Introduction
Bladder cancer (BC) is one of the most common
malignancies of the urinary tract, accounting for over 70,000 new
cases and 16,000 cancer-related deaths in the USA each year
(1). Furthermore, based on the
formation pathway, BC can be divided into two groups:
Non-muscle-invasive BC and muscle-invasive BC (2). Currently, radical cystectomy combined
with chemotherapy is the mainstream treatment for muscle-invasive
BC. However, patients with BC often respond poorly to standard
chemotherapeutic agents (3). Thus,
it is necessary to identify the mechanisms underlying
chemoresistance in BC.
Ferroptosis is a novel form of cell death that is
featured by iron-dependent peroxidation of the lipid membrane
induced by the accumulation of massive amounts of reactive oxygen
species (ROS) (4). Glutathione
peroxidase 4 (GPX4) is an antioxidant that can reduce peroxide,
such as hydrogen peroxide and organic hydroperoxides, by
cooperating with the antioxidant glutathione (GSH) (5). Therefore, during the process of
ferroptosis, inhibition of GPX4 is one of the key events (5). Since ferroptosis is biochemically
different from other forms of cell death, induction of ferroptosis
may be a promising strategy for overcoming chemoresistance in
various cancer cells.
Staphylococcal nuclease and tudor domain containing
1 (SND1) is recognized as a novel oncoprotein that is upregulated
in numerous types of cancer cells (6). SND1 was identified as a transcription
factor that is involved in diverse post-transcriptional regulation
activities, such as mRNA splicing, RNA stability and editing
(7). Furthermore, SND1 is
ubiquitously expressed and highly conserved in mammals and plays
essential physiological roles in various biological activities,
such as cell proliferation, differentiation and death (6). SND1 has also been revealed to be
correlated with chemoresistance to cisplatin in non-small cell lung
cancer (NSCLC) cells (8). At
present, there is little knowledge concerning the role of SND1 in
BC.
The present study aimed to identify the factors that
contribute to the resistance of cisplatin in BC cells. The results
demonstrated that the inhibition of SND1 could overcome
chemoresistance in BC cells by promoting ferroptosis.
Mechanistically, SND1 was revealed to bind to the 3′UTR region of
GPX4 mRNA and stabilise it. The findings of the present study
suggest that targeting SND1 may be a promising strategy to overcome
chemoresistance in BC.
Materials and methods
Cell culture and chemicals
Human bladder cancer cells T24 (ATTC no. HTB-4),
5637 (ATTC no. HTB-9) and RT-4 (ATTC no. HTB-2) were obtained from
American Type Culture Collection (ATCC). RT-112 (product no.
300324) and CLS-439 (product no. 300150) were obtained from Cell
Lines Service GmbH. The cisplatin-resistant T24/R and 5637/R cells
were created by stepwise escalation method: Parental T24 and 5637
cells were cultured with gradually increasing concentrations of
cisplatin from 2 nM to 1 µM over 8 months. All cells were cultured
in RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS)
and 1% penicillin and streptomycin in humidified air with 5%
CO2 at 37°C. All cells were authenticated by STR
profiling and tested for Mycoplasma contamination by
Shanghai Biowing Applied Biotechnology Co., Ltd. The following
reagents were used in the present study: Pan-capase inhibitor
[Z-VAD-FMK (z-VAD); cat. no. S7023; Selleck Chemicals],
necrostatin-1 (Nec-1; cat. no. S8037; Selleck Chemicals),
Liproxstatin-1 (Lip-1; product no. 950455-15-9; Sigma-Aldrich;
Merck KGaA), ferrostatin-1 (Fer-1; cat. no. S7243; Selleck
Chemicals), deferoxamine (DFO; cat. no. HY-B0988; MedChemExpress),
L-γ-glutamyl-p-nitroanilide (GPNA; cat. no. S6670; Selleck
Chemicals), N-acetylcysteine (cat. no. S1623; Selleck Chemicals),
actinomycin D (ActD; cat. no. S8964; Selleck Chemicals). The
concentrations of the various inhibitors used were as follows:
z-VAD, 20 mM; Nec-1, 10 mM; Lip-1, 10 mM; Fer-1, 10 mM; DFO, 10 mM;
and GPNA, 10 mM. The inhibitors were added into the culture medium
and incubated with cells at 37°C. Cisplatin (product no.
15663-27-1) and all other routine chemicals were purchased from
Sigma-Aldrich; Merck KGaA.
Assessment of ROS
The levels of ROS were assessed using the C11-BODIPY
staining kit (cat. no. D3861; Life Technologies; Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions.
Briefly, the T24/R and 5637/R cells were seeded into a 24-well
plate at a density of 10,000 cells/well. Following various
treatments (transfected with shNC/shSND1 or vector/GPX4 OV for 24
h; then treated with or without cisplatin), the cells were
collected and stained with C11-BODIPY (5 µM) at 37°C for 0.5 h. The
FACSCanto II flow cytometer (BD Biosciences) was used to assess the
results. The results were analyzed using the FlowJo 2.0 software
(Tree Star, Inc.).
Assessment of cell death
T24/R and 5637/R cells were seeded onto a 6-well
plate at a density of 1×106 cells/well. Following
various treatments (transfected with shNC/shSND1 or vector/GPX4 OV
for 24 h; then treated with or without cisplatin), cells were
collected and resuspended with PBS and stained with Annexin
V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) solution
(BD Biosciences) at room temperature for 30 min. The Annexin
V-FITC/PI positive cells were analysed by flow cytometry on BD
FACSCalibur (BD Biosciences). The results were analyzed using the
FlowJo 2.0 software.
Assessment of ferrous ions
(Fe2+), GSH and lipid peroxidation marker,
malondialdehyde (MDA)
The levels of Fe2+, GSH and MDA were
assessed using the Iron Assay Kit (product code ab83366,
colorimetric), GSH assay kit (product code ab239727, colorimetric)
and MDA assay kit (product code ab118970, colorimetric), according
to the manufacturer's guide respectively. All kits were obtained
from Abcam.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from the cells using TRIzol
reagent (Beijing Solarbio Science & Technology Co., Ltd.)
according to the manufacturer's instructions. The mRNA levels of
various genes were assessed using a first-strand cDNA synthesis kit
(cat. no. K1612) and SYBR Green Master Mix kit (cat. no. A25741;
both from Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions. The quantitative real-time PCR
reaction was performed using an ABI 7500 Fast real-time PCR system
(Applied Biosystems; Thermo Fisher Scientific, Inc.). The following
thermocycling conditions were used: Initial denaturation at 95°C
for 10 min followed by 35 cycles at 95°C for 15 sec, 55°C for 30
sec, 72°C for 30 sec and a final extension at 72°C for 5 min. The
expression of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was
used as an internal control. The following primers were used in the
present study: SND1 forward, 5′-GTGGACAGCGTAGTTCGGGA-3′ and
reverse, 5′-CCCACGAGACATTTCCACACAC-3′; SLC7A11 forward,
5′-GGGCAGCGTGGGCATGTCTCT-3′ and reverse
5′-GGACCAAAGACTTCCAAAATA-3′; ACSL4 forward,
5′-GCCCACTTCAGACAAACCTGG-3′ and reverse,
5′-ACAGCTTCTCTGCCAAGTGTGG-3′; SLC3A1 forward,
5′-GGGTGTTGATGGTTTTAGTTTGGAT-3′ and reverse,
5′-GCATTCCCACCTGCGAGGTGGAGAAG-3′; GPX4 forward,
5′-GGGACGACTGGCGCTGTGCGCGCTCC-3′ and reverse,
5′-CTCACTGGGAGGCCACGTTG-3′; GSS forward,
5′-CTTCAACCTGCTAGTGGATGCTGT-3′ and reverse,
5′-TGGAACATGTAGTCTGAGCGATTC-3′; GCL forward,
5′-TTGCCTCCTGCTGTGTGATG-3′ and reverse,
5′-ATCATTGTGAGTCAACAGCTGTATGTC-3′; GAPDH forward,
5′-TGAAGGTCGGAGTCAACGGATTTGGT-3′ and reverse,
5′-CATGTGGGCCATGAGGTCCACCAC-3′. The gene expression levels were
calculated using the 2−ΔΔCq method (9).
Luciferase assay
The wild-type and truncated mutant 3′UTR of GPX4
were inserted into the pGL3-basic vector (Life Technologies; Thermo
Fisher Scientific, Inc.). The empty vector (EV) and SND1 expressing
vector (SND1 OV) and pGL3 vector were co-transfected into the cells
using Lipofectamine 2000 reagent (Life Technologies; Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions. The
cells were collected 48 h after transfection and then assayed by
the Dual-luciferase reporter assay system (Promega Corporation).
The relative firefly activity was normalized to Renilla
luciferase activity according to the manufacturer's
instructions.
Chromatin IP (ChIP) analysis
ChIP analysis was carried out using a Millipore
EZ-Magna ChIP kit (cat. no. 17-295; EMD Millipore) according to the
manufacturer's protocol. Briefly, the T24/R and 5637/R cells were
seeded into a 6-well plate at a density of 5×106
cells/well. The cells were collected after 24 h of treatment and
fixed with 1% formaldehyde for 10 min at room temperature. After
rehydrating in 0.125 M glycine, the cells were washed in
phosphate-buffered saline (PBS) and resuspended in ChIP lysis
buffer. The cell lysate was centrifuged at 10,000 × g for 10 min at
4°C. Anti-immunoglobulin G (IgG) (product code ab131368; Abcam) or
anti-SND1 antibody (product code ab65078; Abcam) were incubated
with Dynabeads protein G (cat. no. 10003D; Life Technologies;
Thermo Fisher Scientific, Inc.) for 2 h at 4°C, followed by
incubation with precleared chromatin overnight at 4°C. The
precipitated RNA samples and inputs were subjected to RT-qPCR.
Short hairpin RNA (shRNA)
transfection
shRNA against SND1 (shSND1#1,
5′-AAGGCATGAGAGCTAATAATC-3′; and shSND1#2,
5′-GCCACAACCAGAACATTCTG-3′); GPX4 (shGPX4#1,
5′-CATCGTCACCAACGTGGCC-3′; and shGPX4#2
5′-TCAAATTCGATATGTTCAGCAAGA-3′) and negative control shRNA (shNC;
5′-CCATCGCATTCACTATGCTAAG-3′) were purchased from Shanghai
GenePharm Co., Ltd.
shRNAs were subcloned into the pSilencer 4.1-CMV
FANCF shRNA vector (Life Technologies; Thermo Fisher Scientific,
Inc.). The transfection was performed using Lipofectamine 2000
(Life Technologies; Thermo Fisher Scientific, Inc.) according to
the manufacturer's guide. Briefly, the T24/R and 5637/R cells were
seeded into 6-well plates at a density of 2.5×106
cells/well in full RPMI-1640 medium. When the cells reached ~70%
confluence, the culture medium was replaced with 2 ml fresh
complete medium. A total of 2.5 µg of plasmid was then diluted in
100 µl of Opti-MEM medium (Life Technologies; Thermo Fisher
Scientific, Inc.) with 5 µl of Lipofectamine 2000. The solution was
incubated at room temperature for 30 min and added into the cells.
Subsequently, 24 h later, western blotting was used to assess the
efficiency of the transfection.
Overexpression of SND1 and GPX4
To overexpressing SND1 and GPX4, full-length cDNAs
were subcloned into pcDNA3.1 vector (Life Technologies; Thermo
Fisher Scientific, Inc.) and a blank vector was used as a negative
control. Vectors were transfected into cells using Lipofectamine
2000 (Life Technologies; Thermo Fisher Scientific, Inc.) according
to the manufacturer's instructions. Briefly, 10 µg of plasmids were
diluted into 250 µl Opti-MEM medium (Life Technologies; Thermo
Fisher Scientific, Inc.), and gently mixed at room temperature for
5 min. Subsequently, 5 µl of Lipofectamine 2000 was added into the
mixed Opti-MEM medium and incubated at room temperature for 20 min.
The Opti-MEM medium was added into cells and supplemented with 750
µl of RPMI-1640 medium. A total of 6 h later, the medium was
changed with fresh RPMI-1640 medium. The transfection efficiency
was evaluated 48 h later using western blotting.
Western blotting
Cells were lysed using the radioimmunoprecipitation
assay (RIPA) buffer (Beyotime Institute of Biotechnology), and the
Bradford assay kit was used to measure the protein concentrations.
An equal amount of protein (20 µg) was resolved on a 10%
SDS-polyacrylamide gel electrophoresis (PAGE) and transferred to a
polyvinylidene fluoride (PVDF; BD Biosciences) membrane. The PVDF
membranes were then blocked with 5% skimmed milk at room
temperature for 1 h. Subsequently, the membranes were incubated
with primary antibodies at following dilutions: Caspase-3 (product
no. 14220; dilution 1:1,000; Cell Signaling Technology, Inc.), SND1
(product code ab65078; dilution 1:1,000; Abcam), GPX4 (product no.
52455; dilution 1:1,000; Cell Signaling Technology, Inc.), GAPDH
(product no. HPA040067; 1:10,000; Sigma-Aldrich; Merck KGaA)
overnight at 4°C. The membranes were then incubated with secondary
horseradish peroxidase-conjugated antibodies (sheep-anti-rabbit;
product no. AP510; 1:5,000; Sigma-Aldrich; Merck KGaA). The results
were visualized using the Chemiluminescent Imaging System (Tanon
Science and Technology Co., Ltd.).
RNA sequencing (RNA-seq)
The total cellular RNA was extracted using TRIzol
(cat. no. 15596018; Life Technologies; Thermo Fisher Scientific,
Inc.) according to the manufacturer's instructions. NanoDrop 2000
spectrophotometer (Thermo Fisher Scientific, Inc.) was used to
determine the quality of RNA. The RNA samples that met the
following criteria were used in the following investigations: RNA
integrity number >7.0 and 28S:18S ratio >1.5. Messenger RNA
templates were then isolated with oligo-dT Dynabeads (cat. no.
61005; Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions and fragmented to an average size of
200 nucleotides by incubation in fragmentation buffer (cat. no.
B0330, Enzymatics; Qiagen, Inc.) for 2 min at 70°C. Sequencing
libraries and data analysis were constructed and performed by
BioMarker Technologies Corporation. Briefly, mRNA sequencing
libraries were created using the Illumina TruSeq Stranded mRNA
library Prep Kit (cat. no. RS-122-2101; Illumina, Inc.) according
to the manufacturer's instructions. The quality of the libraries
was verified using Caliper LabChip GX/HT DNA High sensitivity Kit
(cat. no. 50674626; PerkinElmer Inc.). The quantitation of
libraries was verified by Qubit dsDNA HS (cat. no. Q32851; Thermo
Fisher Scientific, Inc.). The paired-end sequencing was performed
using the NovaSeq 6000 sequencing system (Illumina, Inc.) with a
final library concentration of 6 pM as determined by qPCR. The heat
map was designed to visualize the results using ggplots v3.0
(https://cran.r-project.org/web/packages/ggsignif/ggsignif.pdf),
which is a package based on the R language. RNA sequencing data was
uploaded at NCBI Sequence Read Archive (SRA) database (accession
no. PRJNA835161; http://www.ncbi.nlm.nih.gov/bioproject/PRJNA835161).
Enrichment analysis using kyoto
encyclopedia of genes and genomes (KEGG)
KEGG pathway analysis (10) was conducted to analyze the
overlapping differentially expressed genes (DEGs). KEGG pathway
analysis revealed biological pathways associated with DEGs. A
P-value <0.05 was used as the cutoff standard.
Statistical analysis
All statistical analyses were performed using
SPSS12.0 (SPSS, Inc.). Data are presented as the mean ± standard
error. All experiments were repeated at least three times.
Statistical differences between two groups were determined using
the unpaired Student's t-test or one-way analysis of variance
followed by post hoc Tukey's test for multiple comparisons.
P<0.05 was considered to indicate a statistically significant
difference.
Results
SND1 regulates the sensitivity of BC
cells to cisplatin
First, the IC50 values of the response of
different BC cells to cisplatin were determined (Fig. 1A). Because T24 and 5637 cells were
more sensitive than other BC cells, they were selected to create
cisplatin-insensitive BC cells. Cell viability assays revealed that
T24/R and 5637/R cells were markedly less sensitive to cisplatin
than the parental T24 and 5637 cells (Fig. 1B). To reveal the potential
mechanisms underlying the resistance to cisplatin in BC cells, RNA
seq was conducted in T24 and T24/R cells. It was determined that
SND1 was significantly upregulated in T24/R cells compared with T24
cells (Fig. 1C). RT-qPCR confirmed
that SND1 mRNA was significantly increased in T24/R and 5637/R
cells compared with T24 and 5637 cells (Fig. 1D). Western blotting also revealed
that the protein levels of SND1 were upregulated in
cisplatin-insensitive T24/R and 5637/R cells compared with
cisplatin-normal T24 and 5637 cells (Fig. 1E). Furthermore, the expression of
SND1 was assessed in various cancer cells. It was found that the
mRNA and protein levels of SND1 were negatively associated with the
sensitivity to cisplatin in BC cells (Fig. 1F and G). To evaluate whether SND1
could affect the sensitivity of BC cells to cisplatin, two shRNAs
were introduced into T24/R and 5637/R cells to knockdown SND1
(Fig. 1H and I). Cell viability
assays demonstrated that silencing of SND1 significantly inhibited
the cell viability of T24/R and 5637/R cells when treated with
cisplatin (Fig. 1J). Collectively,
these data indicated that silencing of SND1 overcame resistance to
cisplatin in BC cells.
 | Figure 1.Silencing of SND1 increases the
sensitivity of BC cells to cisplatin. (A) The IC50
values of the response of BC cells to cisplatin. (B) Treatment of
T24/R, T24, 5637/R and 5637 cells with various concentrations of
cisplatin for 24 h, and assessment of their cellular viabilities.
(C) RNA sequencing analysis of gene expression in T24/R and T24
cells. (D) mRNA levels of SND1 were assessed by reverse
transcription-quantitative polymerase chain reaction in T24, T24/R,
5637 and 5637/R cells. (E) Western blotting was used to assess the
protein levels of SND1. (F) mRNA levels of SND1 in various BC
cells. (G) Protein levels of SND1 in various BC cells. (H) T24/R
and 5637/R cells were transfected as indicated, and the mRNA levels
of SND1 were assessed. (I) T24/R and 5637/R cells were transfected
as indicated, and protein levels of SND1 were determined. (J) T24/R
and 5637/R cells were transfected as indicated, and treated with
various concentrations of cisplatin for 24 h, and their cellular
viabilities were assessed. Data are presented as the mean ±
standard deviation. **P<0.01. SND1, staphylococcal nuclease and
tudor domain containing 1; BC, bladder cancer; sh, short hairpin
RNA; NC, negative control. |
Silencing of SND1 promotes ferroptosis
induced by cisplatin in BC cells
Next, it was verified whether silencing of SND1
could affect cell death induced by cisplatin in BC cells. Flow
cytometry revealed that knockdown of SND1 significantly promoted
cell death caused by cisplatin in T24/R and 5637/R cells (Fig. 2A). To confirm the cell death caused
by cisplatin and inhibition of SND1, various cell death inhibitors
were applied. Notably, it was found that apoptosis inhibitor
(z-VAD) and necrosis inhibitor (Nec-1) had little effect on cell
death caused by cisplatin in T24/R and 5637/R cells following
knockdown of SND1 (Fig. 2B). In
addition, ferroptosis inhibitors Lip-1 and Fer-1 significantly
attenuated cell death induced by cisplatin in T24/R and 5637/R
cells after knockdown of SND1 (Fig.
2B). To further confirm that the form of cell death was
ferroptosis and not any other form, several markers of ferroptosis
were used. It was revealed that cisplatin induced a significant
increase of Fe2+ in T24/R and 5637/R cells after
knockdown of SND1 (Fig. 2C), and
this effect could be reversed by the iron chelator DFO and
glutamine transporter inhibitor GPNA (Fig. 2C). Additionally, cisplatin
treatment combined with the knockdown of SND1 induced accumulation
of ROS which could be terminated by the ROS scavenger
N-acetylcysteine in T24/R and 5637/R cells (Fig. 2D). Assessment of GSH indicated that
knockdown of SND1 combined with cisplatin significantly reduced the
levels of GSH in T24/R and 5637/R cells (Fig. 2E). In addition, MDA assays showed
that the knockdown of SND1 combined with cisplatin significantly
promoted the levels of MDA in T24/R and 5637/R cells (Fig. 2F). These findings indicated that
knockdown of SND1 overcame the resistance to cisplatin by promoting
ferroptosis in cisplatin-resistant BC cells.
 | Figure 2.Silencing of SND1 overcomes the
resistance to cisplatin by promoting ferroptosis. (A) T24/R and
5637/R cells were transfected as indicated, and treated with or
without cisplatin for 24 h, and cellular death was then assessed.
(B) T24/R and 5637/R cells were treated as indicated for 24 h, and
the cellular death was assessed. (C) T24/R and 5637/R cells were
treated as indicated for 24 h, and the iron levels were determined.
(D) T24/R and 5637/R cells were treated as indicated for 12 h, and
the reactive oxygen species levels were assessed. (E) T24/R and
5637/R cells were treated as indicated for 24 h, and the
glutathione levels were determined. (F) T24/R and 5637/R cells were
treated as indicated for 24 h, and the MDA levels were assessed.
Data are presented as the mean ± standard deviation. *P<0.05 and
**P<0.01. SND1, staphylococcal nuclease and tudor domain
containing 1; sh, short hairpin RNA; NC, negative control; ROS,
reactive oxygen species; GSH, glutathione; MDA,
malondialdehyde. |
SND1 physically binds the 3′UTR of
GPX4 mRNA and promotes its stability
To better understand the mechanism of the regulatory
effects of SND1 on ferroptosis in BC cells treated with cisplatin,
an attempt was made to identify the downstream targets of SND1.
According to the KEGG database (http://www.genome.ad.jp/kegg/), several
ferroptosis-related genes (SLC7A11, ACSL4, SLC3A1, GSS, GPX4
and GCL) were selected as candidates (Fig. 3A). The mRNA levels of the
aforementioned candidate genes were assessed after the knockdown of
SND1. Compared with the control group (shNC), the relative
expression of GPX4 was significantly decreased while other genes
were less affected (Fig. 3A).
Western blotting also revealed that the protein levels of GPX4 in
cisplatin-insensitive cells were increased compared with their
parental cells (Fig. 3B).
Knockdown of SND1 also led to the downregulation of GPX4 in T24/R
and 5637/R cells (Fig. 3C). SND1
has been reported as a transcription factor that was able to bind
with mRNAs (11). It was
hypothesized that SND1 may bind with the mRNA of GPX4 and stabilise
it. To verify this, first, an RNA immunoprecipitation ChIP assay
was performed and high enrichment of GPX4 mRNA in the SND1-RNA
binding complex, was determined using anti-IgG as a negative
control (Fig. 3D). Furthermore, a
vector that successfully overexpressed SND1 (SND1 OV) was
constructed in BC cells (Fig. 3E).
The sequences of the 5′-UTR, coding sequence (CDS) and 3′-UTR were
cloned into the pGL3 vectors and transfected into the BC cells
along with EV or SND1 OV to perform dual-luciferase activity assay.
It was observed that the overexpression of SND1 markedly increased
the relative luciferase activity in BC cells bearing the 3′-UTR but
no other sequences (Fig. 3F). To
further confirm the interaction between SND1 and the 3′-UTR of GPX4
mRNA, vectors expressing the wild-type 3′UTR (WT) and two truncated
mutant forms (Mut 1; Mut 2) were constructed (Fig. 3G). Notably, it was found that SND1
OV increased the luciferase activity in BC cells transfected with
the WT and Mut 1 but not the Mut 2 (Fig. 3G). These findings further
demonstrated that SND1 binds to the 3′UTR region of GPX4 at the
position from nucleotide 765 to nucleotide 865. ActD (2.5 µg/ml)
was also used to inhibit the synthesis of new RNAs in either shNC-
or shSND1-transfected cells and the mRNA decay at various
time-points was assessed. It was determined that ~10 h after ActD
treatment, the abundance of GPX4 mRNA was markedly decreased in
shSND1-transfected BC cells compared with shNC-transfected cells
(Fig. 3H). Collectively these
findings indicated that SND1 positively regulated the mRNA
stability of GPX4.
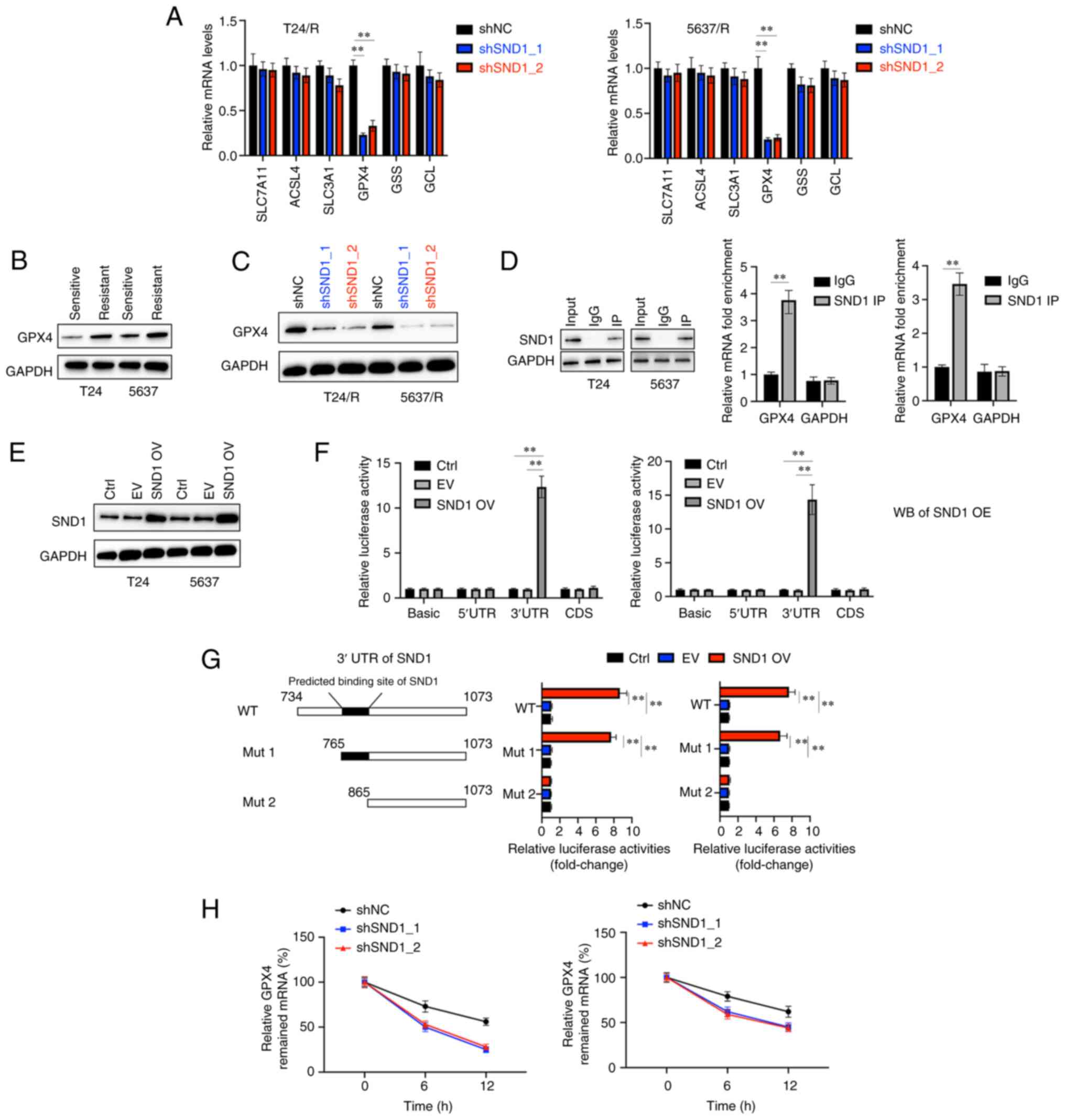 | Figure 3.SND1 directly binds to the 3′UTR of
GPX4 mRNA and stabilises it. (A) T24/R and 5637/R cells were
transfected as indicated, and the mRNA levels of indicated genes
were determined. (B) The protein levels of GPX4 were assessed in
T24, T24/R, 5637 and 5637/R cells. (C) T24/R and 5637R cells were
transfected as indicated, and the protein levels of GPX4 were
assessed. (D) T24/R and 5637/R cells were subjected to
immunoprecipitation with SND1 antibody or control IgG and GAPDH
followed by immunoblotting analysis (left). Reverse
transcription-quantitative polymerase chain reaction analysis of
the relative enrichment of GPX4 mRNA in SND1-RNA binding complexes,
using anti-IgG as a negative control (right). (E) T24 and 5637
cells were transfected as indicated, and the protein levels of SND1
were measured. (F) 5′UTR, 3′UTR and CDS of GPX4 were cloned into a
luciferase reporter vector and co-transfected with a vector that
expressed SND1 in the T24 and 5637 cells, and the relative
luciferase activities were measured. (G) Wild-type or truncated
3′UTR sequences (Mut 1, Mut 2) of GPX4 3′UTR were co-transfected
with SND1-expressing vector into T24 and 5637 cells, and the
relative luciferase activities were measured. (H) GPX4 mRNA
abundance in shNC or shSND1_1 or shSND1_2 transfected cells after
actinomycin D (2.5 µg/ml) administration at different time-points
(10 and 24 h). Data are presented as the mean ± standard deviation.
**P<0.01. SND1, staphylococcal nuclease and tudor domain
containing 1; GPX4, glutathione peroxidase 4; CDS, coding sequence;
sh, short hairpin RNA; NC, negative control; Ctrl, control; EV,
empty vector; OV, overexpression. |
Knockdown of GPX4 overcomes
chemoresistance of BC cells to cisplatin by promoting
ferroptosis
The role of GPX4 in overcoming chemosensitivity in
BC cells was investigated. First, two shRNAs were used to knockdown
both the mRNA and protein levels of GPX4 in T24/R and 5637/R cells
(Fig. 4A and B). As expected,
silencing of GPX4 significantly decreased the viabilities of both
T24/R and 5637/R cells under the treatment of various
concentrations of cisplatin for 24 h (Fig. 4C). Flow cytometric analysis also
demonstrated that silencing of GPX4 significantly increased cell
death induced by cisplatin in T24/R and 5637/R cells (Fig. 4D). Additionally, it was also
observed that silencing of GPX4 led to the significant upregulation
of ROS, Fe2+ and MDA and downregulation of GSH compared
with the shNC group in T24/R and 5637/R cells (Fig. 4E-H). These data indicated that the
SND1-GPX4 axis regulated the sensitivity of BC cells to cisplatin
and knockdown of GPX4 overcame resistance to cisplatin in BC
cells.
 | Figure 4.Knockdown of GPX4 overcomes the
resistance to cisplatin in BC cells. (A) T24/R and 5637/R cells
were transfected as indicated, and the GPX4 mRNA levels were
assessed. (B) T24/R and 5637/R cells were transfected as indicated,
and the protein levels of GPX4 were determined. (C) T24/R and
5637/R cells were transfected as indicated, and the cells were
treated with the indicated concentrations of cisplatin for 24 h,
and the cellular viabilities were assessed. (D) T24/R and 5637/R
cells were treated as indicated, and cellular death was detected.
(E) T24/R and 5637/R cells were treated as indicated, and the
reactive oxygen species levels were assessed. (F) T24/R and 5637/R
cells were treated as indicated, and the iron levels were
determined. (G) T24/R and 5637/R cells were treated as indicated,
and the glutathione levels were detected. (H) T24/R and 5637/R
cells were treated as indicated, and lipid peroxidation levels were
assessed. Data are presented as the mean ± standard deviation.
*P<0.05 and **P<0.01. GPX4, glutathione peroxidase 4; BC,
bladder cancer; sh, short hairpin RNA; NC, negative control; ROS,
reactive oxygen species; GSH, glutathione; MDA,
malondialdehyde. |
Overexpression of GPX4 attenuates
ferroptosis caused by cisplatin and knockdown of SND1 in BC
cells
Furthermore, the role of the SND1-GPX4 axis in
regulating the sensitivity of BC cells to cisplatin was
investigated. T24/R and 5637/R cells were transfected with a vector
expressing GPX4 that successfully overexpressed the mRNA and
protein levels of GPX4 (Fig. 5A and
B). Cell viability assays revealed that overexpression of GPX4
significantly promoted cellular viabilities under the treatment of
cisplatin and knockdown of SND1 (Fig.
5C). Flow cytometry also indicated that overexpression of GPX4
protected cells from cell death induced by cisplatin and knockdown
of SND1 (Fig. 5D). Furthermore,
the combined effects of cisplatin and knockdown of SND1 on the
levels of ROS, Fe2+, GSH and MDA could also be abrogated
by forced expression of GPX4 in T24/R and 5637/R cells (Fig. 5E-H). These data confirmed that
inhibition of SND1 promoted ferroptosis induced by cisplatin and
the knockdown of GPX4.
 | Figure 5.Overexpression of GPX4 reverses the
effects of silencing of SND1 on the sensitivity of BC cells to
cisplatin. (A) T24/R and 5637/R cells were transfected as indicated
for 24 h, and the mRNA levels of GPX4 were determined. (B) The
protein levels of GPX4 were assessed. (C) T24/R and 5637/R cells
were treated as indicated, and the cellular viabilities were
detected. (D) Cellular death was assessed. (E) The reactive oxygen
species levels were determined. (F) Th iron levels were detected.
(G) The glutathione levels were assessed. (H) Malondialdehyde
levels were determined. Data are presented as the mean ± standard
deviation. *P<0.05 and **P<0.01. GPX4, glutathione peroxidase
4; SND1, staphylococcal nuclease and tudor domain containing 1; BC,
bladder cancer; sh, short hairpin RNA; NC, negative control; EV,
empty vector; OV, overexpression; ROS, reactive oxygen species;
GSH, glutathione; MDA, malondialdehyde. |
Discussion
Although great progress has been made in the
treatment of BC, chemoresistance often hinders the clinical
outcomes (12). Furthermore, only
half the patients with BC respond to cisplatin-based chemotherapy
(13). Recently, evidence has
revealed that induction of ferroptosis is a promising strategy to
overcome chemoresistance (14). In
the present study, it was determined that upregulation of SND1
confered resistance to cisplatin, whereas knockdown of SND1
overcame resistance to cisplatin by promoting ferroptosis.
SND1 gene is located on chromosome 7q31.3 and
encodes an evolutionarily conserved protein with five highly
conserved domains (15). SND1 was
initially identified as a transcription co-factor that interacts
with Epstein-Barr nuclear antigen 2 in lymphocytes (16). SND1 has also been reported to be
able to form a complex with the splicing factor SAM68 and regulate
its splicing activity (11). SND1
has been identified as an oncogene in various cancers such as liver
carcinoma, cervical cancer and breast cancer (17–19).
However, there is still little knowledge concerning the role of
SND1 in BC. A study based on The Cancer Genome Atlas and Gene
Expression Omnibus data revealed that high expression of SND1 was
correlated with poor prognosis of BC (20). SND1 contributes to the development
of cancer through multiple mechanisms, such as transcriptional
activation of oncogenes, degradation of tumour suppressor proteins
and post-transcriptional inhibition of the mRNA of tumour
suppressors (21). SND1 has also
been demonstrated to be involved in the chemoresistance of various
cancers. For instance, upregulation of SND1 resulted in blocking of
apoptosis and conferring resistance to cisplatin, oxaliplatin and
5-fluorouracil (5-FU) in NSCLC (8). Downregulation of SND1 enhanced
apoptosis induced by 5-FU in hepatocellular carcinoma cells
(22). In line with the previous
studies, the present study also revealed that overexpression of
SND1 conferred resistance to cisplatin and inhibition of SND1
overcame this resistance in BC cells. Notably, it was found that
SND1 affected cell death caused by cisplatin mainly by regulating
ferroptosis and not apoptosis.
Ferroptosis is different from other forms of cell
death such as apoptosis, necrosis and autophagy, and has a
characteristic accumulation of ROS caused by iron accumulation and
lipid peroxidation (4). Strategies
to trigger ferroptosis have been demonstrated as a promising
anticancer strategy and can mitigate the chemoresistance of tumour
cells that escape apoptosis (23).
Interestingly, cisplatin-resistant tumour cells are prone to
ferroptosis (24). In the present
study, it was determined that cisplatin combined with the silencing
of SND1 induced cell death mainly by ferroptosis, as evidenced by
the upregulation of ROS, Fe2+, MDA and downregulation of
GSH. Mechanistically, it was demonstrated that SND1 positively
regulated the expression of GPX4 by binding and stabilising its
mRNA. GPX4 belongs to the GPX family and was initially identified
to protect liposomes and cellular membranes against iron-catalysed
lipid peroxidation (25). Although
the role of GPX4 in tumorigenesis is still largely elusive,
numerous studies have determined that inhibition of GPX4 can
overcome chemoresistance in various cancer cells. For instance,
inhibition of GPX4 promoted sensitivity to gefitinib in breast
cancer cells (26). Inhibition of
GPX4 was also revealed to overcome resistance to various
chemotherapeutic agents, such as lapatinib, palbociclib, cisplatin
and topotecan in lung cancer cells (27). Another key finding of the present
study is that GPX4 mRNA stabilised itself by binding to SND1.
Previous studies have revealed that the translation of GPX4 mRNA is
under the regulation of various RNA binding proteins, such as
guanine-rich sequence-binding factor 1, deleted in
azoospermia-associated protein 1 and SECIS binding protein 2
(28–30). Moreover, GPX4 mRNA was also
revealed to be directly subjected to the regulation of miRNAs such
as miR-214-3p, miR-1224, miR-324-3p and miR-541-3p (31–34).
These findings revealed the complexity of the post-transcriptional
regulation of GPX4.
Nevertheless, there are still some unresolved
issues. First, the upstream events that cause the upregulation of
SND1 are still not fully understood. A previous study suggested
that nuclear factor-κB, specific protein 1 and nuclear factor Y can
act as transcriptional regulators of SND1 (35). Notably, activation of these
transcription factors has also been demonstrated to be correlated
with the resistance to cisplatin (36). In a future study, it would be of
value to assess whether these factors also affect the process of
ferroptosis. Second, the findings of the present study were only
confirmed in cell lines. It would be of interest to confirm the
findings in vivo.
In conclusion, the present study indicated that
silencing of SND1 overcame the resistance to cisplatin in BC cells
by promoting ferroptosis. Mechanistically, investigations revealed
that SND1 binds to the 3′UTR of GPX4 and stabilises it. Therefore,
targeting the SND1-GPX4 axis may be a potential strategy to
overcome chemoresistance in BC cells.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
All the data in the present study are available upon
reasonable request from the corresponding author.
Authors' contributions
YZ performed most of the experiments. PR repeated
the experiments and confirmed the data. ZY performed the
statistical analysis. LW repeated some of the experiments and
statistical analysis. CH designed the study and drafted the
manuscript. YZ and PR confirm the authenticity of all the raw data.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD, Fuchs HE and Jemal
A: Cancer statistics, 2021. CA Cancer J Clin. 71:7–33. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sanli O, Dobruch J, Knowles MA, Burger M,
Alemozaffar M, Nielsen ME and Lotan Y: Bladder cancer. Nat Rev Dis
Primers. 3:170222017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Minato A, Fujimoto N and Kubo T: Squamous
differentiation predicts poor response to cisplatin-based
chemotherapy and unfavorable prognosis in urothelial carcinoma of
the urinary bladder. Clin Genitourin Cancer. 15:e1063–e1067. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Dixon SJ, Lemberg KM, Lamprecht MR, Skouta
R, Zaitsev EM, Gleason CE, Patel DN, Bauer AJ, Cantley AM, Yang WS,
et al: Ferroptosis: An iron-dependent form of nonapoptotic cell
death. Cell. 149:1060–1072. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yang WS, SriRamaratnam R, Welsch ME,
Shimada K, Skouta R, Viswanathan VS, Cheah JH, Clemons PA, Shamji
AF, Clish CB, et al: Regulation of ferroptotic cancer cell death by
GPX4. Cell. 156:317–331. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jariwala N, Rajasekaran D, Srivastava J,
Gredler R, Akiel MA, Robertson CL, Emdad L, Fisher PB and Sarkar D:
Role of the staphylococcal nuclease and tudor domain containing 1
in oncogenesis (review). Int J Oncol. 46:465–473. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Caudy AA, Ketting RF, Hammond SM, Denli
AM, Bathoorn AM, Tops BB, Silva JM, Myers MM, Hannon GJ and
Plasterk RH: A micrococcal nuclease homologue in RNAi effector
complexes. Nature. 425:411–414. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zagryazhskaya A, Surova O, Akbar NS,
Allavena G, Gyuraszova K, Zborovskaya IB, Tchevkina EM and
Zhivotovsky B: Tudor staphylococcal nuclease drives chemoresistance
of non-small cell lung carcinoma cells by regulating S100A11.
Oncotarget. 6:12156–12173. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ogata H, Goto S, Sato K, Fujibuchi W, Bono
H and Kanehisa M: KEGG: Kyoto encyclopedia of genes and genomes.
Nucleic Acids Res. 27:29–34. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cappellari M, Bielli P, Paronetto MP,
Ciccosanti F, Fimia GM, Saarikettu J, Silvennoinen O and Sette C:
The transcriptional co-activator SND1 is a novel regulator of
alternative splicing in prostate cancer cells. Oncogene.
33:3794–3802. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
McKnight JJ, Gray SB, O'Kane HF, Johnston
SR and Williamson KE: Apoptosis and chemotherapy for bladder
cancer. J Urol. 173:683–690. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cai Z, Zhang F, Chen W, Zhang J and Li H:
miRNAs: A promising target in the chemoresistance of bladder
cancer. Onco Targets Ther. 12:11805–11816. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Mou Y, Wang J, Wu J, He D, Zhang C, Duan C
and Li B: Ferroptosis, a new form of cell death: Opportunities and
challenges in cancer. J Hematol Oncol. 12:342019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ochoa B, Chico Y and Martinez MJ: Insights
into SND1 oncogene promoter regulation. Front Oncol. 8:6062018.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tong X, Drapkin R, Yalamanchili R,
Mosialos G and Kieff E: The Epstein-Barr virus nuclear protein 2
acidic domain forms a complex with a novel cellular coactivator
that can interact with TFIIE. Mol Cell Biol. 15:4735–4744. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wright T, Wang Y and Bedford MT: The role
of the PRMT5-SND1 axis in hepatocellular carcinoma. Epigenomes.
5:22021. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhan F, Zhong Y, Qin Y, Li L, Wu W and Yao
M: SND1 facilitates the invasion and migration of cervical cancer
cells by Smurf1-mediated degradation of FOXA2. Exp Cell Res.
388:1118092020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yu L, Liu X, Cui K, Di Y, Xin L, Sun X,
Zhang W, Yang X, Wei M, Yao Z and Yang J: SND1 acts downstream of
TGFβ1 and upstream of smurf1 to promote breast cancer metastasis.
Cancer Res. 75:1275–1286. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cui X, Zhang X, Liu M, Zhao C, Zhang N,
Ren Y, Su C, Zhang W, Sun X, He J, et al: A pan-cancer analysis of
the oncogenic role of staphylococcal nuclease domain-containing
protein 1 (SND1) in human tumors. Genomics. 112:3958–3967. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gutierrez-Beltran E, Denisenko TV,
Zhivotovsky B and Bozhkov PV: Tudor staphylococcal nuclease:
Biochemistry and functions. Cell Death Differ. 23:1739–1748. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cui X, Zhao C, Yao X, Qian B, Su C, Ren Y,
Yao Z, Gao X and Yang J: SND1 acts as an anti-apoptotic factor via
regulating the expression of lncRNA UCA1 in hepatocellular
carcinoma. RNA Biol. 15:1364–1375. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hassannia B, Vandenabeele P and Vanden
Berghe T: Targeting ferroptosis to iron out cancer. Cancer Cell.
35:830–849. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Guo J, Xu B, Han Q, Zhou H, Xia Y, Gong C,
Dai X, Li Z and Wu G: Ferroptosis: A novel anti-tumor action for
cisplatin. Cancer Res Treat. 50:445–460. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Shi Z, Zhang L, Zheng J, Sun H and Shao C:
Ferroptosis: Biochemistry and biology in cancers. Front Oncol.
11:5792862021. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Song X, Wang X, Liu Z and Yu Z: Role of
GPX4-mediated ferroptosis in the sensitivity of triple negative
breast cancer cells to gefitinib. Front Oncol. 10:5974342020.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang X, Sui S, Wang L, Li H, Zhang L, Xu
S and Zheng X: Inhibition of tumor propellant glutathione
peroxidase 4 induces ferroptosis in cancer cells and enhances
anticancer effect of cisplatin. J Cell Physiol. 235:3425–3437.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ufer C, Wang CC, Fähling M, Schiebel H,
Thiele BJ, Billett EE, Kuhn H and Borchert A: Translational
regulation of glutathione peroxidase 4 expression through
guanine-rich sequence-binding factor 1 is essential for embryonic
brain development. Genes Dev. 22:1838–1850. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang Q, Guo Y, Wang W, Liu B, Yang G, Xu
Z, Li J and Liu Z: RNA binding protein DAZAP1 promotes HCC
progression and regulates ferroptosis by interacting with SLC7A11
mRNA. Exp Cell Res. 399:1124532021. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kinzy SA, Caban K and Copeland PR:
Characterization of the SECIS binding protein 2 complex required
for the co-translational insertion of selenocysteine in mammals.
Nucleic Acids Res. 33:5172–5180. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Xie M, Huang P, Wu T, Chen L and Guo R:
Inhibition of miR-214-3p protects endothelial cells from
ox-LDL-induced damage by targeting GPX4. Biomed Res Int.
2021:99197292021. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li G, Jin J, Liu S, Ding K and Qian C:
Inhibition of miR-1224 suppresses hypoxia/reoxygenation-induced
oxidative stress and apoptosis in cardiomyocytes through targeting
GPX4. Exp Mol Pathol. 121:1046452021. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hou Y, Cai S, Yu S and Lin H: Metformin
induces ferroptosis by targeting miR-324-3p/GPX4 axis in breast
cancer. Acta Biochim Biophys Sin (Shanghai). 53:333–341. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Xu Q, Zhou L, Yang G, Meng F, Wan Y, Wang
L and Zhang L: CircIL4R facilitates the tumorigenesis and inhibits
ferroptosis in hepatocellular carcinoma by regulating the
miR-541-3p/GPX4 axis. Cell Biol Int. 44:2344–2356. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Armengol S, Arretxe E, Rodriguez L, Ochoa
B, Chico Y and Martinez MJ: NF-κB, Sp1 and NF-Y as transcriptional
regulators of human SND1 gene. Biochimie. 95:735–742. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chen HH and Kuo MT: Overcoming platinum
drug resistance with copper-lowering agents. Anticancer Res.
33:4157–4161. 2013.PubMed/NCBI
|



















