Introduction
Glioma is the most common adult central nervous
system tumor and a primary brain tumor disease (1). Generally, gliomas are diagnosed and
classified according to histopathology as astrocytoma,
oligodendroglioma, oligodendroastrocytoma and ependymoma. According
to the World Health Organization, glioma is generally divided into
four grades, among which grade IV glioblastoma has the highest
malignancy degree and the worst prognosis. Although patient
survival time has been prolonged with the improvement of medical
treatment, the five-year survival rate remains consistently low
(2,3).
The human genome is widely transcribed, with only
~2% accounting for protein products. Most transcripts do not encode
proteins, and RNAs with lengths >200 nucleotides (nt) are named
long non-coding RNAs (lncRNAs) (4).
Differentiation antagonizing non-protein-coding RNA (DANCR) is an
lncRNA associated with the occurrence and progress of various
tumors (5). DANCR can promote
pancreatic cancer cell proliferation and stemness (6). The proliferation and colony formation
of lung cancer cells were positively correlated with the DANCR
expression level (7). However,
DANCR-mediated autophagy has not been reported in gliomas.
MicroRNA (miRNA) is a class of small non-coding RNA
of ~21–22 nt, which can recognize sites in the 3′-untranslated
region (UTR) of mRNA and adjust its stability. Furthermore, miRNA
usually affects gene expression by targeting mRNA (8,9).
Previous studies have shown that abnormal expression of a variety
of miRNAs in vivo is associated with hidradenitis
suppurativa, bladder and gliomas (10–12).
Reduced expression of miR-33b is also related to the occurrence of
renal cell carcinoma, prostate cancer and medulloblastoma (13–15).
However, the relationship between miR-33b and DANCR expression in
gliomas has not been reported.
Distal-less homeobox 6 (DLX6) is a homologous
transcription factor of DLX5 and the DLX6 gene is expressed
primarily in the brain and skeleton (16). Moreover, DLX6 is involved in the
cortical neuron differentiation of precursor cells (17). In their latest study, Liang et
al (18) found that DLX6 plays
a role in the progression of oral squamous cell carcinoma
malignancy promoting cell proliferation and survival. Nevertheless,
the function of DLX6 in glioma and its association with posture in
glioma cells have not been reported.
Autophagy is a molecular mechanism that maintains
the dynamic balance of intracellular ecology, and it can act as a
damage repair mechanism when intracellular organelles are damaged.
A recent study showed that autophagy-related 7 (ATG7) protects the
integrity of human neurons and that its loss leads to degeneration
(19). In numerous cases, autophagy
is a tumor-promoting factor, and tumor cells require higher levels
of autophagy (20,21). ATG7 is an autophagy-associated
protein that affects numerous cancer types. Zhu et al
(22) demonstrated that ATG7 can
stabilize the ARHGDIB mRNA by regulating autophagy, thus promoting
bladder cancer progression. In 2016, Li et al (23) also proposed that ATG7-induced
autophagy enhancement is a glioma cell resistance response to the
killing effect of temozolomide.
In the present study, the DANCR, miR-33b and DLX6
expression levels were detected in HA, U251 and U87MG cells and the
relationship between them was explored. The involvement of the
DANCR/miR-33b/DLX6/ATG7 axis in regulating glioma cell autophagy
was also explained. The results of the present study may provide
novel treatment ideas for glioma.
Materials and methods
Cell culture
The human glioma U87MG (established by J. Ponten and
identified by STR profiling) (cat. no. TcHu138), U251 (cat. no.
TcHu58) and 293T (cat. no. GNHu17) were purchased from the Chinese
Academy of Science (Shanghai, China). The human astrocyte (HA)
cells (cat. no. 1800) were purchased from ScienCell Research
Laboratories. These cells were cultured in Dulbecco's Modified
Eagle Medium (DMEM) (Corning, Inc.) medium containing 10% fetal
bovine serum (Procell Life Science & Technology Co., Ltd.) in a
humidified incubator at 37°C with 5% CO2, and the medium
was replaced every 2–3 days or during cell passage until when the
cell confluence reached 70–90%.
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
In the present study, whole-cell RNA was extracted
using TRIzol® reagent (Beijing Solarbio Science &
Technology Co., Ltd.), RNA concentration was detected using a
Nanodrop Spectrophotometer (Thermo Fisher Scientific, Inc.) at a
wavelength of 260 nm, and intracellular RNA expression levels were
measured using RT-qPCR. DANCR and GAPDH expression was analyzed
using HiScript III RT Supermix for qPCR and ChamQ Universal SYBR
qPCR Master mix (Vazyme Biotech Co., Ltd.) according to the
manufacturer's instructions, Briefly, initial denaturation at 95°C
for 30 sec was followed by 40 cycles of 95°C for 10 sec and 60°C
for 30 sec. MiRNA-33b and U6 expression was quantified using miRNA
1st Strand cDNA Synthesis kit (stem-loop) and miRNA Universal SYBR
qPCR Master mix (Vazyme Biotech Co., Ltd.) according to the
manufacturer's instructions. MiRNA-33b primers were designed by
software provided by the manufacturer of the miRNA reverse
transcription kit, and the universal reverse primer in the kit was
used. All RT-qPCR reactions were performed using the 7500 fast
RT-PCR System (Thermo Fisher Scientific, Inc.). Finally, each
expression was calculated through the 2−ΔΔCq method
(24). The primers used in the
present study are listed in Table
SI.
Cell transfection
Before transfection, 5×105 cells were
seeded into 24-well plates, and 1 µg plasmid vector and
Lipofectamine 3000 reagent (Thermo Fisher Scientific, Inc.) were
mixed according to the manufacturer's instructions and added to the
24-well plates containing cells. After 24 h, the culture medium was
replaced, and G418 (500 µg/ml) or puromycin (2 µg/ml) was added to
screen the cells containing the plasmid vector, according to the
transfection reagent instructions. A full-length DANCR plasmid was
constructed with pcDNA 3.1. DANCR [small hairpin (sh-DANCR)] and
DLX6 (sh-DLX6) silencing plasmids were constructed with pGPU6.
MiR-33b overexpression (pre-mir-33b) and miR-33b interference
(anti-miR-33b) plasmids were prepared by Shanghai GenePharma Co.,
Ltd. All plasmids and negative controls were purchased from
Shanghai GenePharma Co., Ltd. The sequences used by the plasmids in
the present study are shown in Table
SII.
Cell proliferation assay
Cell proliferation assays were performed using the
Cell Counting Kit-8 (CCK-8) assay (APeXBIO Technology LLC) in
accordance with the manufacturer's instructions. U87MG or U251
cells were seeded in 96-well plates and 3.0×103 cells
were added to each well. To determine the OD value, the medium was
replaced with 0.1 ml of fresh medium containing 0.01 ml of CCK-8
solution 3 h prior to detection at 24, 48 and 72 h. All of the
aforementioned samples were placed in an incubator that contained
5% CO2 and maintained at 37°C. Then, the absorbance
value at 450 nm was detected with microplate reader (BioTek
Instruments, Inc.).
Dual-luciferase reporter assay
Wild-type DANCR (DANCR-wt) and DLX6 (DLX6-wt)
sequences containing miR-33b binding sites or mutant DANCR
(DANCR-mut) and DLX6 (DLX6-mut) sequences without miR-33b binding
sites were inserted into the vector. 293T cells were co-transfected
with DANCR-wt + pre-miR-33b, DANCR-wt + pre-NC, DANCR-mut +
pre-miR-33b, DANCR-NC + pre-NC, DLX6-wt + pre-miR-33b, DLX6-wt +
pre-NC, DLX6-mut + pre-miR-33b, or DLX6-NC + pre-NC in 96-well
plates using Lipofectamine 3000 reagent (Thermo Fisher Scientific,
Inc.). After 48 h, the dual-Luciferase activity was detected using
the Dual-Luciferase® Reporter Assay System (Promega
Corporation) according to the manufacturer's instructions. The
Renilla luciferase activity was used as internal
control.
Western blotting (WB)
RIPA buffer (Beyotime Institute of Biotechnology)
supplemented with 1% protease inhibitor (added 2 min before use)
was added to the cells and placed on ice for 30 min. Nucleic acids
were crushed with an ultrasonic crusher and centrifuged at 13,800 ×
g at 4°C for 30 min. After detection of protein concentration using
the bicinchoninic acid (BCA) assay, the loading buffer (5X) was
added to the supernatant and heated at 100°C for 5 min. Proteins
were separated using 12 and 10% SDS-PAGE and transferred to 0.22-µm
polyvinylidene difluoride (PVDF) membranes (MilliporeSigma). The
PVDF membranes were blocked by placing them in Tris-buffered saline
containing 0.1% Tween 20 and 5% non-fat milk at room temperature
for 120 min. The membranes were incubated with primary antibodies
including DLX6 (1:2,000; cat. no. 23216-1-AP), LC3 (1:5,000; cat
no. 14600-1-AP; both from ProteinTech Group, Inc.), p62 (1:1,000;
cat. no. 23214; Cell Signaling Technology, Inc.), beclin-1
(1:8,000; cat. no. 66665-1-Ig), ATG7 (1:10,000; cat. no.
67341-1-Ig) and GAPDH (1:20,000; cat. no. 10494-1-AP; all from
ProteinTech Group, Inc.) at 4°C for 14 h. The PVDF membrane was
incubated with HRP-conjugated Affinipure Goat Anti-Mouse IgG (H+L)
(1:10,000; cat. no. SA00001-1; ProteinTech Group, Inc.) or
HRP-conjugated Affinipure Goat Anti-Rabbit IgG (H+L) (1:10,000;
cat. no. SA00001-2; ProteinTech Group, Inc.) at room temperature
for 2 h. The immunoblot bands were detected using an enhanced
chemiluminescence detection kit (ECL kit; Beyotime Institute of
Biotechnology), and images were captured using a chemiluminescence
imaging analysis system (Bio-Rad Laboratories, Inc.). Finally, the
ImageJ 1.53K software (National Institutes of Health) was used to
calculate the relative gray value.
Flow cytometry
The required cells were collected according to the
manufacturer's instructions (Dojindo Laboratories, Inc.). Briefly,
100 µl of 1X Annexin V binding buffer containing 5 µl of Annexin
V-FITC and 5 µl PI was added to the cells, and incubation followed
at room temperature for 15 min. Then, 400 µl of 1X Annexin V
binding buffer was added to terminate the reaction and the assay
was performed on Beckman DxFLEX (Beckman Coulter, Inc.).
CytExpert1.1 software (Beckman Coulter, Inc.) was used for
analysis. FITC+/PI− cells indicated early
apoptosis, and FITC+/PI+ cells indicated late
apoptosis.
Transmission electron microscopy
Cells were treated with trypsin and centrifuged into
clumps, followed by electron microscope fixative (Wuhan Servicebio
Technology Co., Ltd.) at room temperature for 30 min, then at 4°C
overnight. Next, they were pre-embedded with agarose, post-fixed,
and dehydrated at room temperature. This was followed by resin
penetration and embedding using EMBed 812, polymerization,
ultrathin sectioning and staining. Briefly, 2% uranium acetate
saturated alcohol solution (avoiding light) was used for staining
at room temperature for 8 min, then 2.6% Lead citrate (avoiding
CO2) was used for staining at room temperature for 8
min; subsequently, they were put into the grids board and dried
overnight at room temperature. Finally, observation and image
capturing via transmission electron microscopy was performed
(Hitachi, Ltd.).
Chromatin immunoprecipitation (ChIP)
assay
A ChIP assay was performed using a Simple Chip
Enzymatic Chromatin IP kit (Cell Signaling Technology, Inc.),
according to the manufacturer's instructions. Briefly, 1%
formaldehyde was added to the cells at room temperature for 10 min
to cross-link the protein to the DNA, which was then worked with an
ultrasonic crusher (VCX-130, Sonics & Materials, Inc.) at 25%
power for 15 sec and repeated 10 times to truncated into fragments
of ~600 bp in length. The samples were incubated with anti-DLX6
rabbit antibodies (1:25; cat. no. sc-517058; Santa Cruz
Biotechnology, Inc.) or normal rabbit IgG antibodies (1:250; cat.
no. 30000-0-AP; ProteinTech Group, Inc.). After DNA cross-links
were reversed, and the desired DNA fragment was purified, 2X Tap
Master Mix for PAGE (Vazyme Biotech Co., Ltd.) was used to amplify
the DNA fragment. The primers used for PCR were as follows: PCR1
forward, 5′-TTCAGCAAGCCACCTAAC-3′, and reverse,
5′-AACAACAAGAGCGATGACT-3′; and PCR2 forward,
5′-TGACAGAGCGAGACTCCGT-3′ and reverse, 5′-GGCTGGTCTTGAACTACTGGA-3′.
Finally, agarose electrophoresis was used to verify the PCR
products using 3% agarose gels and using SYBR Green staining.
Tumor xenografts in nude mice
First, glioma cells (U87MG) stably expressing
sh-DANCR, pre-miR-33b, sh-DLX6 and sh-DANCR + pre-miR-33b + sh-DLX6
were established (n=6). BALB/c nude mice (female; 5 weeks-old;
weight, 14–16 g) (Hua FuKang, Beijing, China) were used. Mice were
housed under SPF conditions (The temperature of the SPF animal
house was between 24–26°C, the humidity was ~70%, the air exchange
rate was 15 times/h, and the light/dark cycle was 12/12 h) and the
adequacy of food and water was checked daily to ensure that the
mice had access to food and water ad libitum at all times.
Each mouse was injected with 100 µl PBS containing 1×106
cells subcutaneously under the right armpit. Tumor volumes were
recorded on day 7 after injection and then measured every three
days. At day 28, the mice were sacrificed by cervical dislocation
after inhalation of 3% isoflurane anesthesia and death of mice was
confirmed by respiratory arrest. Tumor weights were also compared.
The present experiment was approved (approval no. 2022PS021K) by
the Medical Ethics Committee of Shengjing Hospital (Shenyang,
China).
Immunohistochemistry
After the tumor was removed and fixed with 4%
paraformaldehyde at 4°C for 24 h, it was embedded in paraffin and
cut into 5-µm thin sections. After dewaxing with xylene,
rehydration (absolute ethanol, 95% ethanol, 90% ethanol, 80%
ethanol and 70% ethanol were used for 10 min each in turn), antigen
repair, incubation with anti-ATG7 antibodies (1:200; cat. no.
67341-1-Ig; ProteinTech Group, Inc.) at 4°C overnight, and
incubation with biotin-labeled sheep anti-mouse/rabbit IgG polymer
(cat. no. KIT-9710; Maxim Biotech, Inc.) at room temperature for 30
min, the tumor was finally stained with diaminobenzidine (Maxim
Biotech, Inc.) for 5 min and images were captured under a light
microscope (Nikon Corporation).
Bioinformatic analysis
GEPIA (http://gepia.cancer-pku.cn/) was used to analyze the
difference in DANCR levels between glioma and normal tissues with a
P-value Cutoff of 0.01. The starbase website (https://starbase.sysu.edu.cn) was used to predict
potential sites of connection between DANCR and miR-33b. The
potential binding sites of DLX6 to the promoter binding region of
ATG7 were predicted by JASPAR (https://jaspar.genereg.net).
Statistical analysis
All experiments were repeated at least three times.
GraphPad Prism 8 software (GraphPad Software, Inc.) was used for
all experimental data, and the results were presented as the mean ±
SD. Unpaired student's t-test was used to conduct the comparison
between two groups. More than two groups of data were analyzed
through a one-way analysis of variance followed by LSD post hoc
test. P<0.05 was considered to indicate a statistically
significant difference.
Results
DANCR is upregulated in glioma
To identify the relationship between DANCR and
gliomas, GEPIA (gepia.cancer-pku.cn) was used to preliminarily
verify the DANCR expression level in glioma. A higher expression of
DANCR was identified in glioma tissues compared with normal tissue
(Fig. 1A). Then, the relative
expression of DANCR was detected in HA, U251 and U87MG cells using
RT-qPCR. It was revealed that the DANCR expression level was higher
in the glioma cell lines U251 and U87MG than in HA cells (Fig. 1B).
DANCR regulates autophagy,
proliferation and apoptosis in glioma cells
To study the function of DANCR in glioma cells,
DANCR-overexpressing U251 and U87MG cell lines were constructed via
transfection of the pcDNA3.1-DANCR plasmid vectors into U251 and
U87MG cells. Meanwhile, DANCR-silenced U251 and U87MG cell lines
were constructed via transfection of the sh-DANCR plasmid vectors
in U251 and U87MG cells. First, DANCR expression levels were in
DANCR-overexpressing or DANCR-silenced U251 and U87MG cell lines
using RT-qPCR to verify the effectiveness of the transfected
plasmid vectors. Compared with the pcDNA3.1-NC group, DANCR
expression increased significantly in U251 and U87MG cells
transfected with pcDNA3.1-DANCR. Meanwhile, compared with the sh-NC
group, DANCR expression in U251 and U87MG cells transfected with
sh-DANCR decreased significantly (Fig.
2A and B). Then, the CCK-8 assay showed that upregulated DANCR
expression could promote U251 and U87MG cell proliferation, while
its inhibition slowed the rate of U251 and U87MG cell proliferation
(Fig. 2C and D). Flow cytometric
analysis demonstrated that the increased DANCR expression in U251
and U87MG cells reduced their apoptotic levels, while decreased
DANCR expression exerted the opposite effect (Fig. 2E). Furthermore, the WB assay showed
that the ATG7, beclin-1 and LC3 II/I protein levels increased and
p62 levels decreased in DANCR-overexpressing U251 and U87MG cells.
However, decreasing DANCR expression in U251 and U87MG cells
decreased the ATG7, beclin-1 and LC3 II/I protein expression and
increased p62 expression (Fig. 2F).
Electron microscopic observation showed that the number of
autophagosomes increased in DANCR-overexpressing U251 and U87MG
cells compared with that in the control group. On the contrary,
DANCR inhibition significantly reduced the number of autophagosomes
in U251 and U87MG cells (Fig. 2G).
In conclusion, DANCR can enhance glioma cell proliferation and
autophagy and inhibit their apoptosis.
MiR-33b directly targets DANCR,
regulating glioma cell autophagy, proliferation and apoptosis
To explore how DANCR functions in glioma cells, a
bioinformatics database was used (starbase.sysu.edu.cn). MiR-33b was a predicted target
miRNA of DANCR (Fig. 3C). Then, the
targeted binding and binding site of DANCR with miR-33b were
verified through a dual-luciferase reporter assay. The relative
luciferase intensity of cells in the experimental group transfected
with DANCR-wt + pre-miR-33b was significantly lower than that in
other groups (Fig. 3D). In the
present study, as revealed in Fig.
3A, miR-33b expression in U251 and U87MG cells was
downregulated, compared with that in HA cells. MiR-33b expression
levels were then detected in miR-33b-overexpressing or
miR-33b-silenced U251 and U87MG cell lines using RT-qPCR to verify
the effectiveness of the transfected plasmid vectors. Compared with
the pre-NC group, miR-33b expression increased significantly in
U251 and U87MG cells transfected with pre-miR-33b. Meanwhile,
compared with the anti-NC group, miR-33b expression in U251 and
U87MG cells transfected with anti-miR-33b decreased significantly
(Fig. 3B). Intracellular miR-33b
expression levels were upregulated in U251 and U87MG cells and it
was found that the cell proliferation slowed down (Fig. 3E and F), intracellular autophagy
levels decreased, apoptotic levels increased (Fig. 3G and H), and miR-33b expression was
inhibited, showing opposite effects. Hence, miR-33b may be a
regulator of cell proliferation and autophagy.
 | Figure 3.MiR-33b function in U251 and U87MG
cells; miR-33b as a target gene of DANCR. (A) The relative miR-33b
expression in HA, U251 and U87MG cells was verified using RT-qPCR
(n=3). (B) Validation of pre-miR-33b and anti-miR-33b transfection
in U251 and U87MG cells (n=3). (C and D) Presentation of the
putative binding site of DANCR and miR-33b and relative luciferase
activities of cells co-transfected with pre-miR-33b and DANCR-wt or
DANCR-mut (n=3). (E and F) The effects of pre-miR-33b and
anti-miR-33b on U251 and U87MG cell proliferation were verified
using a Cell Counting Kit-8 assay (n=3); *P<0.05 vs. pre-NC and
#P<0.05 vs. anti-miR-33b. (G) Flow cytometric
analysis was used to verify the effects of pre-miR-33b and
anti-miR-33b on apoptosis in U251 and U87MG cells (n=3). (H)
Western blotting was used to analyze the effects of pre-miR-33b and
anti-miR-33b on the ATG7, beclin-1, LC3 II/I and p62 protein
expression in U251 and U87MG cells (n=3); *P<0.05 vs.
pcDNA3.1-NC and #P<0.05 vs. sh-NC. Data are presented
as the mean ± SD. *P<0.05, **P<0.01 and ***P<0.001. DANCR,
differentiation antagonizing non-protein coding RNA; miR, microRNA;
NC, negative control; HA, human astrocytes; ATG7, autophagy-related
7; wt, wild-type; mut, mutant. |
miR-33b mediates the effects of DANCR
on glioma cells and its function
It was explored whether miR-33b regulates the
function of DANCR in glioma cells. Through co-transfection of the
desired plasmid (pcDNA3.1-DANCR + pre-NC, pcDNA3.1-DANCR +
pre-miR-33b, sh-DANCR + anti-NC, or sh-DANCR + anti-miR-33b) in
U87MG and U251 cells, CCK-8, flow cytometry, and WB assays
demonstrated that pre-miR-33b and anti-miR-33b could respectively
inhibit the accelerated proliferation and decreased apoptosis or
decreased proliferation and increased apoptosis caused by
pcDNA3.1-DANCR or sh-DANCR. Furthermore, they respectively enhanced
or slowed proliferation (Fig. 4A and
B) and increased or decreased autophagy (Fig. 4C and D). These findings suggested
that miR-33b is involved in regulating DANCR in glioma cells.
 | Figure 4.MiR-33b exerts opposite effects to
DANCR and can reverse the changes caused by DANCR
overexpression or knockdown. (A and B) Cell Counting Kit-8 assay
was used to detect the change in proliferation due to DANCR
overexpression via transfection of pre-NC or pre-miR-33b, as well
as the effects of anti-NC and anti-miR-33b on
DANCR-knockdown cells (n=3). (C) Flow cytometric analysis
was conducted to determine the apoptosis ratio of U251 and U87MG
cells co-transfected with DANCR and miR-33b (n=3). (D) Changes in
autophagy levels in U251 and U87MG cells co-transfected with DANCR
and miR-33b were verified via western blotting of the
autophagy-related proteins ATG7, beclin-1, LC3 II/I, and p62 (n=3);
*P<0.05 vs. pcDNA3.1-DANCR + pre-miR-NC; #P<0.05
vs. sh-DANCR + anti-miR-33b. Data are presented as the mean ± SD.
*P<0.05, **P<0.01 and ***P<0.001. miR, microRNA; DANCR,
differentiation antagonizing non-protein coding RNA; NC, negative
control; ATG7, autophagy-related 7; sh-, short hairpin. |
DLX6 knockdown restrains cell
proliferation and autophagy while promoting apoptosis
Next, to research the function of DLX6 in glioma
cells, WB was used to verify its protein expression level in U251,
U87MG and HA cells. DLX6 expression level in the glioma cell lines
U251 and U87MG was significantly higher than that in HA cells
(Fig. 5A). Then, a stable
sh-DLX6-transfected cell line was established and its effectiveness
was verified (Fig. 5E). CCK-8 and
flow cytometry showed that cell proliferation slowed down and
apoptosis increased (Fig. 5B-D).
Meanwhile, the WB assay verified that inhibiting DLX6 expression
also decreased cell autophagy levels (Fig. 5E). These results suggested that
DLX6 knockdown slows down the progression of glioma cell
malignancy.
 | Figure 5.DLX6 function in U251 and U87MG
cells. (A) DLX6 expression verified via western blotting was
upregulated in U251 and U87MG cells, compared with that in HA cells
(n=3). (B-D) Cell Counting Kit-8 and flow cytometric assays were
used to verify the effect of DLX6 knockdown on proliferation
and apoptosis in U251 and U87MG cells (n=3). (E) The effectiveness
of sh-DLX6 was verified, and the ATG7, beclin-1, LC3 II/I, and p62
protein expression in each group was detected via western blot
assays (n=3). Data are presented as the mean ± SD. *P<0.05 vs.
sh-NC. *P<0.05, **P<0.01 and ***P<0.001. DLX6, distal-less
homeobox 6; HA, human astrocytes; ATG7, autophagy-related 7; NC,
negative control; sh-, short hairpin. |
DLX6 directly targets miR-33b and
restores miR-33b-induced proliferation and autophagy reduction
while reducing apoptosis
Firstly, by altering the DANCR or miR-33b levels in
U251 and U87MG cells, it was found that DANCR upregulation or
miR-33b downregulation increased the expression level of DLX6,
while DANCR downregulation or miR-33b upregulation decreased the
expression level of DLX6 (Fig. 6A and
B). Then, through co-transfection of the desired plasmid
(pcDNA3.1-DANCR + pre-NC, pcDNA3.1-DANCR + pre-miR-33b, sh-DANCR +
anti-NC, or sh-DANCR + anti-miR-33b) in U87MG and U251 cells, it
was revealed that the increased expression of DLX6 induced by DANCR
was reversed by pre-miR-33b, and the decreased expression of dlx6
induced by DANCR was also reversed by anti-miR-33b (Fig. 6C). To examine the relationship
between DLX6 and miR-33b, a dual-luciferase reporter assay was
first employed to verify the targeted binding effect between them.
The relative luciferase intensity of cells in the experimental
group transfected with DLX6-WT and pre-miR-33b was significantly
lower than that in other groups (Fig.
6D and E). Then, U87 and U251 glioma cells were co-transfected
with anti-miR-33b + sh-DLX6, and cells co-transfected with
anti-miR-33b + sh-NC were set as a negative control. CCK-8 and flow
cytometric assays showed that DLX6 knockdown could reverse
the increase in the degree of malignant glioma cells caused by the
decreased miR-33b expression (Fig.
6F-H). In addition, WB also verified that DLX6 knockdown
reversed the increase in autophagy caused by decreased miR-33b
expression (Fig. 6I). The
aforementioned results demonstrated that DLX6 is involved in the
regulation of miR-33b in the progression of glioma cell
malignancy.
 | Figure 6.The effects of DLX6 on DLX6 and
sh-DLX6 expression could reverse the effects of anti-miR-33b on
U251 and U87MG cell proliferation, autophagy and apoptosis. (A-C)
The effects of DANCR and miR-33b alone and in combination on DLX6
protein expression in U251 and U87MG cells were verified via
western blotting (n=3). (D) Presentation of the putative binding
site of DLX6 and miR-33b. (E) Relative luciferase activities of
cells co-transfected with pre-miR-33b and DLX6-wt or DLX6-mut
(n=3). (F and G) Cell Counting Kit-8 assay was used to detect the
change in proliferation of DLX6-knockdown U251 and U87MG
cells transfected with anti-NC or anti-miR-33b (n=3). (H) Flow
cytometric analysis was conducted to determine the apoptosis ratio
of U251 and U87MG cells co-transfected with sh-DLX6 and
anti-miR-33b (n=3). (I) The protein expression of ATG7, beclin-1,
LC3 II/I and p62 in each group were detected using western blot
assays (n=3). *P<0.05 vs. anti-miR-33b + sh-NC. Data are
presented as mean ± SD. *P<0.05, **P<0.01 and ***P<0.001.
DLX6, distal-less homeobox 6; sh-, short hairpin; DANCR,
differentiation antagonizing non-protein coding RNA; miR, microRNA;
wt, wild-type; mut, mutant; NC, negative control; ATG7,
autophagy-related 7. |
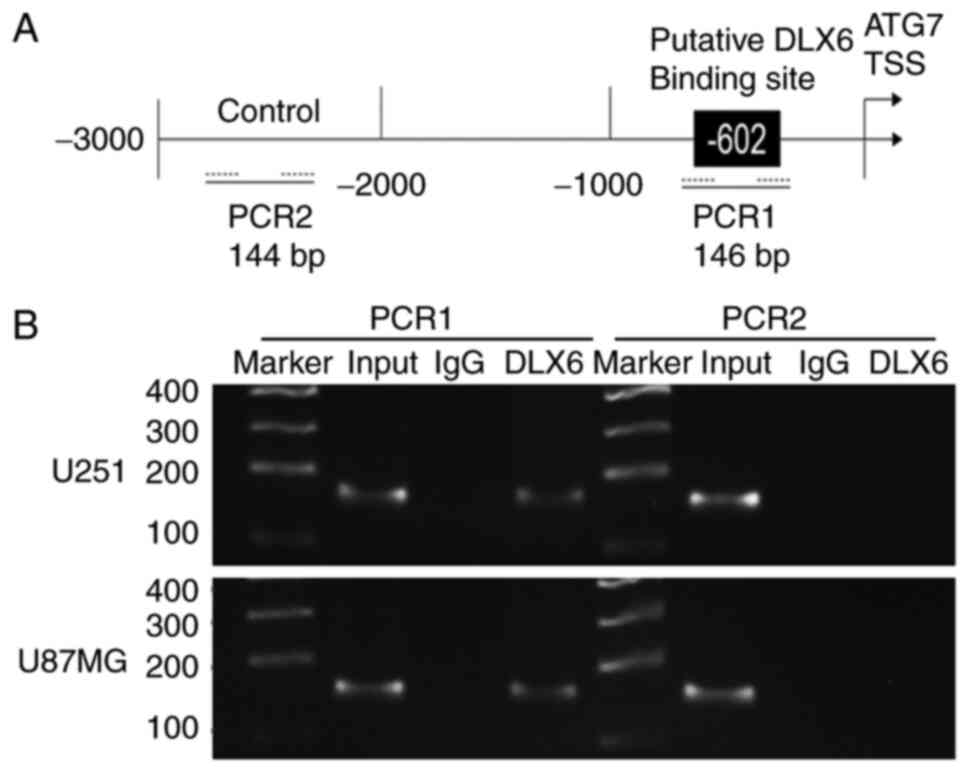
ATG7 is involved in DLX6-mediated
autophagy in glioma cells
Jaspar predicted DLX6 as an ATG7 transcription
factor, the predicted binding site was located between 2,000 base
pairs (bp) upstream and 1,000 bp downstream of the transcription
start site (TSS), and the control sequence was between 2,000 and
3,000 bp upstream of the TSS (Fig.
7A). A ChIP assay was used to verify whether DLX6 directly
binds to the promoter sequence of ATG7 and regulates its
transcription, and the same conclusion was obtained (Fig. 7B). Meanwhile, a WB assay verified
that ATG7 expression level could be inhibited by DLX6
knockdown in U251 and U87MG cells (Fig.
5E). Thus, DLX6 is a transcription factor of ATG7 and promotes
its expression.
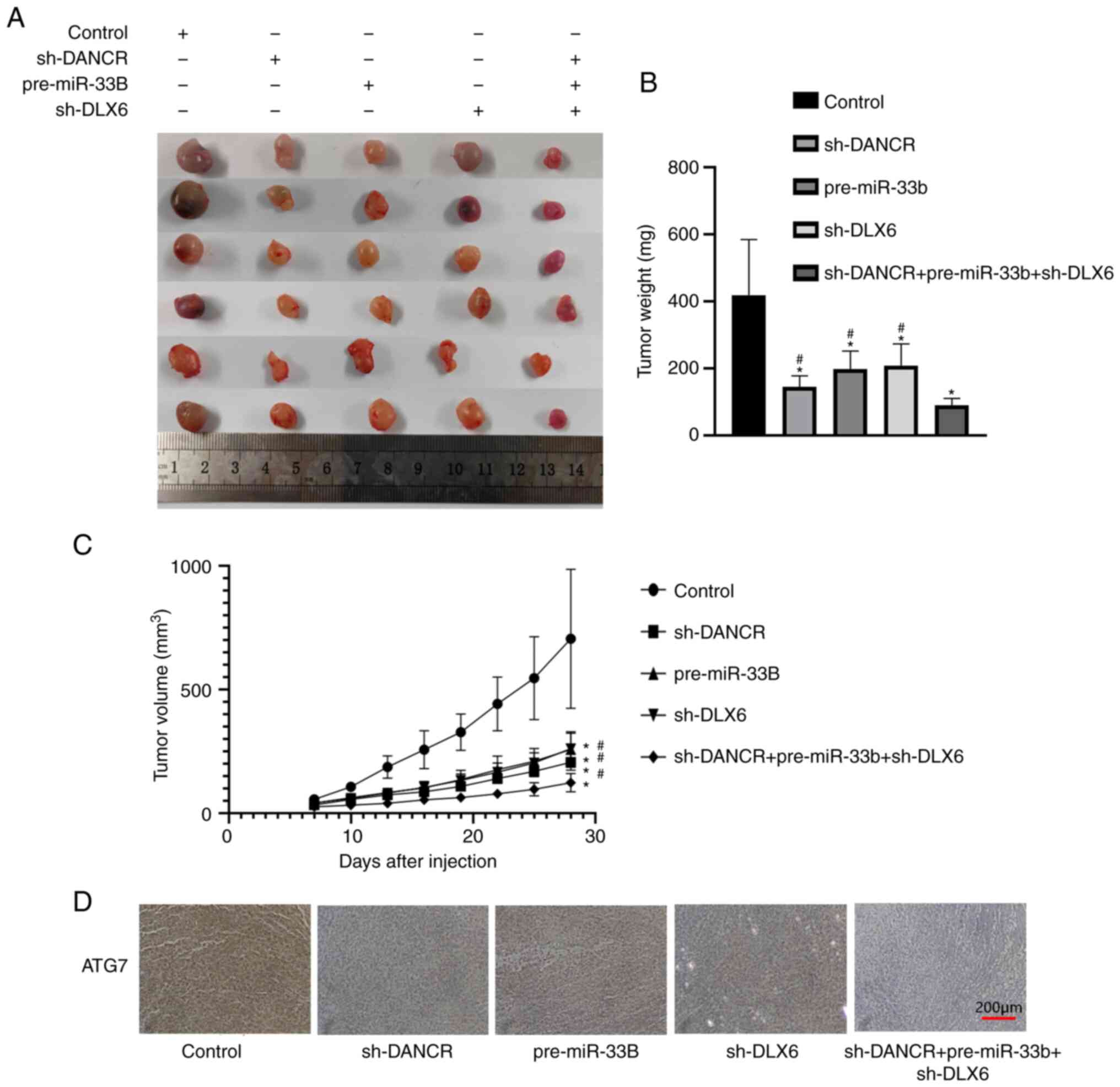
DANCR and DLX6 knockdown combined with
miR-33b overexpression inhibit ectopic tumor growth and ATG7
expression in mice
To further confirm the aforementioned conclusion,
in vivo experiments were conducted. The results showed that
the tumor volumes in the sh-DANCR, pre-miR-33b, sh-DLX6 and
sh-DANCR + pre-miR-33b + sh-DLX6 groups were smaller than that in
the control group. The tumor volume was smallest in the group with
DANCR and DLX6 knockdown combined with miR-33b
overexpression (Fig. 8A-C). When
ATG7 protein levels were measured in vitro via
immunohistochemistry, it was found that they were lower in the
sh-DANCR, pre-miR-33b, sh-DLX6 and sh-DANCR + pre-miR-33b + sh-DLX6
groups than in the control group, and ATG7 protein levels were the
lowest in the sh-DANCR + pre-miR-33b + sh-DLX6 group (Fig. 8D).
Discussion
In the present study, it was found that DANCR
expression is upregulated in glioma cells. By regulating DANCR
expression in glioma cells, it was revealed that proliferation and
autophagy levels in glioma cells were positively associated with
DANCR expression, while the apoptosis ratio showed an inverse
association. By contrast, miR-33b plays a tumor suppressor role in
gliomas. MiR-33b could bind with the 3′-UTR of DLX6 and inhibit its
function. DLX6 is a transcription factor of ATG7 and can
promote its transcription. Therefore, DANCR indirectly promotes
DLX6 expression and ATG7 transcription by reducing the
targeted connection between miR-33b and DLX6 3′-UTR by sponging
miR-33b. Finally, DANCR and DLX6 knockdown combined
with simultaneous miR-33b overexpression showed the strongest
inhibitory effect of tumor growth in the present study.
As a common non-coding RNA, lncRNA involvement has
been demonstrated in the progression of various tumors (25). DANCR, as an lncRNA, has also been
reported to promote the progression of various malignant tumors
(5). In the present study, DANCR
was upregulated in glioma cells and promoted malignant glioma
progression by promoting proliferation and autophagy and inhibiting
apoptosis. Previous studies have reported that DANCR is highly
expressed in glioma tissues and cells and plays a function in the
progression of glioma malignancy and cisplatin resistance (26,27).
DANCR regulates colon cancer cell desensitization to cisplatin by
sponging miR-125B-5p as a competing endogenous RNA (ceRNA), and
sponging miR-335 regulates the lymphatic metastasis of bladder
cancer (28,29). By inhibiting DANCR expression, the
progression of glioma cell malignancy can be inhibited, and its
effect is exerted through miR-135a sponging (30). DANCR enhances cytarabine resistance
in acute myeloid leukemia by promoting autophagy through miR-874-3p
sponging (31). The aforementioned
conclusions were also verified by the present experimental results.
DANCR can increase the level of autophagy and progression of glioma
cell malignancy and interact with miRNA as a ceRNA.
There is increasing evidence that miRNAs play an
important role in the progression of cancer malignancy. MiRNA can
act as a tumor suppressor in lung, prostate and breast cancers
(32–34). Evidence also shows that miR-93 plays
a role in inhibiting autophagy in glioma (10). Other researchers have shown that
miR-33b plays a role in prostate cancer and renal cell carcinoma,
where it also acts as a prognostic factor; in addition, low miR-33b
expression can promote peritoneal metastasis of ovarian cancer
cells and aggravate cancer progression (13,14,35).
In the present study, miR-33b was highly expressed in glioma cells,
which was also observed in the results of Qi and Gao (36). MiR-33b overexpression can inhibit
proliferation and autophagy and promote apoptosis in glioma cells.
Furthermore, it can inhibit enhanced proliferation and autophagy
and reduce apoptosis in glioma cells caused by DANCR
overexpression. On the contrary, miR-33b inhibition can reverse the
decreased proliferation, autophagy and apoptosis in glioma cells
caused by DANCR inhibition. The present study revealed miR-33b as a
possible ceRNA of DANCR, which regulated its function.
In the present study, it was identified that miR-33b
can bind to the 3′-UTR of DLX6 and regulate its expression. Liang
et al (18) showed that DLX6
can promote oral squamous cell carcinoma cell proliferation and
survival. However, the relationship between DLX6 and glioma has not
been reported. The present study found that DLX6 expression was
upregulated in glioma, and inhibiting it could inhibit the
proliferation and autophagy of glioma cells and promote apoptosis.
Moreover, inhibition of DLX6 could reverse the inhibition of
miR-33b-induced proliferation and autophagy enhancement and
apoptosis reduction in glioma. DLX6 was revealed to be a
transcription factor of ATG7 that could directly regulate
its expression. However, whether DLX6 is involved in the
transcription of other autophagy-related transgenes needs further
investigation. In autophagy, ATG7 plays an important role in
processes, such as the transformation of LC3 I to LC3 II and the
transport of autophagic vesicles in the cytoplasm (37). In addition, Atg12-Atg5 and
LC3-lipid/membrane ubiquitin-like conjugation systems are the core
machinery for autophagosome formation (38). Lung cancer studies have shown that
ATG7 expression promotes the progression of lung cancer malignancy
by promoting autophagy and drug resistance in lung cancer cells
(39–41). However, the drug resistance of
glioma was not studied in this study, so whether DANCR can affect
drug resistance by regulating ATG7-induced autophagy needs to be
further studied. A recent study in mice showed that ATG7-related
autophagy promoted anti-tumor immunity, thereby promoting
tumor-cell survival (42). Hypoxia
in glioma cells can induce intracellular protective autophagy, thus
affecting glioma progression (43).
Glioma cells can meet the intracellular capacity demand through
autophagy-mediated lipid metabolism, thereby promoting their
malignancy (44). The
aforementioned studies indirectly support the findings of the
present study.
In summary, it was demonstrated that DANCR
expression is upregulated in glioma cells and that DLX6 expression
level is upregulated via inhibition of the targeted sponge
interaction between miR-33b and DLX6. DLX6 activates ATG7
expression by binding to its promoter region, enhancing
intracellular autophagy and affecting the progression of glioma
cell malignancy. In conclusion, DANCR upregulates ATG7 protein
expression through the miR-33b/DLX6 pathway, thereby enhancing
intracellular autophagy and promoting progression in glioma cell
malignancy, inhibiting tumor proliferation and protecting autophagy
while promoting tumor cell apoptosis. In addition, it was
identified that miR-33b was negatively regulated by the miRNA
sponge-like role of DANCR. Mir-33b could bind to the 3′-UTR of DLX6
and suppress ATG7 expression through negative regulation of DLX6,
thus inhibiting the progression of glioma cell malignancy.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the Liaoning Science and
Technology Plan Project (grant no. 2018225094) and the Natural
Science Foundation of Liaoning Province (grant no.
2020-MS-165).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XXL designed the experiments and preliminary
validation. WY conducted the experiments. LM analyzed and processed
the data. XXL, WY and LM wrote and modified the manuscript. All
authors read and approved the final version of the manuscript and
confirm the authenticity of all the raw data.
Ethics approval and consent to
participate
The present study was approved (approval no.
2022PS021K) by the Medical Ethics Committee of Shengjing Hospital
(Shenyang, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
lncRNA
|
long non-coding RNA
|
|
DANCR
|
differentiation antagonizing
non-protein coding RNA
|
|
HA
|
human astrocyte
|
|
DLX6
|
distal-less homeobox 6
|
|
ATG7
|
autophagy-related 7
|
|
UTR
|
untranslated region
|
|
RT-qPCR
|
reverse transcription-quantitative
PCR
|
|
WB
|
western blot analysis
|
|
ChIP
|
chromatin immunoprecipitation
|
|
PVDF
|
polyvinylidene difluoride
|
|
TSS
|
transcription start site
|
|
ceRNA
|
competing endogenous RNA
|
References
|
1
|
Lapointe S, Perry A and Butowski NA:
Primary brain tumours in adults. Lancet. 392:432–446. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen R, Smith-Cohn M, Cohen AL and Colman
H: Glioma subclassifications and their clinical significance.
Neurotherapeutics. 14:284–297. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ostrom QT, Bauchet L, Davis FG, Deltour I,
Fisher JL, Langer CE, Pekmezci M, Schwartzbaum JA, Turner MC, Walsh
KM, et al: The epidemiology of glioma in adults: A ‘state of the
science’ review. Neuro Oncol. 16:896–913. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
ENCODE Project Consortium, . An integrated
encyclopedia of DNA elements in the human genome. Nature.
489:57–74. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wang M, Gu J, Zhang X, Yang J, Zhang X and
Fang X: Long non-coding RNA DANCR in cancer: Roles, mechanisms, and
implications. Front Cell Dev Biol. 9:7537062021. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hu X, Peng WX, Zhou H, Jiang J, Zhou X,
Huang D, Mo YY and Yang L: IGF2BP2 regulates DANCR by serving as an
N6-methyladenosine reader. Cell Death Differ. 27:1782–1794. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhen Q, Gao LN, Wang RF, Chu WW, Zhang YX,
Zhao XJ, Lv BL and Liu JB: LncRNA DANCR promotes lung cancer by
sequestering miR-216a. Cancer Control. 25:10732748187698492018.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hill M and Tran N: miRNA interplay:
Mechanisms and consequences in cancer. Dis Model Mech.
14:dmm0476622021. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Carthew RW and Sontheimer EJ: Origins and
mechanisms of miRNAs and siRNAs. Cell. 136:642–655. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Huang T, Wan X, Alvarez AA, James CD, Song
X, Yang Y, Sastry N, Nakano I, Sulman EP, Hu B and Cheng SY: MIR93
(microRNA-93) regulates tumorigenicity and therapy response of
glioblastoma by targeting autophagy. Autophagy. 15:1100–1111. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cao HL, Liu ZJ, Huang PL, Yue YL and Xi
JN: lncRNA-RMRP promotes proliferation, migration and invasion of
bladder cancer via miR-206. Eur Rev Med Pharmacol Sci.
23:1012–1021. 2019.PubMed/NCBI
|
|
12
|
Hessam S, Sand M, Skrygan M, Gambichler T
and Bechara FG: Expression of miRNA-155, miRNA-223, miRNA-31,
miRNA-21, miRNA-125b, and miRNA-146a in the inflammatory pathway of
hidradenitis suppurativa. Inflammation. 40:464–672. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Huang G, Lai Y, Pan X, Zhou L, Quan J,
Zhao L, Li Z, Lin C, Wang J, Li H, et al: Tumor suppressor
miR-33b-5p regulates cellular function and acts a prognostic
biomarker in RCC. Am J Transl Res. 12:3346–3360. 2020.PubMed/NCBI
|
|
14
|
Zhao M, Qi M, Li X, Hu J, Zhang J, Jiao M,
Bai X, Peng X and Han B: CUL4B/miR-33b/C-MYC axis promotes prostate
cancer progression. Prostate. 79:480–488. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lv SQ, Kim YH, Giulio F, Shalaby T,
Nobusawa S, Yang H, Zhou Z, Grotzer M and Ohgaki H: Genetic
alterations in microRNAs in medulloblastomas. Brain Pathol.
22:230–239. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Simeone A, Acampora D, Pannese M,
D'Esposito M, Stornaiuolo A, Gulisano M, Mallamaci A, Kastury K,
Druck T, Huebner K, et al: Cloning and characterization of two
members of the vertebrate Dlx gene family. Proc Natl Acad Sci USA.
91:2250–2254. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Batista-Brito R, Machold R, Klein C and
Fishell G: Gene expression in cortical interneuron precursors is
prescient of their mature function. Cereb Cortex. 18:2306–2317.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liang J, Liu J, Deng Z, Liu Z and Liang L:
DLX6 promotes cell proliferation and survival in oral squamous cell
carcinoma. Oral Dis. 28:87–96. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Collier JJ, Oláhová M, McWilliams TG and
Taylor RW: ATG7 safeguards human neural integrity. Autophagy.
17:2651–2653. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Klionsky DJ, Petroni G, Amaravadi RK,
Baehrecke EH, Ballabio A, Boya P, Bravo-San Pedro JM, Cadwell K,
Cecconi F, Choi AMK, et al: Autophagy in major human diseases. EMBO
J. 40:e1088632021. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
White E: The role for autophagy in cancer.
J Clin Invest. 125:42–46. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhu J, Tian Z, Li Y, Hua X, Zhang D, Li J,
Jin H, Xu J, Chen W, Niu B, et al: ATG7 promotes bladder cancer
invasion via autophagy-mediated increased ARHGDIB mRNA stability.
Adv Sci (Weinh). 6:18019272019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li J, Tang C, Li L, Li R and Fan Y:
Quercetin blocks t-AUCB-induced autophagy by Hsp27 and Atg7
inhibition in glioblastoma cells in vitro. J Neurooncol. 129:39–45.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Peng WX, Koirala P and Mo YY:
LncRNA-mediated regulation of cell signaling in cancer. Oncogene.
36:5661–5667. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ma Y, Zhou G, Li M, Hu D, Zhang L, Liu P
and Lin K: Long noncoding RNA DANCR mediates cisplatin resistance
in glioma cells via activating AXL/PI3K/Akt/NF-κB signaling
pathway. Neurochem Int. 118:233–241. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li J and Zhou L: Overexpression of lncRNA
DANCR positively affects progression of glioma via activating
Wnt/β-catenin signaling. Biomed Pharmacother. 102:602–607. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ping Q, Shi Y, Yang M, Li H, Zhong Y, Li
J, Bi X and Wang C: LncRNA DANCR regulates lymphatic metastasis of
bladder cancer via the miR-335/VEGF-C axis. Transl Androl Urol.
10:1743–1753. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Shi H, Li K, Feng J, Liu G, Feng Y and
Zhang X: LncRNA-DANCR interferes with miR-125b-5p/HK2 axis to
desensitize colon cancer cells to cisplatin vis activating
anaerobic glycolysis. Front Oncol. 10:10342020. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Feng L, Lin T, Che H and Wang X: Long
noncoding RNA DANCR knockdown inhibits proliferation, migration and
invasion of glioma by regulating miR-135a-5p/BMI1. Cancer Cell Int.
20:532020. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang H, Liu L, Chen L, Liu H, Ren S and
Tao Y: Long noncoding RNA DANCR confers cytarabine resistance in
acute myeloid leukemia by activating autophagy via the
miR-874-3P/ATG16L1 axis. Mol Oncol. 15:1203–1216. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Sur S, Steele R, Shi X and Ray RB:
miRNA-29b inhibits prostate tumor growth and induces apoptosis by
increasing bim expression. Cells. 8:14552019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Su WZ and Ren LF: MiRNA-199 inhibits
malignant progression of lung cancer through mediating RGS17. Eur
Rev Med Pharmacol Sci. 23:3390–3400. 2019.PubMed/NCBI
|
|
34
|
Kong X, Duan Y, Sang Y, Li Y, Zhang H,
Liang Y, Liu Y, Zhang N and Yang Q: LncRNA-CDC6 promotes breast
cancer progression and function as ceRNA to target CDC6 by sponging
microRNA-215. J Cell Physiol. 234:9105–9117. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang X, Yung MMH, Sharma R, Chen F, Poon
YT, Lam WY, Li B, Ngan HYS, Chan KKL and Chan DW: Epigenetic
silencing of miR-33b promotes peritoneal metastases of ovarian
cancer by modulating the TAK1/FASN/CPT1A/NF-κB axis. Cancers
(Basel). 13:47952021. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Qi Y and Gao Y: Clinical significance of
miR-33b in glioma and its regulatory role in tumor cell
proliferation, invasion and migration. Biomark Med. 14:539–548.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Nishida Y, Arakawa S, Fujitani K,
Yamaguchi H, Mizuta T, Kanaseki T, Komatsu M, Otsu K, Tsujimoto Y
and Shimizu S: Discovery of Atg5/Atg7-independent alternative
macroautophagy. Nature. 461:654–658. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Xie Z and Klionsky DJ: Autophagosome
formation: Core machinery and adaptations. Nat Cell Biol.
9:1102–1109. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Pan X, Chen Y, Shen Y and Tantai J:
Knockdown of TRIM65 inhibits autophagy and cisplatin resistance in
A549/DDP cells by regulating miR-138-5p/ATG7. Cell Death Dis.
10:4292019. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wang H, Guo M, Ding D, Yang F and Chen Z:
Long non-coding RNA NNT-AS1 contributes to cisplatin resistance via
miR-1236-3p/ATG7 axis in lung cancer cells. Onco Targets Ther.
13:3641–3652. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Pan R and Zhou H: Exosomal transfer of
lncRNA H19 promotes erlotinib resistance in non-small cell lung
cancer via miR-615-3p/ATG7 axis. Cancer Manag Res. 12:4283–4297.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Arensman MD, Yang XS, Zhong W, Bisulco S,
Upeslacis E, Rosfjord EC, Deng S, Abraham RT and Eng CH: Anti-tumor
immunity influences cancer cell reliance upon ATG7. Oncoimmunology.
9:18001622020. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Feng X, Zhang H, Meng L, Song H, Zhou Q,
Qu C, Zhao P, Li Q, Zou C, Liu X and Zhang Z: Hypoxia-induced
acetylation of PAK1 enhances autophagy and promotes brain
tumorigenesis via phosphorylating ATG5. Autophagy. 17:723–742.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wang C, Haas MA, Yeo SK, Paul R, Yang F,
Vallabhapurapu S, Qi X, Plas DR and Guan JL: Autophagy mediated
lipid catabolism facilitates glioma progression to overcome
bioenergetic crisis. Br J Cancer. 124:1711–1723. 2021. View Article : Google Scholar : PubMed/NCBI
|