Introduction
Vitamin D is an essential nutrient for the human
body and is not only crucial for regulation of calcium metabolism
but also serves an important role in homeostasis (1–4).
1,25-dihydroxyvitamin D (1α,25(OH)2D3), also
known as calcitriol, is the active form of vitamin D. Previous
studies have reported that 1α,25(OH)2D3 is a
ligand of nuclear vitamin D receptor (VDR), that contributes to
numerous processes in the body, including cell proliferation,
differentiation and cell viability (5–7).
1α,25(OH)2D3 can act protectively against
cancer by promoting apoptosis (8),
and the relationships between vitamin D deficiency and numerous
types of cancer, such as colorectal cancer and prostate cancer have
been reported in previous studies (9–11).
Furthermore, it has been previously shown that supplementation of
vitamin D suppresses carcinogenesis in numerous organs, such as
breast, prostate, colorectal, and head and neck cancer (12). However, the underlying mechanism
linking tumorigenicity and cellular vitamin D status remains
unknown.
The bioavailability of vitamin D is regulated by a
coordinated balance between 1α,25(OH)2D3
biosynthesis and catabolism, and causally determines cellular
responses to vitamin D (1–3). The vitamin D metabolizing enzyme
cytochrome P450 family 24 subtype A1 (CYP24A1) contributes to the
inactivation of 1α,25(OH)2D3 by converting it
to rapidly excreted derivatives (3,5). This
enzymatic activity restricts the access of
1α,25(OH)2D3 to the transcriptional machinery
and limits vitamin D signaling within cells (5). It has been previously reported that
CYP24A1 expression is elevated in certain types of tumor cells,
such as breast, prostate, colorectal, and head and neck cancers and
that numerous types of cancer cells contain inactive vitamin D
metabolites such as 1α,24,25-(OH)3D3 and
24-oxo-1α,25-(OH)2 D3 (13,14).
Previous studies reported that CYP24A1 has an
oncogenic activity in breast cancer (15); however, the clinical relevance of
vitamin D depletion induced by CYP24A1 in breast cancer remains to
be clarified. In the present study, the expression of CYP24A1 in
surgically resected breast tumor specimens and the effect of
CYP24A1 expression on carcinogenesis in breast carcinoma cells was
evaluated.
Materials and methods
Patients and specimens
Tissue specimens from 136 cases of breast cancer
collected from Sapporo Medical University Hospital (Sapporo, Japan)
during surgical resection from 2011–2014 were used in the present
study. Data were also collected from the pathology file of Sapporo
Medical University Hospital. The mean age of the patients was 59.3
years (range, 26–92 years). Histological type was based on the
World Health Organization (WHO) classification of tumors of the
breast (5th edition) (16). For
intrinsic subtype classification, surrogate molecular breast cancer
classification based on immunohistochemical assessment of the
estrogen receptor (ER), progesterone receptor (PR), human epidermal
growth factor receptor 2 (HER2) and Ki-67 biomarkers was used
according to the WHO classification of tumors (5th edition)
(16). All of the 136 cases were
staged according to the Union for International Cancer Control
stage classification (7th edition) in the WHO classification of
tumors (5th edition) (Table I)
(16). In the staging process of
breast tumors, we categorized tumors using parameters such as pT
factor, which is defined as pathological status of primary tumor,
and pN factor, which is defined as pathological status of lymph
node involvement (16). The present
study was approved by the Ethics Committee (approval no. 4-1-44)
and Institutional Review Board (study no. 312-230) of Sapporo
Medical University (Sapporo, Japan). The Ethics Committee waived
the requirement to obtain written informed consent from the
patients for the use of human tissues owing to the retrospective
nature of the study. The research was performed in accordance with
the Declaration of Helsinki. The researchers involved in this study
had no access to information that could identify individual
participants during or after data collection.
 | Table I.Association between CYP24A1
expression examined by immunohistochemistry and certain
clinicopathological parameters. |
Table I.
Association between CYP24A1
expression examined by immunohistochemistry and certain
clinicopathological parameters.
|
|
| CYP24A1 |
|
|---|
|
|
|
|
|
|---|
| Parameter | Total number of
cases | Positive | Negative | P-value |
|---|
| Histology |
|
|
| 0.0067 |
|
DCIS | 29 | 17 | 12 |
|
|
IDC | 103 | 86 | 17 |
|
|
ILC | 3 | 3 | 0 |
|
|
Paget | 1 | 0 | 1 |
|
| Primary tumor
status |
|
|
| 0.1070 |
|
pT1a | 4 | 3 | 1 |
|
|
pT1b | 13 | 9 | 4 |
|
|
pT1c | 49 | 41 | 8 |
|
|
pT1mi | 4 | 3 | 1 |
|
|
pT2 | 39 | 34 | 5 |
|
|
pT3 | 6 | 4 | 2 |
|
|
pT4b | 1 | 1 | 0 |
|
|
pTis | 20 | 11 | 9 |
|
| Lymph node
involvement |
|
|
| 0.3000 |
|
pN0 | 100 | 76 | 24 |
|
|
pN1a | 23 | 19 | 4 |
|
|
pN1c | 2 | 2 | 0 |
|
|
pN1mi | 2 | 2 | 0 |
|
|
pN2a | 6 | 6 | 0 |
|
|
pN3a | 3 | 1 | 2 |
|
| Stage |
|
|
| 0.0294 |
| 0 | 20 | 11 | 9 |
|
| IA | 52 | 40 | 12 |
|
| IB | 1 | 1 | 0 |
|
|
IIA | 40 | 35 | 5 |
|
|
IIB | 14 | 12 | 2 |
|
|
IIIA | 6 | 6 | 0 |
|
|
IIIC | 3 | 1 | 2 |
|
| Subtype |
|
|
| 0.0047 |
| Luminal
A | 46 | 30 | 16 |
|
| Luminal
B | 68 | 57 | 11 |
|
|
HER2 | 7 | 4 | 3 |
|
| Basal
type | 15 | 15 | 0 |
|
| Age |
|
|
| 0.4320 |
|
20-39 | 7 | 6 | 1 |
|
|
40-59 | 56 | 47 | 9 |
|
|
60-79 | 66 | 48 | 18 |
|
|
80-99 | 7 | 5 | 2 |
|
Immunohistochemical staining
Tissue sections were fixed with 10%-buffered
formalin overnight at room temperature. Fixed tissues were embedded
in paraffin and embedded sections were cut at 5 µm thickness.
Tissue sections were then deparaffinized in xylene, rehydrated
using a decreasing ethanol series and incubated in 3%
H2O2 for 10 min to block endogenous
peroxidase activity. After antigen retrieval by microwave heating
(95°C for 30 min) in 10 mmol/l Tris and 1 mmol/l EDTA buffer, the
sections were incubated overnight at 4°C with primary monoclonal
antibodies against CYP24A1 (1:100; cat. no. sc-365700; Santa Cruz
Biotechnology, Inc.). The sections were then incubated with
EnVision (Dako; Agilent Technologies, Inc.) for 30 min at room
temperature, and 3,3′-diaminobenzidine tetrahydrochloride (Dako;
Agilent Technologies, Inc.) was added as the chromogen. Hematoxylin
was used for counterstaining at room temperature for 3 min.
Analysis of immunohistochemical staining positivity was performed
using a light microscope, based on the staining intensity and the
percentage of positive cells. The intensity scores of staining were
set as follows: 3+, strong; 2+, moderate; 1+, weak; and 0,
negative. The observers were blinded to the clinical data during
the evaluation. Consensus was reached by discussion of discordant
cases.
Cell culture and transfection
The ER positive, breast cancer MCF7 cell line, was
purchased from a local distributor (Summit Pharmaceuticals
International Corporation) of the American Type Culture Collection.
Cells were maintained in Dulbecco's modified Eagle's medium (DMEM,
MilliporeSigma) supplemented with 10% fetal bovine serum (FBS;
Invitrogen; Thermo Fisher Scientific, Inc.) and 5% streptomycin
(MilliporeSigma). The cells were transfected with different types
of CYP24A1-specific small-hairpin RNA (shRNA)-expressing lentivirus
plasmids according to manufacturer's instruction provided
(MISSION® shRNA Plasmid DNA; MilliporeSigma).
Transfection was performed with 5 µg shRNA plasmid, using FuGENE6
(Roche Diagnostics) to generate stable transfectants. Cells were
incubated with plasmid for 48 h at 37°C in a humidified 5%
CO2 atmosphere. The shRNAs used were as follows: CYP24A1
shRNA #1296 (shRNA clone ID: NM_ 000782.2-1296s1c1, MilliporeSigma)
with the plasmid sequence,
5′-CCGGGCAGATTTCCTTTGTGACATTCTCGAGAATGTCACAAAGGAAATCTGCTTTTTG-3′
and CYP24A1 shRNA #1016 (shRNA clone ID: NM_000782.2-1016s1c1,
MilliporeSigma) with the plasmid sequence,
5′-CCGGCGAACTGAACAAATGGTCGTTCTCGAGAACGACCATTTGTTCAGTTCGTTTTTG-3′.
Tran-sfected clones were selected using 1.5 µg/ml puromycin
(MilliporeSigma). Drug-resistant clones were selected after ≥14
days of selection and screened for CYP24A1 expression to measure
their CYP24A1 RNA expressions using reverse transcription (RT)-PCR
analysis. Following screening, the MCF7 cell transfectants, CYP24A1
shRNA #1 and CYP24A1 shRNA #7 were used in subsequent experiments.
We have previously reported that the process of transfection with
CYP24A1 scramble shRNA did not affect the cell phenotype (15). Therefore, wild-type MCF7 cells were
used as the control in the present study.
Semi-quantitative RT-PCR analysis
Total RNA of wild-type MCF7 cells and their
transfectants was isolated using TRIzol® reagent
(Invitrogen; Thermo Fisher Scientific, Inc.), and subsequent RT-PCR
was performed using the Superscript II Reverse Transcriptase kit
(Invitrogen; Thermo Fisher Scientific, Inc.) according to
manufacturer's protocols. Samples were incubated at 42°C for 50 min
followed by incubation at 70°C for 15 min. The complementary DNA
was then mixed with the primers as follows: CYP24A1 forward (F),
5′-GCAGCCTAGTGCAGATTTCC-3′ and reverse (R),
5′-ACCAGGGTGCCTGAGTGTAG-3′; and GAPDH F,
5′-GTCTCCTCTGACTTCAACAGCG-3′ and R, 5′-ACCACCCTGTTGCTGTAGCCAA-3′,
as well as 0.5 U of Taq DNA polymerase (Takara Bio, Inc.) to
amplify the genes of interest. The thermocycling conditions used
were as follows: 40 cycles at 96°C for 30 sec, 30 sec at 55°C, and
1 min at 72°C, followed by a final elongation stage at 72°C for 7
min. RNA expression was examined by loading on 1.5% agarose gels
and visualized by ethidium bromide staining. As a loading control,
50 ng of 200 bp DNA ladder (Takara Bio, Inc.) was used. Finally,
RNA expression levels were semi-quantified using ImageJ software
(version 1.52; National Institutes of Health) and normalization to
GAPDH expression.
Immunofluorescent assay
Cells were seeded in 35 mm dishes (10,000
cells/dish) containing 15 mm glass coverslips (AGC Techno Glass
Co., Ltd.) and incubated with DMEM containing 10% FBS (Invitrogen;
Thermo Fisher Scientific, Inc.). Glass coverslips were pre-coated
with 1:1 rat tail collagen overnight at room temperature
(Invitrogen; Thermo Fisher Scientific, Inc). The cells on the
coverslips were fixed at 20°C for 10 min with a fixing solution
(acetone:ethanol, 1:1). The cells were incubated with a primary
monoclonal anti-CYP24A1 antibody (1:100, cat. no. sc-365700, Santa
Cruz Biotechnology, Inc.) at 4°C overnight, and then treated with
Alexa Fluor 488 (green)-conjugated anti-rabbit IgG (1:200, cat. no.
A-11008, Invitrogen; Thermo Fisher Scientific, Inc.) for 1 h at
room temperature. The nuclei in the cells were counterstained using
4′,6-diamidino-2-phenylindole at room temperature for 5 min
(MilliporeSigma). The samples were imaged using an epifluorescence
microscope (Olympus Corporation).
Treatment of cells
To evaluate cell viability, cells were seeded in
12-well dishes (40,000 cells/well) and the cells were counted by
manual cell counting using trypan blue dye exclusion test (cells
were stained with trypan blue at room temperature for 1 min) in a
time and dose-dependent manner (dependent on the treatment
conditions) using a light microscope (Olympus). To assess cell
viability under different levels of cell stress, cells were treated
with H2O2 (0, 25, 50 or 75 µM) for 4, 8 and
12 h to induce oxidative stress at 37°C in a humidified, 5%
CO2 atmosphere. For the assessment of cell proliferation
with and without treatment with vitamin D (1 µM, MilliporeSigma)
and/or ketoconazole (2 µM, MilliporeSigma), cells were seeded in
12-well dishes (1,000 cells/well). The cells were treated with 1 µM
vitamin D and 2 µM ketoconazole and incubated for 48 h at 37°C in a
humidified, 5% CO2 atmosphere.
For the assessment of drug sensitivity, cells were
seeded in 96-well plates (5,000 cells/well) and treated with
cisplatin (0–100 nM, Adipogen AG) and gefitinib (0–100 nM; Cayman
Chemical Company). The viability of cells treated with cisplatin
was assessed every 24 h until 96 h of incubation and the viability
of cells treated with gefitinib was assessed after 48 h. Cell
viability was analyzed using a Cell Counting Kit-8 assay kit
(Dojindo Laboratories, Inc.) according to the manufacturer's
protocols. Absorbance at a wavelength of 450 nm was quantified
using a Varioskan™ LUX microplate reader (Thermo Fisher Scientific,
Inc.).
Immunocytochemistry of cell
blocks
Cells were treated with H2O2
(0, 100 or 750 µM) for 24 h at 37°C in a humidified, 5%
CO2 atmosphere to induce oxidative stress. Cells were
harvested from culture dishes using a cell lifter and then
collected by centrifugation at 300 × g for 3 min at room
temperature. The collected cells were solidified using 10% agarose
gel and fixed in 10% buffered formalin overnight at 4°C. These
tissues were paraffin-embedded and the tissue sections were cut at
5 µm thickness. Immunostaining was performed using primary
antibodies against cleaved caspase-3 (1:50, cat. no. 9664; Cell
Signaling Technology, Inc.) and Ki-67 (MIB-1 clone; 1:200, cat. no.
AM297-5M, BioGenex Laboratories) at room temperature for 1 h. The
sections were then incubated with EnVision (1:1, cat. no. K400311,
Dako; Agilent Technologies, Inc.) at room temperature for 30 min.
After washing with PBS, 3,3′-diaminobenzidine tetrahydrochloride
(1:1, cat. no. GE001, Dako; Agilent Technologies, Inc.) was added
as the chromogen at room temperature for 5 min.
Colony formation
Cells were seeded in 12-well plates (2,500
cells/well). After incubation of cells for 7 days at 37°C in a
humidified 5% CO2 atmosphere, the cells were fixed using
10% buffered formalin for 15 min at room temperature and stained
using 0.04% crystal violet for 15 min at room temperature. Cell
clusters >50 µm in diameter were defined as positive; colonies
were counted using phase-contrast microscopy (Olympus Corporation)
under low magnification (×100) and were quantified using ImageJ
software (version 1.52; National Institutes of Health).
Statistical analysis
At least 3 independent experiments were performed
for each analysis and all data were presented as mean ± standard
deviations. All data from each experiment were analyzed with either
unpaired Student's t-test, Fisher's exact test or the
Kruskal-Wallis test to determine significance. Bonferroni's
post-hoc test was used where appropriate. Survival curves were
constructed, and the Kaplan-Meier method and log-rank test were
used to calculate the survival rate. R (version 4.0.3; RStudio,
Inc.) was used for all statistical analyses. P<0.05 was
considered to indicate a statistically significant difference.
Results
CYP24A1 is expressed in primary breast
neoplasia
Previous studies reported that CYP24A1 is highly
expressed in different types of cancer (13–15).
In the present study, the correlations between CYP24A1 expression
and the clinicopathological parameters of breast cancer were
evaluated using immunohistochemistry (Table I and Fig. 1A-C). CYP24A1 was strongly expressed
but its expression area was limited in normal ductal and acinar
cells (data not shown). In non-invasive breast carcinoma, samples
were positive for CYP24A1 expression 58.6% (17/29) of the cases.
Consistent with previous reports, the CYP24A1-positive rates were
83.5% (86/103) and 100% (3/3) in invasive ductal carcinoma and
invasive lobular carcinoma, respectively (P=0.0067, Fisher's exact
test). No significant association of CYP24A1 expression with
primary tumor status and lymph node involvement was demonstrated;
however, tumor stage was significantly positively associated with a
high expression level of CYP24A1 (P=0.0294, Kruskal-Wallis
test).
Based on immunohistochemical assessment of the
biomarkers ER, PR, HER2 and Ki-67, breast tumors can be classified
into four major immunohistochemically surrogate intrinsic subtypes
as follows: Luminal A (ER+, PR+/-, HER2-), Luminal B (ER+, PR+/-,
HER2-, higher Ki-67 expression), HER2 (ER-, PR-, HER2+) and basal
(ER-, PR-, HER2-) (16). Although
some are overlapping, the prognosis of breast cancer patients has
been reported to become worse in the order of luminal A, luminal B,
HER2-overespressed type, and basal subtype (15–19).
In the present study, intrinsic surrogate subtype was associated
with CYP24A1 expression, with the expression of CYP24A1 being
significantly higher in subtypes with poor prognosis. Indeed, a
significant increase in the expression of CYP24A1 was noted in
hormone receptor negative cancer (P=0.0047, Kruskal-Wallis
test).
The cases were divided into two groups based on the
expression of CYP24A1 assessed by staining intensity and area.
Specimens containing >50% area with staining intensity 3+ were
defined as positive, and specimens containing ≤50% area with
staining intensity 3+ were defined as negative. Kaplan-Meier
survival curves demonstrated that the overall survival rate in the
CYP24A1-positive group was markedly lower than that in the CYP24A
negative group when compared in whole specimens (Fig. 1D). However, a significant
association between the CYP24A1 positivity and overall survival
rate was demonstrated in invasive breast carcinoma (P=0.0266;
Fig. 1E). These results were
consistent with results of previous studies which suggested a
possible oncogenic effect of CYP24A1 in the growth of a breast
neoplasm (13–15).
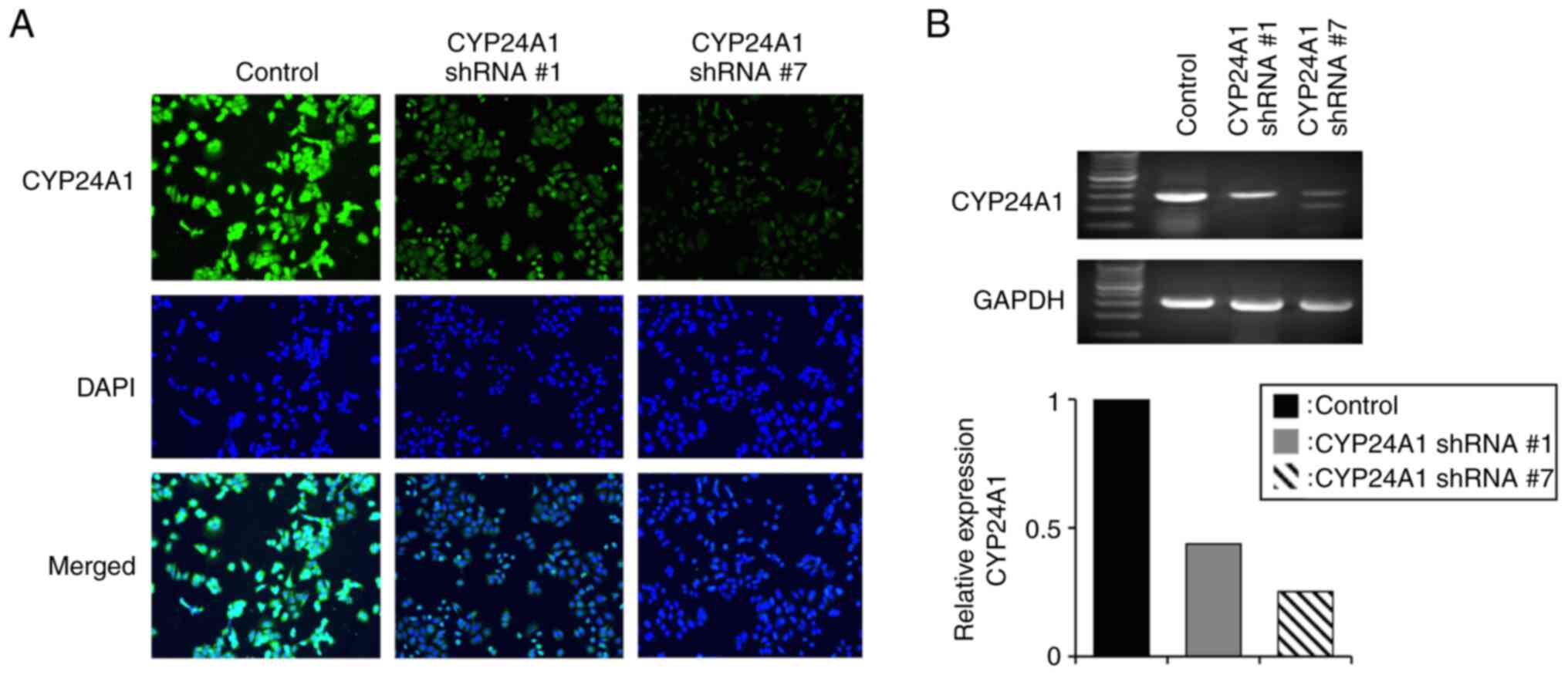
Establishment of CYP24A1 knockdown
cells
For the establishment of MCF7 cells with suppressed
CYP24A1 expression, cells were transfected with shRNAs against
CYP24A1 with two different sequences and the cell lines were
denoted as CYP24A1 shRNA #1 and CYP24A1 shRNA #7. An
immunofluorescent assay demonstrated that the expression of CYP24A1
protein was markedly suppressed in both cell lines (Fig. 2A). RT-PCR analysis demonstrated that
CYP24A1 RNA expression levels were markedly reduced in CYP24A1
shRNA #7 (~75%) and CYP24A1 shRNA #1 cells (~50%) (Fig. 2B).
Effect of CYP24A1 suppression on cell
viability
To evaluate the effect of CYP24A1 knockdown on cell
viability, MCF7 cells were cultured for 12 h with and without
oxidative stress, induced using H2O2
(Fig. 3A). In the absence of
H2O2, no difference in cell viability was
demonstrated among all cell groups. The viability of CYP24A1 shRNA
#7 cells cultured with H2O2 significantly
decreased in a dose and time-dependent manner (Fig. 3B and C). However, there was no
significant difference in the viability of CYP24A1 shRNA #1 cells
cultured with and without H2O2.
Cells were treated with vitamin D and ketoconazole,
a broad-spectrum inhibitor of CYP24A1, and a manual cell count was
performed after 48 h. Although ER-positive cells such as MCF7 cells
are known to express higher levels of VDR than the levels in
ER-negative cells (1,16), vitamin D only demonstrated a marked
decreased in the viability of CYP24A1 shRNA #7 cells. There was no
marked difference in cell viability when ketoconazole was added
(Fig. 3D).
Effect of CYP24A1 suppression on
apoptosis
Cell blocks from cultured cells were established to
assess cell death sensitivity under cell stress conditions and the
number of apoptotic bodies were manually counted (Fig. 4A). Apoptosis was markedly increased
in CYP24A1 knockdown cells. The number of apoptotic cells markedly
increased in cells with suppression of CYP24A1 expression when a
moderate level of oxidative stress (100 µM
H2O2) was added. In controls, the number of
apoptotic cells only increased with a higher level of oxidative
stress (750 µM H2O2) (Fig. 4B). These results suggested that
cells with suppression of CYP24A1 expression had a higher cell
death sensitivity to a cell stressor.
Immunohistochemistry was performed using an antibody
against cleaved caspase-3 to evaluate the effects of altered
CYP24A1 expression on apoptosis (Fig.
4C). The proportion of cleaved caspase-3-positive cells was
significantly increased in CYP24A1 knockdown cells treated with
H2O2 (Fig.
4D).
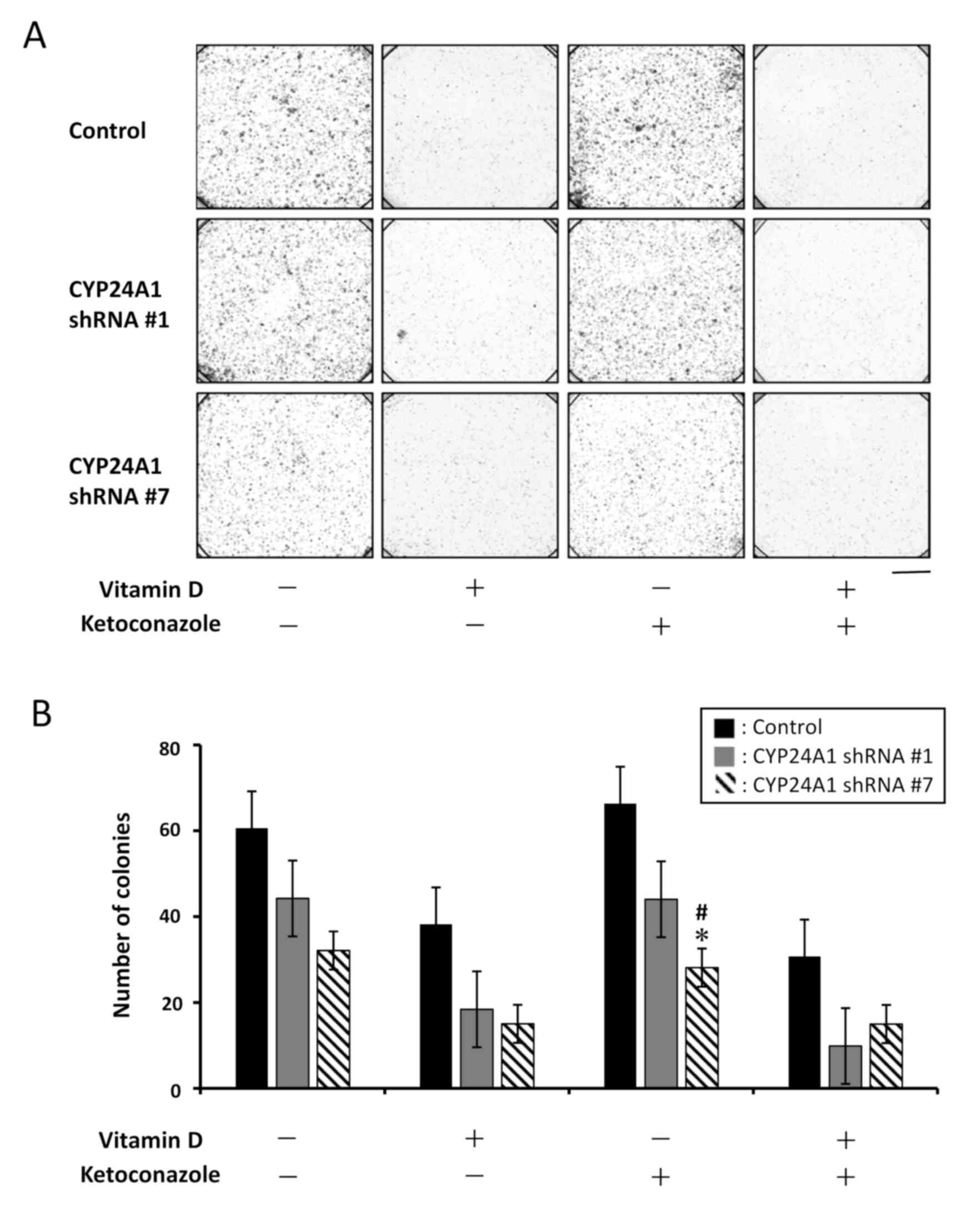
Effect of CYP24A1 suppression on
colony-forming efficacy
To evaluate the effect of CYP24A1 knockdown on
two-dimensional tumorigenicity with and without vitamin D and
ketoconazole treatment, a colony formation assay was performed
(Fig. 5). Compared with the
colony-forming ability of the control cells without treatment, this
ability was markedly suppressed in both CYP24A1 shRNA #1 and shRNA
#7 cells when untreated. Furthermore, colony formation efficacy was
suppressed more in CYP24A1 shRNA #7 cells compared with that by
CYP24A1 shRNA #1 cells. In the presence of vitamin D, the area of
the colonies was markedly decreased in all cell groups. The results
suggested that cellular vitamin D status effected colony formation
efficacy. However, ketoconazole did not affect colony formation
compared with the untreated groups.
Effect of CYP24A1 suppression on cell
death sensitivity to anticancer drugs
To evaluate cell sensitivity to anticancer drugs
with different pharmacological mechanisms (cisplatin and
gefitinib), cells were cultured with each drug and cell viability
was analyzed (Fig. 6). Reduced
expression of CYP24A1 significantly enhanced cell death sensitivity
to both cisplatin and gefitinib, compared with the control.
Discussion
The present study demonstrated for the first time
that increased expression of CYP24A1 leads to a decrease in the
overall survival of patients with invasive primary breast
carcinoma. Furthermore, it was demonstrated that suppression of
CYP24A1 expression inhibited the oncogenic activity of breast
carcinoma cells and enhanced cell sensitivity to anticancer drugs
with different pharmacologic activities. These results suggested
CYP24A1 as a possible therapeutic target in CYP24A1-expressing
breast malignancy.
The prognosis and treatment response differ among
the intrinsic surrogate subtypes of breast cancer, with the basal
subtype having the worst prognosis and Luminal A subtype having the
best prognosis (17–20). Previous studies reported that ER+
breast cancer cell lines were more sensitive to the effects of
calcitriol. In the present study, the protein expression level of
CYP24A1 was higher in the intrinsic subtypes reported to be
associated with a poor prognosis, and particularly in the basal
subtype (Table I), which suggested
that CYP24A1 was a possible prognostic marker in breast cancer.
This hypothesis was supported by the overall survival rate of
patients with a strong expression of CYP24A1 in invasive ductal
carcinoma, which was significantly lower compared with that in
patients with only moderate or no expression of CYP24A1 (Fig. 1).
Previous studies have reported that
1α,25(OH)2D3 serves as a ligand for VDR.
Vitamin D suppresses carcinogenesis and serves an important role in
tissue homeostasis by the regulation of the expression of genes
affecting cell proliferation, differentiation and apoptosis
(15,21–23).
1α,25(OH)2D3 serves an important role in the
promotion of apoptosis by the regulation of calcium signaling
through calcium channels linked to the membrane VDR (1–3,23). In
the process of apoptosis, the concentration of intracellular
calcium increases and interacts with molecular calcium-dependent
targets within cells, including calcium-activated apoptotic
effectors (1–3,15,20).
CYP24A1 converts 1α,25(OH)2D3 into rapidly
excreted inactive derivatives and restricts the access of
1α,25(OH)2D3 to the transcriptional
machinery, which limits vitamin D signaling within cells (15,23).
These reports suggest that CYP24A1 has as a desensitizing effect on
apoptosis-inducing factors through calcium signaling mediated
apoptotic inducers.
The present study demonstrated that suppression of
CYP24A1 expression significantly increased apoptosis in breast
tumor cells under different types of cell stressors such as
oxidative stress mediated by H2O2 and
chemotherapeutic drugs. The results of the present study
demonstrated that supplementation of vitamin D markedly decreased
cell viability and colony-forming efficacy (24); however, the effect was not
statistically significant. Furthermore, the addition of
ketoconazole did not affect the viability and colony-forming
efficacy of MCF7 cells (25,26),
although the suppression of CYP24A1 expression itself markedly
decreased these effects (Fig. 5).
These results suggested that CYP24A1 has an as-yet-unrecognized
activity independent of vitamin D metabolism. It has been
previously reported that CYP24A1 expression is elevated in various
types of tumors and correlates with poor prognosis (13–15).
Therefore, a CYP24A1-specific inhibitor might be able to
effectively inhibit the tumorigenicity of CYP24A1-expressing breast
malignancy.
The results of the present study demonstrated that
suppression of CYP24A1 expression in breast cancer cells increased
cell sensitivity to two anticancer drugs with different
pharmacological mechanisms. The first anticancer drug used was
cisplatin, a chemotherapeutic drug that induces cell apoptosis in
cancer cells by crosslinking with the purine bases on DNA which
disrupts the DNA repair mechanism (27). The second anticancer drug was
gefitinib, which is a tyrosine-kinase inhibitor used for of the
treatment of numerous types of cancers including HER2-positive
breast cancer (28). The results of
the present study indicated that CYP24A1 enhanced cell death
activity in response to cell death inducers with different
mechanisms of action. Therefore, inhibition of CYP24A1 activity
could be a possible therapeutic approach in breast malignancy.
A limitation of the present study is that animal
experiments were not included and that only one type of breast
cancer cell line (MCF7) was used. In our preliminary study, animal
experiments were performed; however, the effects of CYP24A1
suppression on tumor growth were not statistically significant
(data not shown). Furthermore, to evaluate the role of CYP24A1 in
other cell lines with different expression levels of ER, PR and
HER2, our preliminary study attempted to establish CYP24A1
knockdown cells with the T47D (Luminal A), ZR-75 (Luminal B) and
SK-BR-3 (HER2) cell lines; however, none of the cells survived when
CYP24A1 was knocked down using two different shRNA constructs. The
cell line that was used in the present study, MCF7, is a good
candidate for the evaluation of the effect of vitamin D on breast
cancer cells as vitamin D deficiency is known to be associated with
poor outcomes in patients with luminal-type breast cancer such as
MCF7 cells (29). Furthermore,
ER-positive cells have been reported to express higher levels of
VDR compared with the levels in ER-negative cells (16,22)
and the results of previous studies also showed that dietary intake
of vitamin D reduces the risk of ER-positive breast cancer
(30–32). Together with the results of our
previously published study (15),
it can be suggested that CYP24A1 is indispensable for the survival
of those breast cancer cell lines. If CYP24A1 has the same effect
on those breast tumor cells with different hormone receptor status
as it has on MCF7 cells, CYP24A1 inhibiting therapy might have an
even greater impact on those cells. Further studies using different
cell lines with various expression levels of ER, PR and HER2 are
needed.
The results of the present study demonstrated that
high level expression of CYP24A1 in invasive breast cancer led to a
significant decrease in the overall survival rate of patients with
breast carcinoma. Furthermore, it was demonstrated that suppression
of CYP24A1 expression in vitro decreased the tumorigenicity
of breast carcinoma cells and increased cell sensitivity to
differently acting anticancer drugs. In conclusion, the results of
the present study suggest that CYP24A1 is a possible therapeutic
target for breast malignancy with constitutive CYP24A1
expression.
Acknowledgements
Certain parts of this study were included in the
Japanese language PhD thesis of the author SK at Sapporo Medical
University School of Medicine.
Funding
No specific funding was received. This study was supported in
part by the education and research funds of Sapporo Medical
University School of Medicine.
Availability of data and materials
The datasets generated and/or analyzed during the
present study are available from the corresponding author on
reasonable request.
Authors' contributions
SK and MO substantially contributed the conception
and design of this study. SK and YN performed the cell culture
experiments and immunohistochemistry. AT, KT, DK, YO and KM
performed histological examination of the breast cancer. SK, YN and
MO confirm the authenticity of all the raw data obtained. SK and MO
were major contributors to data analysis and interpretation of the
data. SK and MO contributed to manuscript drafting and critical
revisions on the intellectual content. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
The present study was reviewed and approved by the
Institutional Ethics Committee (approval no. 4-1-44) and
Institutional Review Board (study no. 312-230) of Sapporo Medical
University. Specimens of 136 cases of breast cancer collected by
surgical resection from 2011–2014 were used in the present study.
The Ethics Committee waived the requirement to obtain written
informed consent from the patients for the use of human tissues
owing to the retrospective nature of the study. The research was
conducted in accordance with the Declaration of Helsinki. The
researchers involved in this study had no access to information
that could identify individual participants during or after data
collection.
Patient consent to participate
Not applicable.
Competing interests
The authors declare that they have no conflicts of
interests.
References
|
1
|
Bikle DD, Feingold KR, Anawalt B, Boyce A,
Chrousos G, de Herder WW, Dhatariya K, Dungan K, Hershman JM,
Hofland J, et al: Vitamin D: production, metabolism and mechanisms
of action. 2021, Wilson DP: South Dartmouth (MA): MDText.com;
|
|
2
|
Bikle DD: Vitamin D metabolism, mechanism
of action, and clinical applications. Chem Biol. 21:319–329. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Christakos S, Dhawan P, Verstuyf A,
Verlinden L and Carmeliet G: Vitamin D: Metabolism, molecular
mechanism of action, and pleiotropic effects. Physiol Rev.
96:365–408. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Alshahrani F and Aljohani N: Vitamin D:
Deficiency, sufficiency and toxicity. Nutrients. 5:3605–3616. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Saponaro F, Saba A and Zucchi R: An update
on vitamin D metabolism. Int J Mol Sci. 21:65732020. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
DeLuca HF: Vitamin D: Historical overview.
Vitam Horm. 100:1–20. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Trump DL and Aragon-Ching JB: Vitamin D in
prostate cancer. Asian J Androl. 20:244–252. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Carlberg C and Muñoz A: An update on
vitamin D signaling and cancer. Semin Cancer Biol. 79:217–230.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Song ZY, Yao Q, Zhuo Z, Ma Z and Chen G:
Circulating vitamin D level and mortality in prostate cancer
patients: A dose-response meta-analysis. Endocr Connect.
7:R294–R303. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Dou R, Ng K, Giovannucci EL, Manson JE,
Qian ZR and Ogino S: Vitamin D and colorectal cancer: Molecular,
epidemiological and clinical evidence. Br J Nutr. 115:1643–1660.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhou J, Ge X, Fan X, Wang J, Miao L and
Hang D: Associations of vitamin D status with colorectal cancer
risk and survival. Int J Cancer. 149:606–614. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Negri M, Gentile A, de Angelis C, Montò T,
Patalano R, Colao A, Pivonello R and Pivonello C: Vitamin D-induced
molecular mechanisms to potentiate cancer therapy and to reverse
drug-resistance in cancer cells. Nutrients. 12:17982020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mimori K, Tanaka Y, Yoshinaga K, Masuda T,
Yamashita K, Okamoto M, Inoue H and Mori M: Clinical significance
of the overexpression of the candidate oncogene CYP24 in esophageal
cancer. Ann Oncol. 15:236–241. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shiratsuchi H, Wang Z, Chen G, Ray P, Lin
J, Zhang Z, Zhao L, Beer D, Ray D and Ramnath N: Oncogenic
potential of CYP24A1 in lung adenocarcinoma. J Thorac Oncol.
12:269–280. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Osanai M and Lee GH: CYP24A1-induced
vitamin D insufficiency promotes breast cancer growth. Oncol Rep.
36:2755–2762. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
WHO Classification of Tumours Editorial
Board (ed), . Breast tumours. Lyon (France): International Agency
for Research on Cancer; 2019, (WHO classification of tumours
series, 5th edition, volume 2).
|
|
17
|
Dai X, Li T, Bai Z, Yang Y, Liu X, Zhan J
and Shi B: Breast cancer intrinsic subtype classification, clinical
use and future trends. Am J Cancer Res. 5:2929–2943.
2015.PubMed/NCBI
|
|
18
|
Ades F, Zardavas D, Bozovic-Spasojevic I,
Pugliano L, Fumagalli D, de Azambuja E, Viale G, Sotiriou C and
Piccart M: Luminal B breast cancer: Molecular characterization,
clinical management, and future perspectives. J Clin Oncol.
32:2794–2803. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yin L, Duan JJ, Bian XW and Yu SC:
Triple-negative breast cancer molecular subtyping and treatment
progress. Breast Cancer Res. 22:612020. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang X, Harbeck N, Jeschke U and
Doisneau-Sixou S: Influence of vitamin D signaling on hormone
receptor status and HER2 expression in breast cancer. J Cancer Res
Clin Oncol. 143:1107–1122. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Estébanez N, Gómez-Acebo I, Palazuelos C,
Llorca J and Dierssen-Sotos T: Vitamin D exposure and risk of
breast cancer: A meta-analysis. Sci Rep. 8:90392018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hossain S, Beydoun MA, Beydoun HA, Chen X,
Zonderman AB and Wood RJ: Vitamin D and breast cancer: A systematic
review and meta-analysis of observational studies. Clin Nutr ESPEN.
30:170–184. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Voutsadakis IA: Vitamin D receptor (VDR)
and metabolizing enzymes CYP27B1 and CYP24A1 in breast cancer. Mol
Biol Rep. 47:9821–9830. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Manson JE, Cook NR, Lee IM, Christen W,
Bassuk SS, Mora S, Gibson H, Gordon D, Copeland T, D'Agostino D, et
al: Vitamin D supplements and prevention of cancer and
cardiovascular disease. N Engl J Med. 380:33–44. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Park JW, Kim KA and Park JY: Effects of
ketoconazole, a CYP4F2 inhibitor, and CYP4F2*3 genetic polymorphism
on pharmacokinetics of vitamin K1. J Clin Pharmacol. 59:1453–1461.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Czyrski A, Resztak M, Świderski P, Brylak
J and Główka FK: The overview on the pharmacokinetic and
pharmacodynamic interactions of triazoles. Pharmaceutics.
13:19612021. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Dasari S and Tchounwou PB: Cisplatin in
cancer therapy: Molecular mechanisms of action. Eur J Pharmacol.
740:364–378. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Segovia-Mendoza M, González-González ME,
Barrera D, Díaz L and García-Becerra R: Efficacy and mechanism of
action of the tyrosine kinase inhibitors gefitinib, lapatinib and
neratinib in the treatment of HER2-positive breast cancer:
Preclinical and clinical evidence. Am J Cancer Res. 5:2531–2561.
2015.PubMed/NCBI
|
|
29
|
Kim HJ, Lee YM, Ko BS, Lee JW, Yu JH, Son
BH, Gong JY, Kim SB and Ahn SY: Vitamin D deficiency is correlated
with poor outcomes in patients with luminal-type breast cancer. Ann
Surg Oncol. 18:1830–1836. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Blackmore KM, Lesosky M, Barnett H, Raboud
JM, Vieth R and Knight JA: Vitamin D from dietary intake and
sunlight exposure and the risk of hormone-receptor-defined breast
cancer. Am J Epidemiol. 168:915–924. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
McCullough ML, Rodriguez C, Diver WR,
Feigelson HS, Stevens VL, Thun MJ and Calle EE: Dairy, calcium, and
vitamin D intake and postmenopausal breast cancer risk in the
cancer prevention study II nutrition cohort. Cancer Epidemiol
Biomark Prev. 14:2898–2904. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kawase T, Matsuo K, Suzuki T, Hirose K,
Hosono S, Watanabe M, Inagaki M, Iwata H, Tanaka H and Tajima K:
Association between vitamin D and calcium intake and breast cancer
risk according to menopausal status and receptor status in Japan.
Cancer Sci. 101:1234–1240. 2010. View Article : Google Scholar : PubMed/NCBI
|