Introduction
Hedgehog (HH) signaling is a developmentally
conserved pathway in numerous embryonic tissues and has been shown
to be dysregulated in multiple cancers (1,2). The
Sonic Hedgehog cascade involves the Sonic hedgehog (Shh) ligand
binding to Patched (PTCH), a 12-pass transmembrane protein. When
the ligand is absent, PTCH represses the activity of the
neighboring 7-pass membrane protein Smoothened (SMO). This
inhibition is released upon Shh binding. The ensuing activation of
SMO triggers a chain of events that lead to the release of GLI1-3
effector proteins from the Suppressor of Fused (SuFu) and their
subsequent translocation to the nucleus (2). The activated HH/GLI pathway has been
linked to a number of types of human cancers and causes accelerated
proliferation and survival and an enhanced rate of metastasis. HH
also supports the self-renewal of cancer stem cells (CSCs), a
subpopulation of tumor cells with inherent resistance to therapy
(3,4). HH activity can be regulated in a
noncanonical manner, and does not require the initial Shh binding
to the receptor. A number of pathways, such as RAS (5), MAPK (6), AKT (7)
and EGFR (8), have been shown to
activate GLI factors directly in tumor cells.
The HH pathway has been shown to be essential for
the oncogenic properties of melanoma (6). Moreover, blunting GLI1 and GLI2
restores sensitivity to vemurafenib in vemurafenib-resistant
melanoma cells harboring BRAF mutations (9). SOX2 is crucial for the self-renewal of
CSCs in melanoma and is regulated by GLI1 and GLI2, thus mediating
HH signaling (10). GLI1 and GLI2
also transcriptionally regulate several genes involved in positive
regulation of the cell cycle, such as E2F1, cdk1 and cyclin B
(11).
The transcription factor Slug, the protein product
of the SNAIL2 gene, belongs to the Snail family of
zinc-finger transcription factors (12). As early as 1998, the human Slug
protein was described to contain 268 amino acids and its molecular
weight is ~30 kDa (13). Slug is
expressed during embryogenesis and is critical for the development
of the neural crest (14). Notably,
Slug and Snail significantly contribute to the maintenance of CSCs
(15). Slug is an antiapoptotic
protein that contributes to the transcriptional repression of
E-cadherin in epithelial tumors, thus contributing to
epithelial-to-mesenchymal transition (EMT) in tumor cells (16). This is achieved by upregulating the
expression of Zeb1, which is then engaged in repressing E-cadherin
(17). Furthermore, Twist1
upregulates Slug which mediates Twist1-induced changes during EMT
(18). Cooperatively, these
transcription factors repress epithelial markers, enabling cell
detachment and migration during early stages of EMT, which is a key
phenomenon underlying cancer progression and invasion (19). Moreover, increased Slug expression
is found in patients suffering from a number of types of cancers
(20). Increased Slug levels are
linked to metastasis, tumor recurrence and poor prognosis and play
a role in the maintenance of CSCs (21). The CSC-like properties of tumor
cells promote tumor initiation, expansion, EMT, metastasis, and
tumor relapse and confer resistance to chemotherapy and
radiotherapy in multiple types of cancer (22). Downregulation of Slug results in
inhibition of the proliferation of cancer cell lines, and its
overexpression leads to accelerated proliferation (21).
In melanoma, SLUG has been considered to be a
pro-oncogenic gene contributing to EMT (17). Slug expression in melanoma cells has
been reported to be regulated by osteonectin (SPARC). PI3/Akt
kinase signaling acts upstream of SPARC, as its blockade hinders
induction of the SLUG gene by SPARC, cell migration, and
EMT-like changes (23). The protein
Nodal is involved in the expression of SNAIL and SLUG
genes and activation of ALK/Smads and PI3K/AKT pathways (24,25).
Slug silencing has also been shown to increase the radiosensitivity
of melanoma cells (26).
Despite these findings, the precise mechanisms of
Slug expression and its role in EMT remain to be elucidated in
melanoma. Gupta et al (27)
described the necessity of Slug for the development of melanoma
metastases in a mouse model. By contrast, Slug protein expression
has been observed to be diminished in human metastases (28). Slug, together with Zeb2, was notably
found to be downregulated during EMT in melanoma. Slug and Zeb2
transcription factors have been reported to drive a melanocytic
differentiation program and behave as oncosuppressive proteins,
whereas Zeb1 and Twist1 repress differentiation and have oncogenic
properties (29). Similar
conclusions were reported by Gunarta et al (30) after ablating GLI1 function in
melanoma. GLI1-knockdown cells exhibited reduced invasion ability
accompanied by downregulation of the EMT factors Snail1, Zeb1 and
Twist1 but not Snail2 or Zeb2. As SLUG is one of the genes
contributing to CSC maintenance, a central question for
understanding the acquisition of the mesenchymal state and CSC
renewal is how the expression of genes involved in EMT is
regulated. In brief, inconsistent results have been reported
regarding SLUG gene function in melanoma cells, and the
mechanisms of its expression have not been extensively studied. The
present study investigated the transcriptional regulation of
SLUG in human melanoma cells and observed that Slug
expression is controlled by the HH/GLI pathway, particularly the
GLI2 transcription factor.
Materials and methods
Cell culture
The present study used eight melanoma cell lines
(listed in Table SI). Their
mutational status (BRAF and NRAS mutations) is shown in Table SII. The origin of cells has been
described previously (31,32). Cells were cultivated in RPMI medium
(MilliporeSigma) supplemented with 10% FCS (Thermo Fisher
Scientific, Inc.), glutamine and antibiotics (MilliporeSigma) at
37°C and 5% CO2 in 100% humidity. All cell lines were
authenticated and tested for mycoplasma using a mycoplasma
detection kit (MP0035; MilliporeSigma). Cells were passaged every
72–96 h using a trypsin-EDTA solution. When plated, cells
(5×105) were seeded from a stable culture into 12-well
plates and incubated for 24 h at 37°C prior to inhibitor treatment
or transfection, unless specified otherwise. Normal human
melanocytes were purchased from Cascade Biologics Inc. and
cultivated according to the manufacturer´s instructions. The
generation of inducible melanoma cell lines in which
melanoma-associated transcription factor (MITF) protein can be
downregulated by the addition of doxycycline (Tet-on system) has
been previously described (32).
Chemical inhibitors
GANT61 (stock prepared in DMSO) and cyclopamine
(stock prepared by dissolving in ethanol) were purchased from
Selleck Chemicals LLC. The chemicals were applied to cells as
indicated in the appropriate figures for 20 h before cell
harvesting if not stated otherwise. The addition of 20 µM GANT61
for 20 h was used after the optimization of both the concentration
and time to follow the changes in expression of GLI-dependent
genes, when no signs of apoptosis had been yet detected in
cells.
Western blot analysis
To obtain whole-cell extracts for immunoblotting
analysis, cells were lysed in RIPA buffer (1% NP-40, 150 mM NaCl, 5
mM EDTA, 0.5% sodium deoxycholate, 50 mM Tris-HCl pH 7.5 and 0.1%
SDS) with the addition of the protease and phosphatase inhibitors
aprotinin, pepstatin and leupeptin at 1 mg/ml each. cOmplete (Roche
Diagnostics) was added as recommended by the supplier. Then, 1 mM
phenylmethylsulfonyl fluoride (MilliporeSigma) and phosphatase
inhibitor PhosSTOP (Roche Diagnostics) were added. Equal amounts of
protein (30 µg; concentration determined by Bradford's assay) were
loaded on 10–12% SDS-polyacrylamide gels, separated by
electrophoresis and transferred onto PVDF membranes. Blots were
blocked in 5% non-fat dry milk Blotto (cat. no. sc-2325, Santa Cruz
Biotechnology, Inc.), and incubated with 1:1,000 diluted primary
antibodies for 2 h, washed, and then incubated at room temperature
for 1 h with 1:4,000 diluted secondary anti-mouse or anti-rabbit
horseradish peroxidase-conjugated antibodies (cat. no. sc-2055 or
cat. no. sc-2030; Santa Cruz Biotechnology, Inc.). Chemiluminescent
detection was used. For western blotting shown in Fig. 5A and B, cells were lysed in PLB
buffer (Promega Corporation), used in dual luciferase measurements
and these extracts were then directly utilized. Primary antibodies
for western blotting were: GLI1 (cat. no. 3538; Cell Signaling
Technology, Inc.), GLI2 (cat. no. TX46056; GeneTex, Inc.), and GLI3
(cat. no. AF3690; R&D Systems, Inc.). Anti-Slug (cat. no.
sc-166476) was purchased from Santa Cruz Biotechnology, Inc. Klf4
(cat. no. LS-C415468) and ALDH1A1 (cat. no. LS-B10149) from LSBio.
Anti-FLAG (M2; cat. no. F1804) and anti-β-actin (A5316) antibodies
were purchased from MilliporeSigma. Anti-livin antibody (cat. no.
sc-30161) was from Santa Cruz Biotechnology, Inc. and MITF (D5)
antibody (cat. no. NBP2451590) was from Thermo Fisher Scientific,
Inc. Brn-2 (cat. no. sc-514295) was from Santa Cruz Biotechnology,
Inc. E- and N-cadherins, vimentin, Zeb1, and Zeb2 antibodies were
from Cell Signaling Technology (cat. no. 9782). Western blot bands
were quantified using ImageJ (v. 1.52j) software (National
Institutes of Health; data not shown).
 | Figure 5.MITF is an imperfect activator of
Slug in 501mel melanoma cells. (A) Left, SLUG promoter
activity is slightly decreased by cotransfection of the MITF
expression vector and activated ~twice by the MITF-Vp16 chimeric
hyperactive MITF vector. Middle, the melastatin promoter is
activated by both the MITF and MITF-Vp16 proteins. pcDNA3 plasmid
served as a control. Three other melanoma cell lines likewise
showed nonsignificant differences in the activity of the
SLUG promoter compared with the control plasmid when
cotransfected with the MITF vector (not shown). Right, western
blotting shows MITF expression after transfection. The control
sample also has a strong MITF signal because the endogenous protein
level is already present in naive 501mel cells. (B) Left,
SLUG promoter activity after cotransfection of FLAG-tagged
versions of the same plasmids as shown in (A). Middle, stimulation
of melastatin promoter activity. Empty pFLAG-CMV-4 plasmid served
as a control. Statistical analysis was as described in Materials
and methods and verified using one-way ANOVA followed by Tukey's
post hoc test and equal results (P-values) were obtained. Right,
Western blotting performed with the anti-FLAG antibody show
expression of FLAG-tagged proteins, the same pattern of expression
was seen after the transfection of melastatin promoter (not shown).
(C) Western blots depicting inducible knockdown of MITF by
tetracycline-regulatable shRNA repressing MITF. Concomitant
expression of Slug and livin proteins was observed in the same
samples, whereas no change in Slug protein levels was observed. Six
melanoma cell lines (left panel) and two control lines (right
panel) were analyzed. Β-actin detection served as a loading control
for whole cell lysates (bottom). This figure is reprinted as a part
of Figure 3 from our previously published paper Vlčková K et
al: Inducibly decreased MITF levels do not affect proliferation
and phenotype switching but reduce differentiation of melanoma
cells (J. Cell. Mol. Med. 22, 2018, 2240-2251) (32), with the permission of the publisher
(Wiley Global Permissions, permissions@wiley.com). MITF,
melanoma-associated transcription factor. *P<0.05,
**P<0.01. |
Plasmids
The 12×GLI-TK-Luc plasmid was obtained from
Professor R Toftgard, Karolinska Institute, Sweden. pGL3-PTCH1 was
kindly donated by Professor Aberger, University of Salzburg,
Austria. The PATCHED gene promoter contains one active
GLI-binding site responsible for its activity (data not shown). The
pGL3-slug-prom-full-length promoter (−5216+112) and
pGL3-slug-prom-Δmiddle (−5216-4635, −2092+112) were cloned as
XhoI-HindIII (New England BioLabs, Inc.) inserts in
the pGL3-basic plasmid (Promega Corporation).
pGL3-slug-prom-Δproximal (−5216-2092) was cloned as the
XhoI-HindIII insert. The-4635+112 promoter was cloned
as the NheI-HindIII insert. pGL3-slug-prom-middle
(−4635-2092) was cloned as the NheI-NheI (New England
BioLabs, Inc.) insert and the pGL3-slug-proximal promoter
(−2092+112) was cloned as the NheI-HindIII insert.
Cloning of all GLI expression vectors has been described previously
(31). Briefly, original GLI1 (GL1;
cat. no. 16419), GLI2 (pCS2-MT GLI2 FL; cat. no. 17648), ΔNGLI2
(pCS2-MT GLI2 delta N; cat. no. #17649) and GLI3 (GLI3 bs-2; cat.
no. 16420) were acquired from the nonprofit plasmid repository
Addgene, Inc. Respective coding sequences were amplified by PCR and
subsequently cloned into the pcDNA3 expression vector or into the
pFLAG-CMV-4 plasmid (MilliporeSigma) background to obtain
FLAG-tagged GLI proteins. The types of GLI expression plasmids were
similarly effective in promoter activation. The melastatin promoter
plasmid with the three MITF-binding sites was constructed as
previously described (33). The
construction of the short hairpin plasmid directed to MITF has been
previously described (32). All
final plasmid inserts were verified by sequencing on both strands
(GATC Biotech).
Transfection and promoter-reporter
assays
Transient cell transfections of the promoter
reporters were performed using 12-well plates at 37°C, seeded and
incubated for 24 h before transfections, and the transfection
reagent Mirus TransIT-2020 (Mirus Bio LLC) following the
manufacturer's instructions and harvested 48 h after transfection.
For detection, a dual luciferase system (Promega Corporation) was
used according to the manufacturer's instructions. The pGL3-basic
vector was used as a negative control. Expression vectors or
controls (pCDNA3 or pFLAG-CMV-4) were cotransfected together with
the reporter plasmids. Cell lysates were used for dual luciferase
assays performed as recommended by the manufacturer's instructions.
The luciferase values were acquired on a Turner Designs 20/20
luminometer (Promega Corporation). Data were normalized to
Renilla luciferase activity (internal control) as arbitrary
units. Statistical analysis of luciferase assay results was
performed using a two-tailed unpaired Student's t test and
SigmaPlot software V10.0 (Systat Software Inc.). Where indicated,
cells were treated with GANT61 or cyclopamine 20 h at 37°C before
harvesting. Tomatidine, a compound inactive in the HH pathway but
structurally similar to cyclopamine, was tested as a negative
control and revealed similar results as vehicle (data not
shown).
Reverse transcription-quantitative
(RT-q) PCR
Total RNA was isolated using TRIzol®
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
supplier´s instructions (3×105 cells per 30 mm well).
Then 2 µg of RNA was reverse transcribed using SuperScript IV
reverse transcriptase (Thermo Fisher Scientific, Inc.), and qPCR
was performed using a TaqMan QuantiTect Probe PCR kit (Qiagen GmbH)
on a ViiA7 Real-Time PCR system (Thermo Fisher Scientific, Inc.)
following the manufacturer's instructions (cycling: 30 sec at 94°C
and 1 min at 60°C). Data analysis was performed by QuantStudio 6
Software (Thermo Fisher Scientific, Inc.). Concurrent results were
obtained in three independent experiments with the following PCR
primers and probe for SLUG: Forward,
5′-AGAACTCACACGGGGGAGAAG-3′; reverse,
5′-CTCAGATTTGACCTGTCTGCAAA-3′; probe,
5′-6-FAM-TTTTTCTTGCCCTCACTGCAACAGAGC-TAMRA-3′. β-Actin was utilized
as an internal standard control: Forward, 5′-ATTGCCGACAGGATGCAGAA,
reverse, 5′-GCTGATCCACATCTGCTGGAA; probe,
6-FAM-CAAGATCATTGCTCCTCCTGAGCGCA-TAMRA. Fluorescent probes were
purchased from Thermo Fisher Scientific, Inc.
Chromatin immunoprecipitation
501mel cell cultures were transfected using Mirus
TransIT-2020 (Mirus Bio LLC) with the pcDNA3-GLI1,
pcDNA3-GLI2 or pcDNA3-GLI3 expression plasmids in 90 mm plates.
After 24 h of treatment, fresh medium was applied. After another 24
h, cells were crosslinked with 1% formaldehyde for 10 min at room
temperature and incubated with glycine solution, and chromatin
immunoprecipitation was performed by using the ChIP-IT High
Sensitivity kit (Active Motif, Inc.) in accordance with the
manufacturer's protocols. Anti-acetylated histone H3 antibody
(MilliporeSigma) was used as the positive control, and rabbit
nonimmune IgG (MilliporeSigma) was used as a negative control. To
detect GLI1 bound to the promoter, a rabbit anti-GLI1 antibody
(cat. no. sc-20687; Santa Cruz Biotechnology, Inc.) was used.
Rabbit anti-GLI2 (cat. no. ab26056; Abcam) was used to precipitate
GLI2. For GLI3, a rabbit anti-GLI3 antibody (cat. no. 3538; Cell
Signaling Technology, Inc.) was used and 2 µg of each antibody was
added in each reaction. To assess the amount of ChIP-generated DNA,
PCR was performed using Phusion High-Fidelity DNA polymerase
(Thermo Fisher Scientific, Inc.). The amplification of the regions
of the proximal SLUG promoter was performed with four alternative
primer pairs: Region A (−2108-1766): sense,
5′-GCATACGTGTTACTCGCTAGCAC-3′, antisense,
5′-TCCTTGTTTCACTCTACACAGTCTATTCAC-3′; region B (−1769-1163): sense,
5′-AGGAAATAATAGCCATGGCGATA-3′, antisense,
5′-CATCTCTGTCCATTGCAGAC-3′; region C (−1182-490): sense,
5′-GTCTGCAATGGACAGAGATGC-3′, antisense, 5′-GGGAAGCGGGAAGACAAAG-3′;
and region D: (−509+112): sense, 5′-CCTTTGTCTTCCCGCTTCCCCCTTCC-3′,
antisense, 5′-ACACGGCGGTCCCTACAGCATCG-3′. PCR products were
resolved on 1% agarose gels. The intensity of the final PCR bands
was quantified using ImageJ (v. 1.52j) software (National
Institutes of Health).
Immunofluorescence
For immunofluorescence, four cell lines (501mel,
Hbl, SK-MEL-5 and SK-MEL-28) were seeded into 8-well Lab-Tek II
Chamber Slides (Thermo Fisher Scientific, Inc.). After 48 h, 20 µM
GANT61 was added for 20 h at 37°C. Vehicle (DMSO) alone was added
to the controls. The cells were fixed with 3% paraformaldehyde at
room temperature for 10 min, washed, permeabilized with 0.1% Triton
X-100, and blocked with 5% goat serum. Slides were then stained
with Slug primary antibody (1:1,000; cat. no. sc-166476; Santa Cruz
Biotechnology, Inc.) at room temperature for 2 h followed by
1:1,000 secondary anti-mouse fluorescein-coupled antibody (cat. no.
FI-2000-1.5, Vector Laboratories, Inc.) and mounted in medium with
DAPI to stain nuclei. The visualization was performed using an
Olympus IX70 microscope with cellSens software V2.2 (Olympus
Corporation).
Immunohistochemical analysis
Formalin-fixed paraffin-embedded tissues (skin,
nevus and melanoma) were retrieved from the archive of the
Department of Pathology and Molecular Medicine, Second Faculty of
Medicine, Charles University, University Hospital Motol, Prague. At
least four samples of each tissue were processed and similar
results were obtained. Deparaffinized, rehydrated in descending
ethanol series, and blocked (with 3% H2O2)
sections were stained with 1:1,000 primary antibodies.
Immunohistochemistry for MITF was performed with the primary
antibody MITF (cat. no. D5; cat. no. NBP2451590, Thermo Fisher
Scientific, Inc.). GLI2 was stained with antibody cat. no. GTX46056
(GeneTex, Inc.) and Slug with anti-Slug (A-7) sc-166476 antibody
(Santa Cruz Biotechnology, Inc.). Detection was performed using an
EnVision+ avidin-biotin detection system (Dako; Agilent
Technologies, Inc.). Each tissue was examined by two pathologists.
Tissues were scored on a scale of 0 to 4 based on the combined
extent and intensity of staining. The final score represented the
predominant staining pattern of both combined parameters. Section
fields were imaged using a BX51 microscope (Olympus Corporation)
equipped with a PROMICAM 3-3CP 3.1 camera and QuickPHOTO CAMERA 3.2
software (Olympus Corporation).
Statistical analysis
Statistical significance (P-values) was assessed
using a two-tailed Student's t test and Mann-Whitney test. Standard
error of the mean values are depicted in graphs as bars within each
column in the reporters and RT-qPCR. Data not significant
(P>0.05) were not labeled, values of 0.01 <P<0.05 are
marked by an asterisk, and values with P<0.01 are marked by two
asterisks. SigmaPlot software V10.0 (Systat Software Inc.) and
GraphPad Prism v.8.4.3 software (Dotmatics) were used to perform
statistical analysis. Statistical analysis was verified using
one-way ANOVA followed by post hoc tests as specified in figure
legends. P<0.05 was considered to indicate a statistically
significant difference.
Results
Slug expression in melanomas is
inhibited by the Hedgehog pathway inhibitor GANT61
Snail1 and Snail2 (Slug) are among the main players
in the tumorigenic program of EMT (19). SLUG gene deletions have been
found in Waardenburg syndrome and piebaldism in humans (34,35),
indicating the involvement of Slug in melanin pigmentation. The
SLUG gene has long been thought to be under the
transcriptional control of MITF (34). To further explore the regulation of
Slug expression in melanoma, the present study examined the SLUG
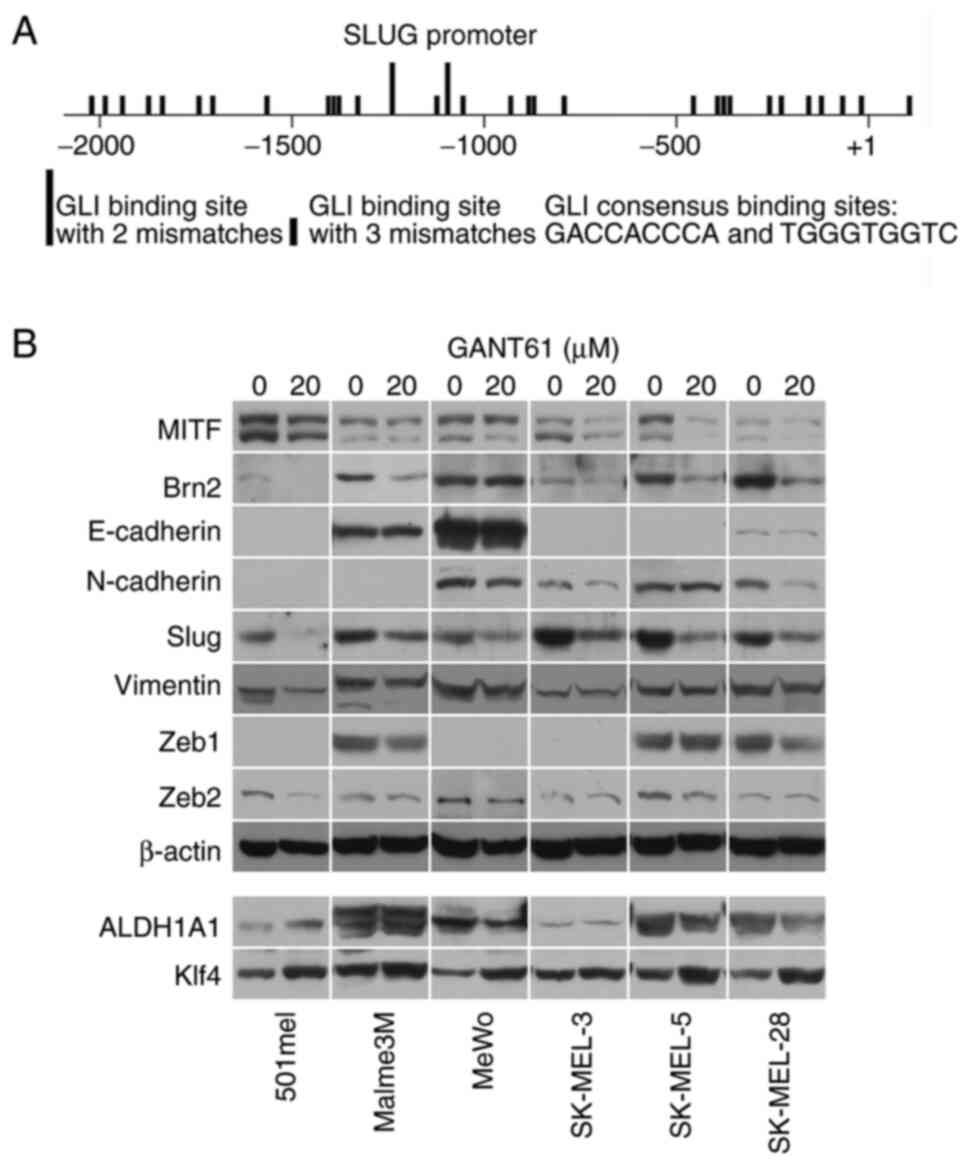
promoter and found 85 GLI-binding sites in the long SLUG promoter
(−5216 +112; the start of translation is +1) and 31 sites in the
proximal promoter (−2092 +112; Fig.
1A and Table SIII). Although
none of the sites was a full consensus, GLI proteins, which are the
final executive factors of the Hedgehog signaling pathway, bind to
and are also active from sites with two or three mismatched
nucleotides. This implies that SLUG gene expression could be
controlled by the Hedgehog pathway.
To further investigate this hypothesis, Slug protein
expression was examined in melanoma cell lines treated with GANT61,
a potent and specific pan-GLI transcriptional inhibitor. It was
found that GANT61 decreased Slug protein in all eight melanoma cell
lines investigated as well as in normal melanocytes (Figs. 1B and SI). BRAF- or NRAS-mutated or wild-type
cells for both oncogenes were present among the analyzed melanoma
cell lines (Table SII). The levels
of other proteins known to be involved in EMT, such as E- and
N-cadherins, vimentin, Zeb1 and Zeb2, were not appreciably changed
after GANT61 treatment in six melanoma cell lines. Likewise, the
CSC markers ALDH1A1 and Klf4 were generally only slightly affected
(Fig. 1B). A mild increase in Klf4
was noted in the cell lines 501mel, MeWo, SK-MEL-5 and SK-MEL-28,
while ALDH1A1 levels were somewhat increased in 501mel cells and
slightly diminished in SK-MEL-28 cells after GANT61 treatment
(Fig. 1B). Only Brn2 (N-Oct-3,
POU3F2) protein, a known repressor of MITF (36), noticeably decreased in five of six
cell lines, suggesting that its transcription may also be regulated
by the HH pathway in most melanomas. MITF was slightly
downregulated in three cell lines (Fig.
1B). Immunofluorescence staining confirmed blunting of Slug
expression after the treatment of melanoma cells with GANT61
(Fig. S2).
SLUG promoter-reporter is activated by
GLI2 and inhibited by GANT61 and cyclopamine
Since the transcriptional inhibitor of GLI factors
GANT61 downregulated Slug protein expression, the activity of the
SLUG promoter was analyzed in reporter assays. Examination
of luciferase expression driven by the long (full-length) promoter
(−5216+112) and its truncated versions revealed that the middle
part of promoter (−4635-2092) was the most active fragment
(Fig. 2A). Its deletion greatly
decreased the activity of the full-length promoter. The short
portion most upstream (−5216-4635) probably performs an inhibitory
function because its presence in the contexts of the full-length
promoter, middle part and proximal promoter (−2092+112) decreased
the luciferase activity (Fig. 2A).
As the proximal promoter also showed appreciable activity, it was
used in the following experiments.
 | Figure 2.Promoter-reporter analysis of the
SLUG gene promoter. (A) The long (full-length) SLUG
promoter (−5216+112) and its truncated versions were assayed for
activity in 501mel cells. Only two-mismatched consensus GLI-binding
sites are shown as blue bars. pGL3-basic was used as a negative
control and exhibited near zero activity (data not shown). The
statistical significance of truncated promoters is related to the
long promoter (100%). (B) Inhibition of the proximal SLUG
promoter by GANT61 and cyclopamine in three cell lines. Bars are
the mean ± standard error of the mean). The statistical
significance is related to the activity of the untreated proximal
promoter. (C) Activation of the 12×GLI-site full consensus promoter
by GLI transcription factors. Left, the control plasmid
(pGL3-basic) had extremely low activity and was negligibly
influenced by GLI factors; right, the massive effect of GLI
expression vectors on a positive control reporter plasmid with 12
full consensus GLI sites. (D) Left, inhibition of the GLI-activated
PATCHED promoter by GANT61; middle left, middle right and
right graphs, GANT61 inhibited three versions of the SLUG
promoter. The most significant inhibition was seen with the
proximal promoter (right). The GLI-mediated effect is compared with
the control (pGL3-basic) in (B-left) and (D). The significance of
GANT61-mediated inhibition is related to the vehicle-treated
control. Statistical analyses were performed using a Student´s t
test. Statistical analysis was verified using one-way ANOVA test,
followed by Dunnett's post hoc test and equal results (P-values)
were obtained. In all reporter assays, two or three independent
experiments were performed (in triplicate) with similar results,
and one representative experiment is shown. In all graphs, ±
standard error of the mean bars are shown. Statistical
significance: no mark, not significant, *P<0.05, **P<0.01.
(E) Western blotting showing similar levels of expression of all
GLI proteins in (C) and (D). GLI, GLI family zinc finger. |
Next, to test whether the SLUG promoter-reporter is
directly activated by cotransfected expression vectors for GLI
factors and inhibited by GANT61, it was first investigated whether
the proximal promoter activity decreased after the addition of HH
pathway inhibitors. Indeed, both GANT61 and cyclopamine lowered the
activity in all three melanoma cell lines tested (Fig. 2B). As a subsequent experiment, each
of the expression vectors for GLI factors (GLI1-3) were
cotransfected with the GLI-responsive promoter containing 12
canonical GLI-binding sites (12×GLI). First, it was ascertained
that there was only a minimal difference between the cotransfected
control (pcDNA3) and GLI vectors with the pGL3-basic empty control
promoter (Fig. 2C, left). By
contrast, all three GLIs greatly increased 12×GLI promoter activity
compared with the negative control plasmid pcDNA3 (Fig. 2C, right). GLI3 showed the weakest
activation of the canonical 12×GLI promoter (but still ~80-fold),
whereas the highest activation (800-fold) was achieved by ΔGLI2, a
truncated version of GLI2 in which the N-terminal repression domain
was removed (37). The activity of
the GLI factors was then tested on a known HH-responsive promoter
of the PATCHED gene, also a component of the HH pathway. The
results were similar to those obtained with the canonical promoter,
but the extent of stimulation was substantially lower, ~6-fold in
the case of the most active ΔGLI2 (Fig.
2D, first graph). Similar experiments were then conducted with
the three types of SLUG promoters, the long (full-length),
Dmiddle and proximal promoters (Fig.
2D, second, third and fourth graphs). In all cases, the DGLI2
construct again showed the best activation. In accordance with the
results in Fig. 2A, the Dmiddle
promoter exhibited the lowest overall activity. When the
PATCHED and SLUG promoters were tested, the addition
of GANT61 more or less decreased the GLI-stimulated promoter
activities (Fig. 2B and D). Western
blotting verified that all GLI proteins were similarly expressed
(Fig. 2E). These results together
indicated that SLUG promoter activity is dependent mainly on
GLI2 in reporter assays and that stimulation by GLI factors can be
inhibited by GANT61.
Slug RNA levels are diminished after
GANT61 treatment
To further evaluate the transcriptional regulation
of Slug by HH/GLI, the present study examined the effect of GANT61
on Slug RNA levels using real-time PCR. RT-PCR was performed first,
followed by qPCR. In all eight melanoma cell lines tested, 20 µM
GANT61 significantly (P<0.01) lowered the mRNA level of Slug.
This indicates that the positive effect of GLI factors on
endogenous SLUG gene expression is mediated through the
activation of transcription (Fig.
3).
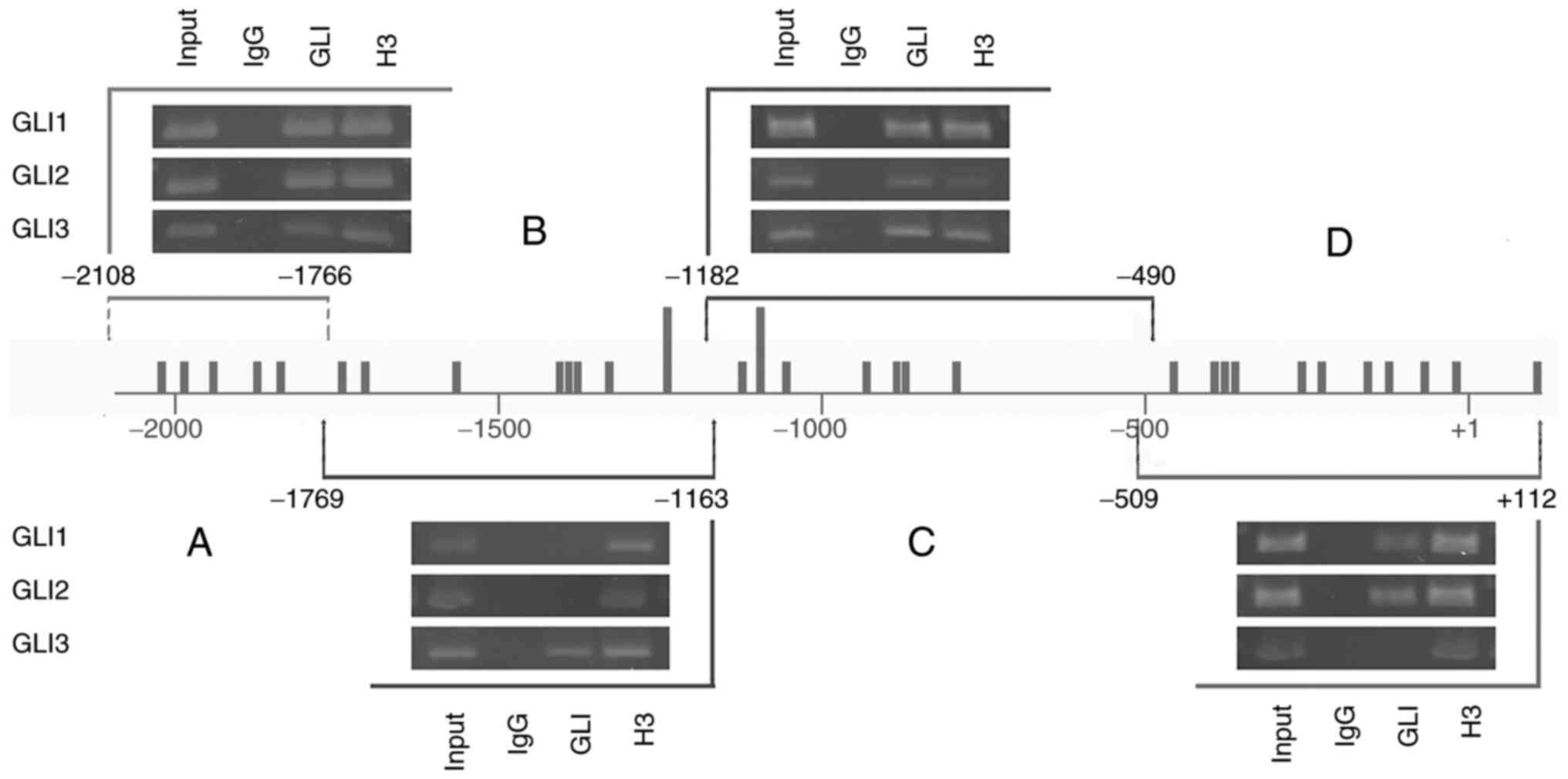
ChIP assays show binding of GLI
factors to the proximal SLUG promoter
The proximal SLUG promoter contains 31
GLI-binding sites, of which only two sites harbor two mismatches
and all other sites have three mismatches (Fig. 2A and Table SIII). To investigate whether these
sites are occupied by GLI proteins in cells, the 501mel cell line
was used to chromatin immunoprecipitate DNA fragments bound to GLIs
using specific anti-GLI1, GLI2 and GLI3 antibodies (Fig. 4). To obtain improved insight into
whether the particular GLI binds to specific regions of the
proximal promoter (−2092+112), the immunoprecipitated purified DNA
was amplified by four primer pairs (see Materials and methods) that
demarcated the four subregions A-D (Fig. 4) of the promoter. Each subregion
contained several GLI-binding sequences. The most distal region A
and a middle region C were bound by all three GLI factors. The
middle region B was remarkably occupied only by the GLI3 factor.
The most proximal fragment D was bound by GLI1 and GLI2 but not
GLI3 (Fig. 4). Thus, regions B and
D showed some preference for GLI occupancy, whereas regions A and C
clearly exhibited enrichment by all three GLI factors. Acetylated
histone H3, which was precipitated by anti-acetylated H3 antibody
as a positive control, showed occupancy at all four subregions.
Nonimmune IgG as a negative control showed no binding in any
experiment. Taken together, these data indicate that GLI
transcription factors are abundantly present at their recognition
DNA sites within the SLUG proximal promoter, further
confirming their role in the transcriptional regulation of Slug
expression through HH signaling. The data quantifying the final DNA
amount formed through PCR are in Table
SIV.
Slug is an imperfect target for MITF
in melanoma cells
The SLUG gene has been demonstrated to be
transcriptionally regulated by MITF in melanocytes (34) and during Xenopus laevis
development (38). However, data
relevant to a possible regulation of Slug expression by MITF in
melanoma cells are lacking. To test whether MITF overexpression
influences the activity of the SLUG gene promoter, we
performed promoter-reporter assays in which the proximal promoter
was cotransfected with the MITF expression construct into 501mel
cells. Additionally, we compared activation by MITF-Vp16, a
hyperactive MITF derivative in which the MITF N-terminal activation
domain (AD) was replaced by a stronger Vp16 AD (39), with native MITF. While MITF had no
effect on promoter activity, MITF-Vp16 increased it ~2-fold
(Fig. 5A left). On the other hand,
the melastatin promoter, a known MITF target (33), was stimulated by both MITF and
MITF-Vp16 ~4-fold and 10-fold, respectively (Fig. 5A middle). Western blotting verified
the total MITF protein level (the control sample also showed the
MITF protein signal because relatively high endogenous MITF protein
is already present in nontransfected 501mel cells; Fig. 5A, right). Only transfected cells
with ectopic MITF or MITF-Vp16 were measured for luciferase
activity, which explained why promoter activity increased more
compared with the overall MITF protein level. The same experiment
was repeated with the FLAG-tagged vectors, and the same results
were obtained. The expression of proteins expressed from
transfected plasmids was verified with the anti-FLAG antibody
(Fig. 5B).
To corroborate these results,
doxycycline-regulatable melanoma cell lines, in which MITF could be
downregulated by inducible expression of shRNA directed at MITF
were used (32). Whereas a decrease
in endogenous MITF protein was achieved after the addition of
doxycycline in all cell lines, Slug expression remained unchanged.
By contrast, the level of livin (ML-IAP), a known MITF target
(40), mirrored the decrease in
MITF protein (Fig. 5C). In
agreement with this, overexpression of MITF did not change the
endogenous level of Slug protein (data not shown). In a control
experiment, inducible control nontargeting shRNA revealed no
changes in the expression of all proteins tested. Thus, endogenous
Slug seems to be expressed independently of MITF protein levels in
human melanoma cells.
To further investigate the relationship between MITF
and Slug and GLI2 vs. Slug protein expression in human samples,
parallel sections of skin, nevus and melanoma metastasis were
compared by immunohistochemical staining (Fig. 6). Positive staining by anti-GLI2 and
anti-Slug antibodies was observed in the epidermis of normal skin
(scored 2–3), consistent with previous observations (41,42).
Only a few strongly MITF-positive (score 4) melanocytes were
present in the epidermis. GLI2 and Slug were also stained in nevus
cells, albeit somewhat less intensively (score 2, rare cells scored
3 in Slug staining). The nevus showed abundant MITF-positive cells
(score 4). Epidermal keratinocytes were, as expected, MITF-negative
in the skin and nevus sections (score 0). In metastatic melanoma,
both GLI2 and Slug staining was positive in ~half of the cell
population (scored 2–4), with Slug staining slightly more
positively than GLI2 staining. MITF-specific staining of metastatic
melanoma was negative (score 0), with only a small number of
positive cells (not shown in the picture). Sections that were
negative or very slightly positive for MITF (scored 0–1) showed
cells stained for both GLI2 and Slug (scores 2–3; Fig. 6). This is also consistent with the
idea that invasive and metastatic cells have low or absent MITF
(43). Notably, all three proteins
were almost exclusively localized to the nucleus. Thus, Slug
staining was associated with GLI2, but not MITF, positive staining
in immunohistochemical sections of melanomas (Fig. 6). This result supports the
aforementioned findings by demonstrating the positive regulation of
Slug expression by GLI2, but not by MITF, in melanoma cells.
Discussion
The present study described the essential role of HH
signaling and the transcription factor GLI2 in the transcription of
the SLUG gene in human melanoma cells. The large number of
GLI-binding sites present in the SLUG promoter led to the
investigation of how GLI factors regulated SLUG gene
expression. The presented data indicated that GLI factors activated
the SLUG promoter in reporter assays and that the promoter
was repressed by the HH signaling inhibitors GANT61 and
cyclopamine. The most potent activator appeared to be GLI2. All
GLI1-3 factors occupied the proximal SLUG promoter.
Chromatin immunoprecipitation data revealed abundant and specific
binding of GLI factors to four contiguous subregions of the
proximal SLUG promoter. RT-qPCR, induced decrease of MITF
and immunohistochemical experiments corroborated the foregoing
data; GANT61 diminished Slug RNA abundance, decreased MITF and did
not change Slug protein levels. In addition, the
immunohistochemical analysis showed that MIFT-negative portions of
metastatic melanomas contained Slug-positive and GLI2-positive
cells. Given that an extremely high number of GLI-binding sites are
present in the SLUG promoter, the possibility that Slug can
also be upregulated by HH in other cancer cell types is worthy of
further investigation. Of course, other transactivation mechanisms
may also be plausible in different tumors beyond melanoma. For
example, c-myb has been shown to regulate Slug expression in colon
carcinoma, chronic myeloid leukemia, and neuroblastoma cells
(44). C-myb-elicited EMT-like
characteristics through SLUG gene activation in these cells.
Homeobox C10 (HOXC10), which is a pro-oncogenic protein in cancer,
has been shown to activate the SLUG promoter in melanoma
through the YAP/TAZ signaling pathway (45). Das et al (46) reported that the oncogenic phenotype
was induced by transcriptional upregulation of osteopontin through
GLI1 in melanoma cells. Increased invasion, proliferation and
migration was relieved by HH inhibitors. On the other hand,
miR-33a-5p downregulates Slug in melanoma (47).
Previously, the essential role for HH signaling in
melanoma has been demonstrated to occur mainly through the
transcription factor GLI1 (6).
Other signaling routes, such as RAS/MAPK and Akt/mTOR, regulate the
nuclear localization of GLI1, not only in melanoma but also in
other cancer cell types (6).
Another report showed that GLI2 is capable of mediating the
invasion and metastatic properties of melanoma (48). Furthermore, Slug is considered to be
regulated by MITF, an essential transcription factor in melanoma
(34). The present study showed
that Slug expression depended mainly on the HH/GLI pathway. MITF
probably does not serve any role in the endogenous expression of
Slug because MITF downregulation had no effect on Slug protein
levels in several melanoma cell lines. In the reporter assays, only
the hyperactive MITF-Vp16 chimera, but not native MITF, activated
the SLUG gene promoter (Fig.
5A).
It has been demonstrated that the oncogene
CRKL is the downstream effector of GLI2 in lung
adenocarcinoma cells (49). Crkl,
which is oncogenic in several types of cancer, is activated by GLI2
through the binding of GLI2 to the site in the second intron of the
CRKL gene. Blunting Crkl by shRNA or CRISPR-Cas9 knockout
weakens the GLI2-elicited positive effect on cell viability,
migration, invasion and colony formation. Thus, in lung
adenocarcinoma, Crkl appears to be necessary for pro-oncogenic GLI2
effects and is itself regarded as an oncogene. For example, Crkl
attenuates the therapeutic effect of several antioncogenic drugs
(49) and has been found to be
amplified in lung adenocarcinoma (50). It remains to be investigated whether
Crkl is a general mediator of GLI factor activity in other tumor
cell types. Although CRKL has been found to be amplified
specifically in acral melanomas (51), no other data are available about the
melanoma-specific role of the Crkl protein.
HH pathway and GLI factors are highly oncogenic and
known to substantially contribute to the maintenance of CSC. In
addition, GLIs are observed to be associated with EMT in various
types of cancer (52–54). On the other hand, little data are
available that GLI factors could be directly involved in the
transcription of typical EMT-associated genes. In melanoma, GLI1
and GLI2 are reported to enhance transcription of Sox2, a stem cell
marker (10). Furthermore, it was
previously observed that EMT process is atypical in melanoma
(55), as suggested in the present
study. The present study clearly defined that GLIs, especially
GLI2, are direct transcriptional regulators of Slug, a hallmark
protein of EMT, in melanoma cells.
In melanoma, therapy is predominantly focused on
targeting the mutated driver oncogenes BRAF and NRAS and/or kinases
in the downstream MAPK signaling pathway. Unfortunately, therapies
for advanced melanoma with low molecular weight inhibitors
targeting these proteins result in acquired resistance. Despite
advances in using a combination of drugs, the concept of targeting
only the MAPK route remains questionable. As there are multiple
mechanisms responsible for resistance to the inhibition of MAPK
signaling in melanoma (56,57), alternative therapies should also be
considered. Targeting HH and Bcl2 protein by the combination of
GANT61 and obatoclax was effective in most melanoma cell lines
tested previously (58). Taken
together, the present study described a new mechanism of Slug
transcription. It stressed the importance of the HH pathway for
melanoma progression and suggested that targeting GLI2 in
combination therapies could be beneficial for treatment, as GLI2 is
a recognized transcriptional activator of a number of oncogenic
proteins, including survivin (31).
Although other mechanisms may play a role in Slug regulation in
various types of cancer, the present study demonstrated that Slug
is another HH/GLI target.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgments
The authors thank Professor F. Aberger (University
of Salzburg) for providing the PATCHED promoter plasmid and
Professor R. Toftgard (Karolinska Institute) for the 12×GLI
reporter plasmid.
Funding
The present study was supported by funding from the
institutional programs PROGRESQ25 and Cooperatio (research areas
SURG, MED BIOCHEM, METABOLISM, and ONCOLOGY) from Charles
University Prague, from the League against Cancer Prague, and from
the Conceptual Development of Research Organization, Motol
University Hospital, Prague, funded by the Ministry of Health,
Czech Republic (grant no. 6028). These funding organizations played
no role in the analysis of the data or the preparation of this
article.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
JV and PH conceived the project. JV and PH confirm
the authenticity of all the raw data. PH, JR, KK, LO, JV and PZ
performed the experiments. JB and JV Jr. performed the
immunohistochemical experiments. PH and KK performed statistical
analyses. PH and KK prepared the figures. PH and JV wrote the
manuscript. All authors read and approved the manuscript.
Ethics approval and consent to
participate
Immunohistochemical analysis of paraffin sections of
human tissues was approved by the Ethical Committee of the
University Hospital Motol, Prague (approval no. EK-36/20). The
study was carried out in accordance with the Declaration of
Helsinki.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
Slug (Snai2, Snail2)
|
snail family zinc finger 2
|
|
Snail1
|
snail family zinc finger 1
|
|
EMT
|
epithelial-to-mesenchymal
transition
|
|
ALDH1A1
|
aldehyde dehydrogenase 1 family member
A1
|
|
Klf4
|
Krüppel-like transcription factor
4
|
|
GLI
|
GLI family zinc finger
|
|
CSC
|
cancer stem cells
|
|
HH
|
Hedgehog signaling pathway
|
|
MITF
|
melanoma-associated transcription
factor
|
References
|
1
|
Teglund S and Toftgård R: Hedgehog beyond
medulloblastoma and basal cell carcinoma. Biochim Biophys Acta.
1805:181–208. 2010.PubMed/NCBI
|
|
2
|
Varjosalo M and Taipale J: Hedgehog:
Functions and mechanisms. Genes Dev. 22:2454–2472. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Marini KD, Payne BJ, Watkins DN and
Martelotto LG: Mechanisms of Hedgehog signalling in cancer. Growth
Factors. 29:221–234. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jeng KS, Chang CF and Lin SS: Sonic
Hedgehog signaling in organogenesis, tumors, and tumor
microenvironments. Int J Mol Sci. 21:7582020. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lauth M, Bergström A, Shimokawa T, Tostar
U, Jin Q, Fendrich V, Guerra C, Barbacid M and Toftgård R:
DYRK1B-dependent autocrine-to-paracrine shift of Hedgehog signaling
by mutant RAS. Nat Struct Mol Biol. 17:718–725. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Stecca B, Mas C, Clement V, Zbinden M,
Correa R, Piguet V, Beermann F, Ruiz IA and Altaba A: Melanomas
require HEDGEHOG-GLI signaling regulated by interactions between
GLI1 and the RAS-MEK/AKT pathways. Proc Natl Acad Sci USA.
104:5895–5900. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Riobó NA, Lu K, Ai X, Haines GM and
Emerson CP Jr: Phosphoinositide 3-kinase and Akt are essential for
Sonic Hedgehog signaling. Proc Natl Acad Sci USA. 103:4505–4510.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mangelberger D, Kern D, Loipetzberger A,
Eberl M and Aberger F: Cooperative Hedgehog-EGFR signaling. Front
Biosci (Landmark Ed). 17:90–99. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Faião-Flores F, Alves-Fernandes DK,
Pennacchi PC, Sandri S, Vicente AL, Scapulatempo-Neto C, Vazquez
VL, Reis RM, Chauhan J, Goding CR, et al: Targeting the hedgehog
transcription factors GLI1 and GLI2 restores sensitivity to
vemurafenib-resistant human melanoma cells. Oncogene. 36:1849–1861.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Santini R, Pietrobono S, Pandolfi S,
Montagnani V, D'Amico M, Penachioni JY, Vinci MC, Borgognoni L and
Stecca B: SOX2 regulates self-renewal and tumorigenicity of human
melanoma-initiating cells. Oncogene. 33:4697–4708. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Pandolfi S, Montagnani V, Lapucci A and
Stecca B: HEDGEHOG/GLI-E2F1 axis modulates iASPP expression and
function and regulates melanoma cell growth. Cell Death Differ.
22:2006–2019. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Nieto MA: The snail superfamily of
zinc-finger transcription factors. Nat Rev Mol Cell Biol.
3:155–166. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cohen ME, Yin M, Paznekas WA, Schertzer M,
Wood S and Jabs EW: Human SLUG gene organization, expression, and
chromosome map location on 8q. Genomics. 51:468–471. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Pérez-Mancera PA, González-Herrero I,
Maclean K, Turner AM, Yip MY, Sánchez-Martín M, García JL, Robledo
C, Flores T, Gutiérrez-Adán A, et al: SLUG (SNAI2) overexpression
in embryonic development. Cytogenet Genome Res. 114:24–29. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang Y, Shi J, Chai K, Ying X and Zhou BP:
The role of snail in EMT and tumorigenesis. Curr Cancer Drug
Targets. 13:963–972. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bolós V, Peinado H, Pérez-Moreno MA, Fraga
MF, Esteller M and Cano A: The transcription factor Slug represses
E-cadherin expression and induces epithelial to mesenchymal
transitions: A comparison with Snail and E47 repressors. J Cell
Sci. 116:499–451. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wels C, Joshi S, Koefinger P, Bergler H
and Schaider H: Transcriptional activation of ZEB1 by Slug leads to
cooperative regulation of the epithelial-mesenchymal
transition-like phenotype in melanoma. J Invest Dermatol.
131:1877–1885. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Casas E, Kim J, Bendesky A, Ohno-Machado
L, Wolfe CJ and Yang J: Snail2 is an essential mediator of
Twist1-induced epithelial mesenchymal transition and metastasis.
Cancer Res. 71:245–254. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Brabletz S, Schuhwerk H, Brabletz T and
Stemmler MP: Dynamic EMT: A multi-tool for tumor progression. EMBO
J. 40:e1086472021. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Pérez-Mancera PA, González-Herrero I,
Pérez-Caro M, Gutiérrez-Cianca N, Flores T, Gutiérrez-Adán A,
Pintado B, Sánchez-Martín M and Sánchez-García I: SLUG in cancer
development. Oncogene. 24:3073–3082. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cobaleda C, Pérez-Caro M, Vicente-Dueñas C
and Sánchez-García I: Function of the zinc-finger transcription
factor SNAI2 in cancer and development. Annu Rev Genet. 41:41–61.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Barbato L, Bocchetti M, Di Biase A and
Regad T: Cancer stem cells and targeting strategies. Cells.
8:9262019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Fenouille N, Tichet M, Dufies M, Pottier
A, Mogha A, Soo JK, Rocchi S, Mallavialle A, Galibert MD, Khammari
A, et al: The epithelial-mesenchymal transition (EMT) regulatory
factor SLUG (SNAI2) is a downstream target of SPARC and AKT in
promoting melanoma cell invasion. PLoS One. 7:e403782012.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Guo Q, Ning F, Fang R, Wang HS, Zhang G,
Quan MY, Cai SH and Du J: Endogenous Nodal promotes melanoma
undergoing epithelial-mesenchymal transition via Snail and Slug in
vitro and in vivo. Am J Cancer Res. 5:2098–2112. 2015.PubMed/NCBI
|
|
25
|
Pearlman RL, Montes de Oca MK, Pal HC and
Afaq F: Potential therapeutic targets of epithelial-mesenchymal
transition in melanoma. Cancer Lett. 391:125–140. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Arienti C, Tesei A, Carloni S, Ulivi P,
Romeo A, Ghigi G, Menghi E, Sarnelli A, Parisi E, Silvestrini R and
Zoli W: SLUG silencing increases radiosensitivity of melanoma cells
in vitro. Cell Oncol (Dordr). 36:131–139. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gupta PB, Kuperwasser C, Brunet JP,
Ramaswamy S, Kuo WL, Gray JW, Naber SP and Weinberg RA: The
melanocyte differentiation program predisposes to metastasis after
neoplastic transformation. Nat Genet. 37:1047–1054. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Shirley SH, Greene VR, Duncan LM, Torres
Cabala CA, Grimm EA and Kusewitt DF: Slug expression during
melanoma progression. Am J Pathol. 180:2479–2489. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Caramel J, Papadogeorgakis E, Hill L,
Browne GJ, Richard G, Wierinckx A, Saldanha G, Osborne J,
Hutchinson P, Tse G, et al: A switch in the expression of embryonic
EMT-inducers drives the development of malignant melanoma. Cancer
Cell. 24:466–480. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gunarta IK, Li R, Nakazato R, Suzuki R,
Boldbaatar J, Suzuki T and Yoshioka K: Critical role of
glioma-associated oncogene homolog 1 in maintaining invasive and
mesenchymal-like properties of melanoma cells. Cancer Sci.
108:1602–1611. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Vlčková K, Ondrušová L, Vachtenheim J,
Réda J, Dundr P, Zadinová M, Žáková P and Poučková P: Survivin, a
novel target of the Hedgehog/GLI signaling pathway in human tumor
cells. Cell Death Dis. 7:e20482016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Vlčková K, Vachtenheim J, Réda J, Horák P
and Ondrušová L: Inducibly decreased MITF levels do not affect
proliferation and phenotype switching but reduce differentiation of
melanoma cells. J Cell Mol Med. 22:2240–2251. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Miller AJ, Du J, Rowan S, Hershey CL,
Widlund HR and Fisher DE: Transcriptional regulation of the
melanoma prognostic marker melastatin (TRPM1) by MITF in
melanocytes and melanoma. Cancer Res. 64:509–516. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sánchez-Martín M, Rodríguez-García A,
Pérez-Losada J, Sagrera A, Read AP and Sánchez-García I: SLUG
(SNAI2) deletions in patients with Waardenburg disease. Hum Mol
Genet. 11:3231–3236. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Sánchez-Martín M, Pérez-Losada J,
Rodríguez-García A, González-Sánchez B, Korf BR, Kuster W, Moss C,
Spritz RA and Sánchez-García I: Deletion of the SLUG (SNAI2) gene
results in human piebaldism. Am J Med Genet A. 122A:125–132. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Goodall J, Carreira S, Denat L, Kobi D,
Davidson I, Nuciforo P, Sturm RA, Larue L and Goding CR: Brn-2
represses microphthalmia-associated transcription factor expression
and marks a distinct subpopulation of microphthalmia-associated
transcription factor-negative melanoma cells. Cancer Res.
68:7788–7794. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Roessler E, Ermilov AN, Grange DK, Wang A,
Grachtchouk M, Dlugosz AA and Muenke M: A previously unidentified
amino-terminal domain regulates transcriptional activity of
wild-type and disease-associated human GLI2. Hum Mol Genet.
14:2181–2188. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kumasaka M, Sato S, Yajima I, Goding CR
and Yamamoto H: Regulation of melanoblast and retinal pigment
epithelium development by Xenopus laevis Mitf. Dev Dyn.
234:523–534. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Vachtenheim J and Drdová B: A dominant
negative mutant of microphthalmia transcription factor (MITF)
lacking two transactivation domains suppresses transcription
mediated by wild type MITF and a hyperactive MITF derivative.
Pigment Cell Res. 17:43–50. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Dynek JN, Chan SM, Liu J, Zha J,
Fairbrother WJ and Vucic D: Microphthalmia-associated transcription
factor is a critical transcriptional regulator of melanoma
inhibitor of apoptosis in melanomas. Cancer Res. 68:3124–3132.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ikram MS, Neill GW, Regl G, Eichberger T,
Frischauf AM, Aberger F, Quinn A and Philpott M: GLI2 is expressed
in normal human epidermis and BCC and induces GLI1 expression by
binding to its promoter. J Invest Dermatol. 122:1503–1509. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Parent AE, Choi C, Caudy K, Gridley T and
Kusewitt DF: The developmental transcription factor slug is widely
expressed in tissues of adult mice. J Histochem Cytochem.
52:959–965. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Müller J, Krijgsman O, Tsoi J, Robert L,
Hugo W, Song C, Kong X, Possik PA, Cornelissen-Steijger PD, Geukes
Foppen MH, et al: Low MITF/AXL ratio predicts early resistance to
multiple targeted drugs in melanoma. Nat Commun. 5:57122014.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Tanno B, Sesti F, Cesi V, Bossi G,
Ferrari-Amorotti G, Bussolari R, Tirindelli D, Calabretta B and
Raschellà G: Expression of Slug is regulated by c-Myb and is
required for invasion and bone marrow homing of cancer cells of
different origin. J Biol Chem. 285:29434–29445. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Miao Y, Zhang W, Liu S, Leng X, Hu C and
Sun H: HOXC10 promotes growth and migration of melanoma by
regulating Slug to activate the YAP/TAZ signaling pathway. Discov
Oncol. 12:122021. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Das S, Harris LG, Metge BJ, Liu S, Riker
AI, Samant RS and Shevde LA: The hedgehog pathway transcription
factor GLI1 promotes malignant behavior of cancer cells by
up-regulating osteopontin. J Biol Chem. 284:22888–22897. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zhang ZR and Yang N: MiR-33a-5p inhibits
the growth and metastasis of melanoma cells by targeting SNAI2.
Neoplasma. 67:813–824. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Alexaki VI, Javelaud D, Van Kempen LC,
Mohammad KS, Dennler S, Luciani F, Hoek KS, Juàrez P, Goydos JS,
Fournier PJ, et al: GLI2-mediated melanoma invasion and metastasis.
J Natl Cancer Inst. 102:1148–1159. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Liu X, Hu Y, Yu B, Peng K and Gan X: CRKL
is a critical target of Hh-GLI2 pathway in lung adenocarcinoma. J
Cell Mol Med. 25:6280–6288. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Kim YH, Kwei KA, Girard L, Salari K, Kao
J, Pacyna-Gengelbach M, Wang P, Hernandez-Boussard T, Gazdar AF,
Petersen I, et al: Genomic and functional analysis identifies CRKL
as an oncogene amplified in lung cancer. Oncogene. 29:1421–1430.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Weiss JM, Hunter MV, Cruz NM, Baggiolini
A, Tagore M, Ma Y, Misale S, Marasco M, Simon-Vermot T, Campbell
NR, et al: Anatomic position determines oncogenic specificity in
melanoma. Nature. 604:354–361. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Jiang L, Huang J, Hu Y, Lu P, Luo Q and
Wang L: Gli promotes tumor progression through regulating
epithelial-mesenchymal transition in non-small-cell lung cancer. J
Cardiothorac Surg. 15:182020. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Chun HW and Hong R: Significance of the
hedgehog pathway-associated proteins Gli-1 and Gli-2 and the
epithelial-mesenchymal transition-associated proteins Twist and
E-cadherin in hepatocellular carcinoma. Oncol Lett. 3:1753–1762.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Wang L, Jin JQ, Zhou Y, Tian Z, Jablons DM
and He B: Gli is activated and promotes epithelial-mesenchymal
transition in human esophageal adenocarcinoma. Oncotarget.
9:853–865. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Kim JE, Leung E, Baguley BC and Finlay GJ:
Heterogeneity of expression of epithelial-mesenchymal transition
markers in melanocytes and melanoma cell lines. Front Oncol.
4:972013.
|
|
56
|
Davies MA and Kopetz S: Overcoming
resistance to MAPK pathway inhibitors. J Natl Cancer Inst.
105:9–10. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Vachtenheim J and Ondrušová L: Many
distinct ways lead to drug resistance in BRAF- and NRAS-mutated
melanomas. Life (Basel). 11:4242021.PubMed/NCBI
|
|
58
|
Vlčková K, Réda J, Ondrušová L, Krayem M,
Ghanem G and Vachtenheim J: GLI inhibitor GANT61 kills melanoma
cells and acts in synergy with obatoclax. Int J Oncol. 49:953–960.
2016. View Article : Google Scholar : PubMed/NCBI
|