google Scholar
|
|
30
|
Pillai-Kastoori L, Schutz-Geschwender AR
and Harford JA: A systematic approach to quantitative western blot
analysis. Anal Biochem. 593:1136082020. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kirshner ZZ and Gibbs RB: Use of the
REVERT® total protein stain as a loading control
demonstrates significant benefits over the use of housekeeping
proteins when analyzing brain homogenates by western blot: An
analysis of samples representing different gonadal hormone states.
Mol Cell Endocrinol. 473:156–165. 2018. View Article : Google Scholar
|
|
32
|
Poondla N, Chandrasekaran AP, Kim KS and
Ramakrishna S: Deubiquitinating enzymes as cancer biomarkers: New
therapeutic opportunities? BMB Rep. 52:181–189. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Pozhidaeva A and Bezsonova I: USP7:
Structure, substrate specificity, and inhibition. DNA Repair
(Amst). 76:30–39. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ashton NW, Valles GJ, Jaiswal N, Bezsonova
I and Woodgate R: DNA polymerase ι interacts with both the
TRAF-like and UBL1-2 domains of USP7. J Mol Biol. 433:1667332021.
View Article : Google Scholar
|
|
35
|
Sheng Y, Saridakis V, Sarkari F, Duan S,
Wu T, Arrowsmith CH and Frappier L: Molecular recognition of p53
and MDM2 by USP7/HAUSP. Nat Struct Mol Biol. 13:285–291. 2006.
View Article : Google Scholar
|
|
36
|
Georges A, Marcon E, Greenblatt J and
Frappier L: Identification and characterization of USP7 targets in
cancer cells. Sci Rep. 8:158332018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lai KP, Chen J and Tse WKF: Role of
deubiquitinases in human cancers: Potential targeted therapy. Int J
Mol Sci. 21:25482020. View Article : Google Scholar
|
|
38
|
Yang XD and Sun SC: Targeting signaling
factors for degradation, an emerging mechanism for TRAF functions.
Immuno Rev. 266:56–71. 2015. View Article : Google Scholar
|
|
39
|
Bhattacharya S and Ghosh MK: HAUSP
regulates c-MYC expression via de-ubiquitination of TRRAP. Cell
Oncol (Dordr). 38:265–277. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Bryant JP, Levy A, Heiss J and
Banasavadi-Siddegowda YK: Review of PP2A tumor biology and
antitumor effects of PP2A inhibitor LB100 in the nervous system.
Cancers (Basel). 13:30872021. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Haesen D, Abbasi Asbagh L, Derua R, Hubert
A, Schrauwen S, Hoorne Y, Amant F, Waelkens E, Sablina A and
Janssens V: Recurrent PPP2R1A mutations in uterine cancer act
through a dominant-negative mechanism to promote malignant cell
growth. Cancer Res. 76:5719–5731. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Ruvolo PP: The broken ‘Off’ switch in
cancer signaling: PP2A as a regulator of tumorigenesis, drug
resistance, and immune surveillance. BBA Clin. 6:87–99. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Tinsley SL and Allen-Petersen BL: PP2A and
cancer epigenetics: A therapeutic opportunity waiting to happen.
NAR Cancer. 4:zca0022022. View Article : Google Scholar
|
|
44
|
Cristóbal I, Manso R, Rincón R, Caramés C,
Senin C, Borrero A, Martínez-Useros J, Rodriguez M, Zazo S,
Aguilera O, et al: PP2A inhibition is a common event in colorectal
cancer and its restoration using FTY720 shows promising therapeutic
potential. Mol Cancer Ther. 13:938–947. 2014. View Article : Google Scholar
|
|
45
|
Vera J, Jaumot M, Estanyol JM, Brun S,
Agell N and Bachs O: Heterogeneous nuclear ribonucleoprotein A2 is
a SET-binding protein and a PP2A inhibitor. Oncogene. 25:260–270.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
O'Connor CM, Perl A, Leonard D, Sangodkar
J and Narla G: Therapeutic targeting of PP2A. Int J Biochem Cell
Biol. 96:182–193. 2018. View Article : Google Scholar
|
|
47
|
Perrotti D and Neviani P: Protein
phosphatase 2A: A target for anticancer therapy. Lancet Oncol.
14:e229–e238. 2013. View Article : Google Scholar
|
|
48
|
Liu X, Zhang Y, Wang Y, Yang M, Hong F and
Yang S: Protein phosphorylation in cancer: Role of nitric oxide
signaling pathway. Biomolecules. 11:10042021. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Jiang C, Wu Y, Xia Q and Huang Q: Novel
molecular targets in malignant diseases of digestive system.
Gastroenterol Res Pract. 2013:5682802013. View Article : Google Scholar
|
|
50
|
Arriazu E, Vicente C, Pippa R, Peris I,
Martinez-Balsalobre E, Garcia-Ramirez P, Marcotegui N, Igea A,
Alignani D, Rifón J, et al: A new regulatory mechanism of protein
phosphatase 2A activity via SET in acute myeloid leukemia. Blood
Cancer J. 10:32020. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Galarreta A, Valledor P, Ubieto-Capella P,
Lafarga V, Zarzuela E, Muñoz J, Malumbres M, Lecona E and
Fernandez-Capetillo O: USP7 limits CDK1 activity throughout the
cell cycle. EMBO J. 11:e996922021.
|