Introduction
Colorectal cancer (CRC) occurs in the large
intestine and is the third most common cancer worldwide. CRC is the
second leading cause of cancer-related death (1). In addition, the recurrence rate of CRC
after curative resection is 30–50% (1,2). The
survival and mortality rates of patients with CRC are closely
related to cancer recurrence and metastasis. In the United States,
the 5-year relative survival rate of patients with CRC ranges from
90% in patients diagnosed with local disease to 14% in patients
diagnosed with distant-stage disease (3). Prediction of CRC recurrence and
metastasis is key for determining treatment methods for patients
with CRC. However, reliable prognostic molecular biomarkers for
evaluating CRC recurrence and metastasis are lacking. Thus, despite
progress made towards the discovery of novel therapeutic methods,
the mortality of patients with CRC remains relatively high.
Lymphocyte antigen 6 (LY6) family (LY6D, LY6E, LY6H
and LY6K) is found in human lymphoid cells (4–6) and
participates in T cell activation and the function of the
complement membrane attack complex (7). Members of the LY6 family are
low-molecular-weight glycoproteins with conserved cysteine residues
bound to the cell membrane via a C-terminal
glycosylphosphatidylinositol moiety. Although the biological
function of LY6 family is not well-understood, these proteins are
involved in signaling and cell activation (8,9). The
LY6 family is associated with common copy number gain mutations in
various types of cancer (10,11).
Recently, numerous researchers have shown interest in the LY6
family, which plays a multifaceted role in cancer pathogenesis,
stem cell maintenance, and immune regulation and its association
with aggressive and intractable cancers. In particular, the mRNA
expression levels of several genes in the LY6 family were
significantly higher in lung, brain, breast, and head and neck
cancers than in normal tissues (4,12).
Lymphocyte antigen 6 complex, locus E (LY6E) is a member of the LY6
superfamily in lymphostromal cells (13–15).
LY6E is a glycosylphosphatidylinositol-anchored cell-surface
protein that regulates the proliferation, oncogenesis,
differentiation, immunological regulation, and activation of
T-lymphocytes (16). Previous
studies identified LY6E as a common surface marker in gastric and
pancreatic cancer (12,17). Additionally, LY6E is associated with
tumor immune escape and drug resistance in breast cancer (16). However, the expression of LY6E in
lymphostromal cells of cancer tissues has not been reported so
far.
In the present study, the effects of LY6E
downregulation on cellular function in CRC cell lines were
evaluated. Using siRNA-based technology, some roles of LY6E protein
in CRC carcinogenesis were investigated. The clinical data of the
present study suggested that LY6E is a prognostic marker for
CRC.
Materials and methods
Immunohistochemistry and data
analysis
Archived CRC tissues were obtained from 100 patients
in 2005 and another 10 samples were obtained in 2018. Written
informed consent was obtained from all patients for use of the
specimens. The present study was approved (approval no.
2018-07-061) by the Institutional Review Board of Soonchunhyang
University Cheonan Hospital (Cheonan, Korea). Immunohistochemistry
(IHC) for LY6E was performed on paraffin-embedded tissues using
4-µm tissue sections mounted on slides. For immunohistochemistry
staining of LY6E, all slides were blocked with serum and incubated
with the LY6E primary antibody at 4°C, overnight (1:200; cat. no.
GTX101567; GeneTex, Inc.). The slides were washed and then
incubated with EnVision+ System- HRP-labelled polymer anti-rabbit
(cat. no. K4002; Dako; Agilent Technologies, Inc.) for 30 min at
room temperature. To evaluate the immunohistochemistry results, the
antibodies were stained with 70 µl 3′-3-diaminobenzidine (DAB; cat.
no. K3468; Dako; Agilent Technologies, Inc.) and visualized under a
light microscope. The expression of LY6E in CRC tissues was scored
in a blinded manner by independent investigators, and a consensus
score was determined for each specimen. The expression scoring of
LY6E was graded on a four-point scale based on the intensity and
percentage of LY6E staining: The percentage of staining was scored
as follows: 0, 0–5; 1, 5–25; 2, 25–50; 3, 50–75; and 4, 75–100%,
whereas intensity of staining was scored in the following manner:
0, negative; 1, weak; 2, moderate; 3, strong. The final scores were
calculated by multiplying of two scores: 0 points for negative, 1–3
points for weak, 4–6 points for moderate and 7–12 points for
strong. The LY6E low expression group included negative and weak
categories, and the LY6E high expression group included cases that
scored above 4 points.
Cell culture
Human CRC cell lines (SW480, SW620, HCT116 and HT29)
were provided by the Korean Cell Line Bank (Seoul, Korea) and
cultured in Roswell Park Memorial Institute 1640 (RPMI-1640) medium
(cat. no. SH30027.01; Hyclone; Cytiva) containing 10% fetal bovine
serum (FBS; cat. no. 35-015-CV; Corning, Inc.) and 1%
penicillin/streptomycin (cat. no. 30-022-CI; Corning, Inc.) at 37°C
in a 5% CO2 incubator. The identities of CRC cells were
confirmed by short tandem repeat (STR) profiling by KCLB and all
cell lines were each purchased within the last 5 years.
Transfection of small interfering
RNA
CRC cell lines were transfected with 100 nM of the
appropriate small interfering RNA (siRNA) with 12 µl of HiPerFect
Transfection Reagent (cat. no. 301705; Qiagen GmbH) in 12-well
plates. After 72 h, the cells were used for experiments. The
targeting sequences for LY6E-specific siRNA were siRNA1:
5′-GCAUUGGGAAUCUCGUGACTT-3′, siRNA2: 5′-GCUUCUCCUGCUUGAACCATT-3′,
and siRNA3: 5′-GCCAGAGCUUUCUGUGCAATT-3′. Non-targeting control
siRNAs of AccuTarget™ Negative Control siRNA (cat. no. SN-1002;
Bioneer Corporation) was used as control. The LY6E-specific siRNAs
were used in combination.
Reverse transcription-quantitative
(RT-q) PCR
Total RNA from CRC cell lines was extracted from the
control group and LY6E-siRNA treated group using RiboEx Total RNA
Solution (cat. no. 301-001; GeneAll Biotechnology). Using 1 µg of
the extracted RNA, cDNA was synthesized using the ReverTra Ace™
qPCR RT kit (cat. no.FSQ-101; Toyobo Life Science) according to the
manufacturer's instructions. The PCR reaction temperature
conditions were followed by 35 cycles of pre-denaturation (95°C, 10
min), denaturation (95°C, 30 sec), annealing (55–65, 15 sec) and
extension (72°C, 15 sec). RT-qPCR for LY6E was performed using
primers purchased from Bioneer Corporation. To confirm the
expression of genes for cancer metastasis-related cell growth and
proliferation-related phenotypes, primers were designed for CDKN2A,
IGF1, CXCR4 and MET and determined by RT-qPCR, and GAPDH was used
as an endogenous control (Table I).
The target gene mRNA level was quantified as GAPDH mRNA, and the
gene expression level was analyzed using the 2−ΔΔCq
method (18). The amplicon was
mixed with 10X loading buffer (TaKaRa Bio, Inc.) and then agarose
gel electrophoresis was performed using 2% agarose gel containing
NEOgreen (NeoScience) DNA staining reagent. then FluoroBox
(Neoscience) nucleic acid gel imaging system was used to visualize
the amplicon, and the mRNA expression level was analyzed using
image J 1.53t software (National Institutes of Health).
 | Table I.Primer sequences were used in
reverse-transcription polymerase chain reaction. |
Table I.
Primer sequences were used in
reverse-transcription polymerase chain reaction.
| Gene name | Amplicon size
(bp) | Primer sequence
(5′à3′) |
|---|
| LY6E | 391 | Sense:
AAGTAGCTGACCACAGAGCA |
|
|
| Antisense:
TGTCACGAGATTCCCTGCAT |
| CDKN2A | 384 | Sense:
TCTGACCATTCTGTTCTCTC |
|
|
| Antisense:
CTCAGCTTTGGAAGCTCTCA |
| IGF1 | 134 | Sense:
CTCTTCAGTTCGTGTGTGGAGAC |
|
|
| Antisense:
CAGCCTCCTTAGATCACAGCTC |
| CXCR4 | 127 | Sense:
CTCCTCTTTGTCATCACGCTTCC |
|
|
| Antisense:
GGATGAGGACACTGCTGTAGAG |
| MET | 142 | Sense:
TGCACAGTTGGTCCTGCCATGA |
|
|
| Antisense:
CAGCCATAGGACCGTATTTCGG |
| GAPDH | 131 | Sense:
CCAGCCGAGCCACATCGCTC |
|
|
| Antisense:
ATGAGCCCCAGCCTTCTCCAT |
Western blotting
Total cellular protein was extracted using PRO-PREP
(Intron Biotechnology, Inc.) and quantified using a BCA kit (cat.
no. 23227; Thermo Fisher Scientific, Inc.) and an xMark Microplate
Absorbance Spectrophotometer (Bio-Rad Laboratories, Inc.). The 30
µg protein was analyzed using 10% sodium dodecyl
sulfate-polyacrylamide gel electrophoresis and β-actin was used as
a loading control. Then proteins were transferred to a
polyvinylidene fluoride membrane (cat. no. ISEQ00010;
MilliporeSigma). The polyvinylidene fluoride membrane was incubated
with anti-LY6E antibody (1:100; cat. no. ABIN517581;
Antibodies-online GmbH) overnight at 4°C, and the appropriate
horseradish peroxidase-conjugated secondary antibody (1:2,000; cat.
no. 32260; Thermo Fisher Scientific, Inc.) was reaction for 2 h at
room temperature. Protein was visualized with the addition of
enhanced chemiluminescence (cat. no. 34577; Thermo Fisher
Scientific, Inc.). Western blot results were visualized using a
Molecular Imager ChemiDoc XRS + system (Bio-Rad Laboratories, Inc.)
and quantified using the ImageJ software (National Institutes of
Health).
Migration and invasion assay
The control group and LY6E-siRNA treated group of
cells were collected, and 2×105 cells were resuspended
in 200 µl of medium and inserted into the upper of 6.5 mm
Transwell® with 8.0 µm Pore Polycarbonate Membrane
Insert with serum-free RPMI-1640 medium. In the lower chamber, 600
µl of supplemented medium was added, and the cells were cultured at
37°C in a humidified incubator for 48 h. The remaining cells in the
upper chamber that could not pass through the 8 µm pore-size filter
were removed using a cotton swab. The cells were fixed with 3.7%
neutral-buffered formalin at room temperature for 3 min and stained
with 0.005% crystal violet at room temperature for 20 min. Cell
images were obtained, and cells were counted using an inverted
microscope. Invasion assays were performed as aforementioned for
the migration assay, except that the Transwell chamber was coated
with 50 µl Matrigel 3 h before the experiment and stored at
37°C.
Proliferation assay (WST-1 assay)
Post 48-h transfection with LY6E-inhibited and
scramble control siRNA, nine replicates (1×104
cells/well) were uniformly dispensed in a 96-well plate and were
incubated at 37°C, 5% CO2 for 24, 48 and 72 h. Next,
WST-1 assay reagent (EZ-CYTOX; cat. no. EZ-1000; DogenBio;
http://www.dogenbio.com/) was added 10 µl (5
mg/ml) at every set time and reacted for 1 h. The absorbance was
measured at 562 nm using a microplate reader.
Soft agar colony formation assay
As basement agar, 0.5% agarose was prepared in
RPMI-1640 medium supplemented with 10% FBS. As top agar, the
control and LY6E-siRNA treated groups were plated in six-well
dishes (5×103 cells/well) in 0.35% agarose in RPMI-1640
supplemented with 10% FBS. After 10–14 days, the colonies were
stained for 1 h at room temperature with 0.005% crystal violet and
images were captured after removing the growth medium. Numbers of
colonies with a diameter greater than 30 µm were quantified after 2
weeks and analyzed using Image J 1.53t software (National
Institutes of Health).
Statistical analysis
The data are presented as the average of the values
of three replicates. Analysis was conducted using SPSS v19 software
(IBM Corp.) was used for statistical analysis using unpaired
Student's t-test. Mann Whitney U test and Kruskal-Wallis followed
by Dunn's or Steel-Dwass post hoc tests were used to compare for
analyzing the IHC scores. Cox regression and Kaplan-Meier curves
were used to analyze the associations between LY6E expression and
patient outcomes. P≤0.05 was considered to indicate a statistically
significant difference.
Results
LY6E expression and survival analysis
in patients with CRC
To investigate the genes affecting among the LY6
family in CRC, the expression of LY6 family (LY6D, LY6E, LY6H,
LY6K) was confirmed in colon adenocarcinoma and rectal
adenocarcinoma based on the The Cancer Genome Atlas database
(https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga).
As a result, it was confirmed that only LY6E was specifically
overexpressed among LY6 family in CRC and genes other than LY6E
were rarely expressed in CRC (Fig.
1A). The mRNA expression was analyzed from 275 colon cancer
tissues, 349 relevant normal tissues, 92 rectal cancer tissues, and
318 normal rectum tissues from the Gene Expression Profiling
Interactive Analysis (GEPIA) website (http://gepia.cancer-pku.cn/index.html) to investigate
the role of LY6E in CRC. LY6E was found to show higher mRNA
expression in tumor tissues than in normal tissues (Fig. 1B and C). Immunohistochemical
staining was performed using tissues from 110 patients obtained
from Soonchunhyang University Cheonan Hospital. Low LY6E expression
was detected in 78 (70.9%), while high LY6E expression was found in
32 (29.1%) CRC tissues (Fig. 1D).
However, lymphostromal cells did not express LY6E as revealed by
immunohistochemistry. High LY6E expression was significantly
associated with pN (P<0.0001) and clinical stage (P<0.0001,
Fig. 1E and F, Table II). Univariate and multivariate cox
proportional hazard regression analysis were performed to further
evaluate according to clinical pathological factors whether
expression of LY6E can be used as independent predictive biomarkers
to predict treatment response. Univariate and multivariate analysis
were expressed using hazard ratio (HR) according to
clinicopathological factors (Table
III). The univariate HR values for vascular invasion were 2.622
(1.151-5.974, P=0.022), and the univariate and multivariate HR
values for lymphatic invasion were 3.216 (1.683-6.144, P<0.001),
2.272 (1.019-5.066, P<0.001), and metastasis were 6.860
(2.800-16.804, P<0.001), 6.472 (2.252-18.594, P<0.001). Also,
in a Kaplan-Meier analysis of 110 patients, high LY6E expression
was significantly associated with overall survival. High expression
of LY6E significantly reduced cancer-specific survival in patients
compared with those with low LY6E expression (log-rank test;
P=0.048, Fig. 1G).
 | Table II.Comparison of clinicopathological
factors and LY6E expression in patients with CRC by chi-square
analysis. |
Table II.
Comparison of clinicopathological
factors and LY6E expression in patients with CRC by chi-square
analysis.
|
| Expression level of
LY6E |
|
|
|---|
|
|
|
|
|
|---|
| Clinicopathological
factors | Low (n=42) | High (n=68) | Total (n=110) | P-value |
|---|
| Age, years (mean ±
SD) | 62.1 (13.5) | 63.2 (13.9) | 62.5 (13.5) |
|
| Sex, N (%) |
|
|
| 0.686 |
|
Female | 17 (41.5) | 24 (58.5) | 41 (37.3) |
|
|
Male | 25 (36.2) | 44 (63.8) | 69 (62.7) |
|
| pT stage, N
(%) |
|
|
| 0.765 |
| 1,
2 | 5 (41.7) | 7 (58.3) | 12 (10.9) |
|
| 3,
4 | 37 (37.8) | 61 (62.2) | 98 (89.1) |
|
| pN stage, N
(%) |
|
|
| <0.001 |
| 0 | 27 (57.4) | 20 (42.6) | 47 (42.7) |
|
| 1,
2 | 15 (23.8) | 48 (76.2) | 63 (57.3) |
|
| Vascular invasion,
N (%) |
|
|
| 0.241 |
|
Negative | 39 (40.6) | 57 (59.4) | 96 (87.3) |
|
|
Positive | 3 (21.4) | 11 (78.6) | 14 (12.7) |
|
| Lymphatic invasion,
N (%) |
|
|
| 0.091 |
|
Negative | 37 (42.5) | 50 (57.5) | 87 (79.1) |
|
|
Positive | 5 (21.7) | 18 (78.3) | 23 (20.9) |
|
| Metastasis, N
(%) |
|
|
| 0.081 |
|
Negative | 42 (40.4) | 62 (59.6) | 104 (94.5) |
|
|
Positive | 0 (0.0) | 6 (100.0) | 6 (5.5) |
|
| Clinical stage, N
(%) |
|
|
| <0.001 |
| I and
II | 27 (60.0) | 18 (40.0) | 45 (40.9) |
|
| III and
IV | 15 (23.1) | 50 (76.9) | 65 (59.1) |
|
 | Table III.Cox regression analysis of the
clinicopathological factors in colorectal cancer. |
Table III.
Cox regression analysis of the
clinicopathological factors in colorectal cancer.
|
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|
|---|
| Clinicopathological
factors | Variable | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Age, years | <60 vs. ≥60 | 0.921
(0.479–1.773) | 0.964 | 0.984
(0.477–2.028) | 0.806 |
| Sex | Female vs.
male | 1.154
(0.614–2.170) | 0.436 | 1.315
(0.660–2.618) | 0.657 |
| pT stage | T1, T2 vs. T3,
T4 | 0.532
(0.236–1.200) | 0.038 | 0.405
(0.172–0.949) | 0.128 |
| pN stage | N0 vs. N1, N2 | 1.812
(0.960–3.418) | 0.196 | 3.264
(0.544–19.599) | 0.066 |
| Vascular
invasion | Negative vs.
Positive | 2.622
(1.151–5.974) | 0.365 | 1.594
(0.582–4.364) | 0.022 |
| Lymphatic
invasion | Negative vs.
Positive | 3.216
(1.683–6.144) | 0.045 | 2.272
(1.019–5.066) | <0.001 |
| Metastasis | Negative vs.
Positive | 6.860
(2.800–16.804) | <0.001 | 6.472
(2.252–18.594) | <0.001 |
| Clinical stage | I, II vs. III,
IV | 1.737
(0.920–3.280) | 0.326 | 0.394
(0.061–2.525) | 0.088 |
| Expression level of
LY6E | Low vs. High | 1.912
(0.991–3.692) | 0.397 | 1.385
(0.652–2.944) | 0.053 |
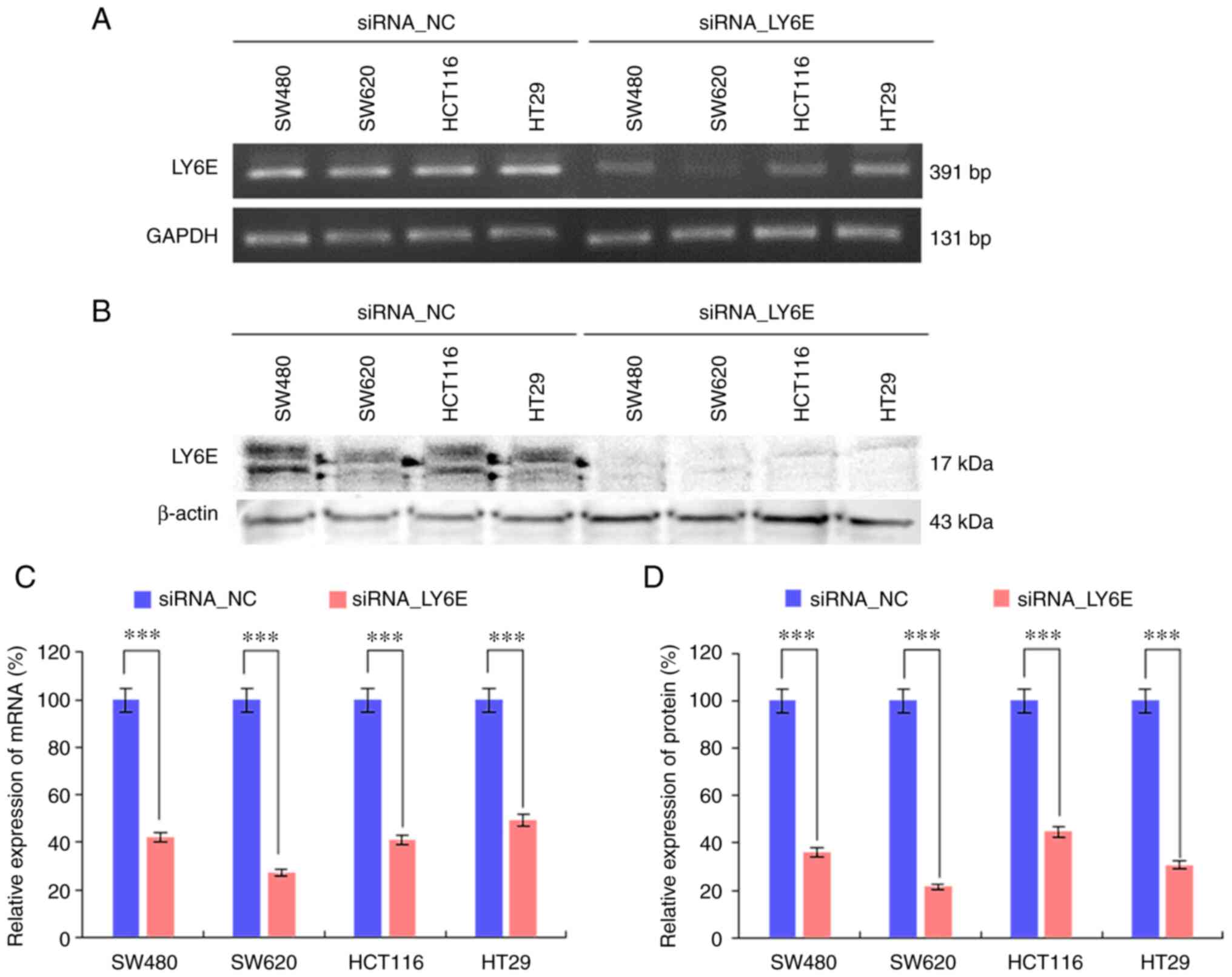
LY6E expression in CRC cells is
reduced by siRNAs
The expression of LY6E was confirmed using RT-qPCR
and immunoblotting in four CRC cell lines (SW480, SW620, HCT116 and
HT29). Then, four CRC cell lines were transfected by LY6E-targeted
siRNA to inhibit LY6E expression. The mRNA level of LY6E decreased
significantly in SW480 (57.8±4.6%), SW620 (72.7±2.5%), HCT116
(58.9±5.3%), and HT29 (50.9±4.6%) cells compared with that in the
control group (Fig. 2A and B,
P<0.001). Additionally, the protein level of LY6E decreased in
SW480 (63.9±3.9%), SW620 (78.5±3.3%), HCT116 (55.3±3.2%) and HT29
(69.3±7.2%) cells compared with that in the control group (Fig. 2C and D, P<0.001). These results
indicated that LY6E expression was successfully reduced at both the
mRNA and protein levels in CRC cells.
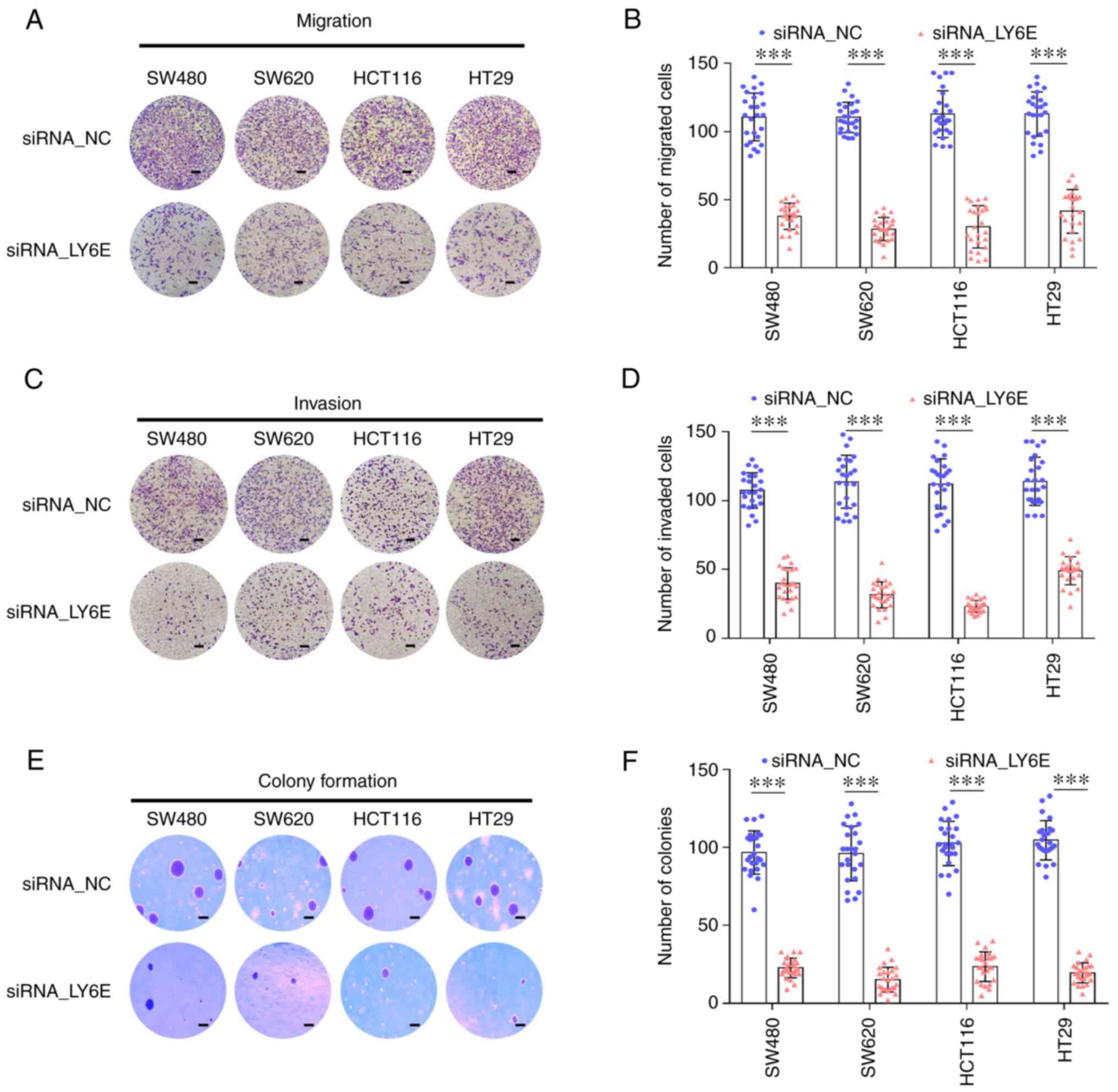
Downregulation of LY6E results in
reduced tumorigenicity, migration and invasiveness of CRC
cells
The effect of LY6E downregulation on cell migration
and invasion was examined. LY6E downregulation notably reduced cell
migration into the lower chamber compared with that of control
cells (SW480, 64.3±8.2%; SW620, 73.5±5.3%; HCT116, 58.3±7.9%; and
HT29, 52.1±8.0%) (Fig. 3A and B,
P<0.001). siRNA_LY6E cells had significantly reduced invasive
properties (SW480, 57.3±7.3%; SW620, 69.3±8.4%; HCT116, 76.7±7.7%;
and HT29, 55.4±7.0%) (Fig. 3C and
D, P<0.001). To confirm whether LY6E plays an important role
in CRC cell colonization, a colony formation assay was performed.
Cells in which LY6E was downregulated showed decreased colony
formation compared with the control group (SW480, 65.3±14.7%;
SW620, 79.2±10.6%; HCT116, 69.5±9.7%; and HT29, 79.8±8.0%)
(Fig. 3E and F, P<0.001).
Inhibition of the LY6E reduces cell
growth and proliferation
Proliferation assay (WST-1 assay) was performed to
determine whether the decreased LY6E gene is involved of cell
proliferation in CRC. Cells in which LY6E was downregulated showed
significant absorbance values compared with the control group
(SW480, 1.81±0.09; SW620, 1.72±0.11; HCT116, 1.86±0.07; and HT29,
1.50±0.12) (Fig. 4A-D, P<0.001).
The expression of genes for phenotypes related to cell growth and
cell proliferation was compared between CRC cells in which LY6E
expression was suppressed and a control. MET, IGF1, CDKN2A and
CXCR4 are cell growth and cell proliferation-related genes that
affect cancer metastasis (19) and
were verified using RT-qPCR. Expression of MET, IGF1, CDKN2A, and
CXCR4 was confirmed in four CRC control cell lines. By contrast,
LY6E-suppressed CRC cells were confirmed that the expression of
MET, IGF1, CDKN2A and CXCR4 was reduced (Fig. 4E and F).
Discussion
Although CRC treatment and diagnostic methods have
improved over the past few decades, the survival rate of patients
with stage IV CRC remains low. The survival rate of these patients
depends on the presence of lymph nodes metastasis, distant
metastasis and recurrence. In the Republic of Korea, the survival
rate of patients with CRC with regional lymph node metastasis is
82.6% but is remarkably decreased by 20.2% in patients with CRC
with distant metastasis (1).
Moreover, the prediction of CRC recurrence is essential for
determining chemotherapeutic or targeted molecular therapeutic
approaches. Several biomarkers correlated with CRC progression have
been identified in molecular studies. The efficacy of conventional
therapies for CRC is increased by targeted therapies. Notably,
prognostic biomarkers can help oncologists optimize therapies for
challenging cases of CRC.
In T-cell among lymphocytes, LY6 functions to
regulate proliferation and differentiation, and immunological
regulation and activity (19). In
addition, LY6E affects the development of cancer by increasing
CTLA4 and tumor-infiltrating T regulatory cells (16). LY6E has been reported to play a
multifaceted role in cancer pathogenesis, stem cell maintenance,
immunomodulation and association with aggressive and refractory
cancers (4,13,16,20).
In breast cancer, it is related to immune evasion and anticancer
drug resistance (16). In
particular, LY6E expression in CRC plays important roles in CRCs by
increasing the expression of PDL1 and has been reported as a
biomarker (16). The present in
vitro studies revealed that LY6E is related to increased
cellular proliferation, invasion, and migration, which are
hallmarks of tumorigenesis. AlHossiny et al (16) reported that high expression of LY6E
is associated with TGF-β signaling maintenance in breast cancer.
TGF-β signaling is involved in cancer cell growth, metastasis,
invasion, and epithelial-to-mesenchymal transition (EMT) (20–24)
and regulates chemotherapeutic drug resistance (25,26).
However, Yeom et al (13)
demonstrated that LY6E downregulates PTEN expression, thereby
activating PI3K-AKT signaling in gastric cancer. Knockdown of LY6E
inhibits the expression of ZEB1, an EMT inducer and E-cadherin
(27). In the present study, LY6E
overexpression was an independent prognostic marker related to poor
overall survival of patients with CRC. It remains unclear whether
LY6E regulates TGF-β or PI3K-AKT signaling and other signaling in
CRC carcinogenesis. Therefore, further studies are needed on the
effects of LY6E, TGF-β and PI3K-AKT signaling in CRC and the direct
correlation between LY6E and T cells. In addition, it is necessary
to elucidate the relationship between the tumorigenesis and
metastasis according to the expression of LY6E through in
vivo studies.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Research
Foundation of Korea (NRF) grant funded by the Korea government
(MSIT, Ministry of Science and ICT) (grant no. 2022R1A2C2010445),
the Basic Science Research Program through the National Research
Foundation of Korea (NRF) funded by the Ministry of Education
(grant no. NRF-2021R1A6A1A03039503) and the Soonchunhyang
University Research Fund.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
DJ and HJK conceptualized the present study. IH, SK,
SR, HK and SO conducted investigation. TSA, DHK and M-JB curated
the data. SK and IH prepared the original draft of the manuscript.
DJ wrote, reviewed, and edited the manuscript and supervised the
study. IH and SK performed visualization. DJ and SK confirm the
authenticity of all the raw data. All authors have read and
approved the final version of the manuscript.
Ethics approval and consent to
participate
The present study was approved (approval no.
2018-07-061) by the Institutional Review Board of Soonchunhyang
University Cheonan Hospital (Cheonan, Korea). Written informed
consent was obtained from all patients for use of the
specimens.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ryuk JP, Choi GS, Park JS, Kim HJ, Park
SY, Yoon GS, Jun SH and Kwon YC: Predictive factors and the
prognosis of recurrence of colorectal cancer within 2 years after
curative resection. Ann Surg Treat Res. 86:143–151. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Siegel RL, Miller KD, Goding Sauer A,
Fedewa SA, Butterly LF, Anderson JC, Cercek A, Smith RA and Jemal
A: Colorectal cancer statistics, 2020. CA Cancer J Clin.
70:145–164. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Luo L, McGarvey P, Madhavan S, Kumar R,
Gusev Y and Upadhyay G: Distinct lymphocyte antigens 6 (Ly6) family
members Ly6D, Ly6E, Ly6K and Ly6H drive tumorigenesis and clinical
outcome. Oncotarget. 7:11165–11193. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Loughner CL, Bruford EA, McAndrews MS,
Delp EE, Swamynathan S and Swamynathan SK: Organization, evolution
and functions of the human and mouse Ly6/uPAR family genes. Human
Genomics. 10:102016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Upadhyay G: Emerging role of lymphocyte
antigen-6 family of genes in cancer and immune cells. Front
Immunol. 10:8192019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Davies A, Simmons DL, Hale G, Harrison RA,
Tighe H, Lachmann PJ and Waldmann H: CD59, an LY-6-like protein
expressed in human lymphoid cells, regulates the action of the
complement membrane attack complex on homologous cells. J Exp Med.
170:637–654. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Eisen A, Schulzer M, MacNeil M, Pant B and
Mak E: Duration of amyotrophic lateral sclerosis is age dependent.
Muscle Nerve. 16:27–32. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Malek TR, Fleming TJ and Codias EK:
Regulation of T lymphocyte function by glycosylphosphatidylinositol
(GPI)-anchored proteins. Semin Immunol. 6:105–113. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Naylor TL, Greshock J, Wang Y, Colligon T,
Yu QC, Clemmer V, Zaks TZ and Weber BL: High resolution genomic
analysis of sporadic breast cancer using array-based comparative
genomic hybridization. Breast Cancer Res. 7:R1186–R1198. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Grisanzio C and Freedman ML: Chromosome
8q24-associated cancers and MYC. Genes Cancer. 1:555–559. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Russ E, Bhuvaneshwar K, Wang G, Jin B,
Gage MM, Madhavan S, Gusev Y and Upadhyay G: High mRNA expression
of LY6 gene family is associated with overall survival outcome in
pancreatic ductal adenocarcinoma. Oncotarget. 12:145–159. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yeom CJ, Zeng L, Goto Y, Morinibu A, Zhu
Y, Shinomiya K, Kobayashi M, Itasaka S, Yoshimura M, Hur CG, et al:
LY6E: A conductor of malignant tumor growth through modulation of
the PTEN/PI3K/Akt/HIF-1 axis. Oncotarget. 7:65837–65848. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Classon BJ and Coverdale L: Mouse stem
cell antigen Sca-2 is a member of the Ly-6 family of cell surface
proteins. Proc Natl Acad Sci USA. 91:5296–5300. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Antica M, Wu L and Scollay R: Stem cell
antigen 2 expression in adult and developing mice. Immunol Lett.
55:47–51. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
AlHossiny M, Luo L, Frazier WR, Steiner N,
Gusev Y, Kallakury B, Glasgow E, Creswell K, Madhavan S, Kumar R
and Upadhyay G: Ly6E/K signaling to TGFβ promotes breast cancer
progression, immune escape, and drug resistance. Cancer Res.
76:3376–3386. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lv Y, Song Y, Ni C, Wang S, Chen Z, Shi X,
Jiang Q, Cao C and Zuo Y: Overexpression of lymphocyte antigen 6
complex, locus E in gastric cancer promotes cancer cell growth and
metastasis. Cell Physiol Biochem. 45:1219–1229. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yin Y, Chen X and Shu Y: Gene expression
of the invasive phenotype of TNF-alpha-treated MCF-7 cells. Biomed
Pharmacother. 63:421–428. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Derynck R, Muthusamy BP and Saeteurn KY:
Signaling pathway cooperation in TGF-β-induced
epithelial-mesenchymal transition. Curr Opin Cell Biol. 31:56–66.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Asundi J, Crocker L, Tremayne J, Chang P,
Sakanaka C, Tanguay J, Spencer S, Chalasani S, Luis E, Gascoigne K,
et al: An antibody-drug conjugate directed against lymphocyte
antigen 6 complex, locus E (LY6E) provides robust tumor killing in
a wide range of solid tumor malignancies. Clin Cancer Res.
21:3252–3262. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Derynck R, Akhurst RJ and Balmain A:
TGF-beta signaling in tumor suppression and cancer progression. Nat
Genet. 29:117–129. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lamouille S, Xu J and Derynck R: Molecular
mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell
Biol. 15:178–196. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Massagué J: TGFβ signalling in context.
Nat Rev Mol Cell Biol. 13:616–630. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Brunen D, Willems SM, Kellner U, Midgley
R, Simon I and Bernards R: TGF-β: An emerging player in drug
resistance. Cell Cycle. 12:2960–2968. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yoo YA, Kim YH, Kim JS and Seo JH: The
functional implications of Akt activity and TGF-beta signaling in
tamoxifen-resistant breast cancer. Biochim Biophys Acta.
1783:438–447. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gupta GP and Massagué J: Cancer
metastasis: Building a framework. Cell. 127:679–695. 2006.
View Article : Google Scholar : PubMed/NCBI
|