Introduction
Bladder cancer (BCa) is the fourth most common
tumour in males, with a high rate of occurrence and recurrence
(1). Currently, the development of
urinary cytology, cystoscopy, laparoscopic surgery and immune
therapy has promoted the diagnosis and treatment of BCa patients.
However, problems such as recurrence, metastasis and drug
resistance still exist and need to be solved (2). Previous studies have suggested that
the progression of BCa closely correlates with the imbalance of
genes and signaling pathways (3).
Hence, there is a need to explore new therapeutic targets for BCa
to obtain improved prognosis.
Circular RNAs (circRNAs) are a kind of non-coding
RNAs with tissue-specific expression patterns. Compared with mRNAs,
the covalently closed structure of circRNAs contribute to the
stability and resistance to degradation (4). The development of sequencing
technology has improved our understanding of circRNAs, and a
growing number of circRNAs have been identified as important
molecules that were involved in in the pathological process of BCa
(5). For example, circRNA BCRC-3
was reported to inhibit BCa proliferation through miR-182-5p/p27
axis, while circLIFR could synergize with MSH2 and attenuate
chemoresistance in BCa (6,7). Moreover, circRNAs have been proven to
possess multiple functions: i) As sponges for miRNA (8); ii) Serving as protein baits or
antagonists (9); iii) Adjusting
alternative splicing (10); and iv)
Translated into polypeptides (11).
The Janus kinase (JAK)/signal transducer and
activator of transcription 3 (STAT3) signaling pathway is closely
related to the cancer progression. The intrinsic activation of
STAT3 has been demonstrated in numerous human solid malignancies
(12). After activation by JAKs,
p-STAT3 could promote the transcription of target genes after
translocating to the nucleus. Additionally, STAT3 is a central
intracellular node that integrates signals from EGFR,
RAS-RAF-mitogen activated protein kinase, C-MET, and TGF-β
pathways, thereby forming a complicate oncogenic signal network
(13). In fact, several studies
have found that STAT3 is extensively involved in the growth,
metastasis and chemotherapy resistance of BCa (14,15).
In the present study, bioinformatics analysis was
used to explore the expression of circRNAs in BCa tissues,
indicating that circRPPH1 was significantly upregulated in BCa
cells. Then, the potential function and mechanism of circRPPH1 in
BCa were further investigated through a series of experiments. The
results indicated that circRPPH1/STAT3 axis plays a key role in BCa
progression. Therefore, CircRPPH1 may be a therapeutic target of
BCa.
Materials and methods
Cell culture
Human BCa cell lines (T24 and 5637) were purchased
from Procell Life Science & Technology Co., Ltd. The human
normal urothelial cell line SV-HUC-1 and 293T cell line were
obtained from the Cell Bank of the Chinese Academy of Science
(Shanghai, China) and maintained in our laboratory. Both BCa cells
were cultured in RPMI-1640 medium (Gibco; Thermo Fisher Scientific,
Inc.). SV-HUC-1 cells were maintained in the F-12 K medium (Gibco;
Thermo Fisher Scientific, Inc.), while 293T cells were cultured in
DMEM (Gibco; Thermo Fisher Scientific, Inc.). The aforementioned
cell mediums were supplemented with 10% fetal bovine serum (Gibco;
Thermo Fisher Scientific, Inc.). All cells were incubated in a
humidified incubator at 37°C with 5% CO2.
Bioinformatics analysis
The differentially expressed circRNA data (GSE92675
and GSE97239) in BCa were downloaded from the GEO database
(https://www.ncbi.nlm.nih.gov/geo/)
(16,17), while hierarchical clustering
analysis was performed by Cluster3.0 software (http://bonsai.hgc.jp/~mdehoon/software/cluster/software.htm)
and visualized with Java TreeView software (http://jtreeview.sourceforge.net). To predict the
potential circRNA-miRNA and mRNA-miRNA interactions, circBank
(18), circinteractome (19), and TargetScan (20) were used according to the
corresponding instructions. Furthermore, RNAfold Web Server
(http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi),
RNACOmposer (http://rnacomposer.ibch.poznan.pl/), Protein Data Bank
(https://www.rcsb.org/), and HDOCK (http://hdock.phys.hust.edu.cn/) were used to
predict the binding between circRPPH1 and FUS protein.
Vector construction
The PCDH-H1 shRNA cloning vector was used to
construct circRPPH1 knockdown (shRNA#1, shRNA#2, shRNA#3) plasmids
and FUS knockdown plasmid. The construction of the PCDH-H1 plasmid
was previously described by the authors (21). Gene-specific shRNA target sequences
were synthesized by Tsingke Biological Technology. These shRNA
sequences all contain the sequences of restriction enzymes. Then,
these shRNA sequences were annealed and cloned into the PCDH-H1
plasmid. In the construction of overexpression plasmids, the
circRPPH1 sequence was cloned into the pLC5-circ vector (Guangzhou
Geneseed Biotech Co., Ltd.). Moreover, a second-generation
lentiviral system was applied for the production of lentiviruses. A
total of 24 µg plasmids were used for lentivirus packaging, and the
ratio of lentivirus plasmid: target plasmid: pHelper 1.0: pHelper
2.0 was 2:1:1. After ~48 h of transfection, the lentiviruses were
collected and used to infect BCa cells (MOI=5). After 72 h, BCa
cells were further cultured with fresh culture medium containing
puromycin (2 µg/ml) for 5 days to establish the stable interference
or overexpression systems in BCa cells. Both pHelper 1.0 plasmid
and pHelper 2.0 plasmid were obtained from GeneChem (Shanghai,
China).
The luciferase reporter assays were performed with
the psiCHECK2 vector (Shanghai GenePharma Co., Ltd.). All of the
wild-type (WT) and mutant (MUT) sequences were directly synthesized
by Tsingke Biotechnology (Beijing, China). Then, each sequence was
cloned into the polyclonal site region of the vector. Besides,
miR-296-5p mimic or inhibitor and their negative controls were
provided by Tsingke Biological Technology. The final working
concentration of miRNA inhibitor, miRNA mimic, or negative controls
was 50 nM. When the cell density reached 70%, the oligonucleotides
were transfected using Lipofectamine® 3000 (Thermo
Fisher Scientific, Inc.) according to the manufacturer's protocol.
Following 48 h of transfection at 37°C, the cells were used for
further detection. All oligonucleosides used in the present study
are provided in Table SI. All
plasmids were verified by sequencing.
Reverse transcription-quantitative
(RT-q) PCR
Total RNA was isolated with TRIzol Reagent (Sangon
Biotech Co., Ltd.), following the manufacturing protocol. For
circRNA and mRNA, the complementary DNA (cDNA) was generated using
the HiScript® III 1st Strand cDNA Synthesis kit (Vazyme
Biotech Co., Ltd.) according to the manufacturing protocol and the
miRNA First Strand cDNA Synthesis kit (Sangon Biotech Co., Ltd.)
was used to synthesize cDNA for miRNA. Next, quantification of RNA
was performed with a SYBR Green PCR kit (Shanghai Yeasen
Biotechnology Co., Ltd.) according to the manufacturing protocol.
The conditions for PCR amplification were as follows: 5 min at 95°C
for one cycle, followed by denaturation for 10 sec at 95°C and
extension for 30 sec at 60°C for 40 cycles. The 2−ΔΔCq
method was used to determine the fold change of expression
(22). All primers were designed by
ourselves and synthesized by Tsingke Biological Technology.
Notably, the design of the circRPPH1-divergent forward primer
required coverage of the back-splice point of circRPPH1. The
sequences of all primers were provided in Table SII. The circRPPH1, STAT3 or
miR-296-5p expression was normalized to GAPDH or U6, respectively.
Additionally, cytoplasmic and nuclear RNA was extracted by Thermo
Fisher BioReagents (Thermo Fisher Scientific, Inc.).
RNase R treatment and Actinomycin D
assay
To evaluate the stability of circRPPH1, RNase R
(Beyotime Institute of Biotechnology) and actinomycin D
(MedChemExpress) were used. Firstly, the 15 U RNase R was used to
treat total RNA (5 µg) at 37°C for 1 h. Then, the levels of
circRPPH1 and linear RPPH1 RNA was detected by RT-qPCR.
Additionally, cells were exposed to actinomycin D (10 µg/ml) for
different time intervals (0, 3, 6, 9 and 12 h). The circRPPH1
expression levels were also determined with RT-qPCR. Notably, PCR
products of circRPPH1 were applied to Sanger sequencing.
Fluorescence in situ hybridization
(FISH)
Using a FISH kit (cat. no. F12201/50; Shanghai
GenePharma Co., Ltd.), FISH assays were performed following the
manufacturer's protocol. In brief, tumor cells were seeded in
confocal petri dishes and cultured to 80% confluence. The dishes
were then washed twice with phosphate-buffered saline (PBS), fixed
with 200 µl 4% paraformaldehyde at room temperature for 15 min, and
permeabilized with 200 µl 0.1% Triton X for 15 min at room
temperature. After washing twice with PBS, 200 µl of 2X sodium
citrate buffer (SSC) solution was added to each dish at 37°C for 30
min. Then, the dishes were incubated in 200-µl denatured probe
mixture in a humidified incubator at 37°C overnight. The next day,
the dishes were washed with a solution of 0.1% Tween-20 in 4X SSC
for 5 min, a solution of 0.1% Tween-20 in 2X SSC for 5 min, and a
solution of 0.1% Tween-20 in 1X SSC for 5 min at 42°C in dark.
After that, DAPI working solution (1:10,000) was added into the
dishes for 15 min in dark. Finally, the dishes were washed twice
with PBS. The pictures were captured directly using an
immunofluorescence microscope. All probes were provided by Shanghai
GenePharma Co., Ltd.
Cell Counting Kit-8 (CCK-8) and colony
formation assay
The proliferation ability of BCa cells was assessed
with different experiments. Firstly, the CCK-8 assay was conducted
with T24 or 5637 cells seeded in a 96-well plate at a density of
2×103 cells per well. After cell attachment to the wall,
10 µl CCK-8 solution (Beyotime Institute of Biotechnology) was
added at different time points (0, 24, 48 and 72 h) and incubated
at 37°C for 1 h. Finally, absorbance was measured at 450 nm.
During the colony formation assay, 2×103
cells were seeded in six-well plates and incubated at 37°C for 14
days. The medium was replaced every 5 days. Then, the colonies
(>50 cells) were sequentially fixed with 100% methanol at room
temperature for 30 min and stained with 0.1% crystal violet at room
temperature for 15 min. The cells were manually counted under a
dissecting microscope and the colony formation efficiency was
calculated (number of colonies/number of cells inoculated
×100%).
Cell invasion and migration assay
Cell invasion or migration abilities were measured
using 8-µm pore size Transwell chambers (Corning, Inc.). For
invasion assays, upper chambers were pre-coated with Matrigel (1:8;
BD Biosciences) for 1 h at room temperature. The upper chamber was
inoculated with a cell suspension (3×105 cells/ml) and
cultured with serum-free medium, while the lower chamber was
supplemented with medium containing 10% FBS. The cells were
incubated at 37°C for 12 h. After 12 h of incubation, cells in the
lower chamber were fixed with 4% paraformaldehyde at room
temperature for 30 min and stained with 0.1% crystal violet at room
temperature for 15 min. Then, pictures were captured by a light
microscope (Olympus Corporation). The number of invasive cells were
counted in 5 randomly selected visual fields in each group.
Luciferase reporter assay
The WT or MUT luciferase reporter plasmids were
co-transfected into 293T cells along with miR-NC and miR-296-5p
mimic. Following transfection for 36 h, the cells were harvested
and lysed, then the supernatant was collected. Dual Luciferase
Reporter Gene Assay kit (Shanghai Yeasen Biotechnology Co., Ltd.)
was used to measure the dual Luciferase activity of each sample
with ~20 µl supernatant. The internal control was firefly
luciferase reporters co-expressed on psiCHECK2 plasmids.
RNA immunoprecipitation (RIP)
assay
An RNA Immunoprecipitation kit (cat. no. P0102;
Guangzhou Geneseed Biotech Co., Ltd.) was used to perform the RIP
assays. In accordance with the manufacturer's protocol, 100 µl
protein A/G beads were first conjugated to antibodies against FUS
(1:50), AGO2 (1:50) or IgG (1:50). After that, the cell extracts
were mixed with A/G protein beads. Finally, qPCR was used to
analyze the precipitated RNAs.
Western blotting,
co-immunoprecipitation (co-IP) and antibodies
The total protein was extracted using RIPA lysis
buffer (Sangon Biotech Co., Ltd.), while the nuclear protein was
extracted using a Nuclear Protein Extraction kit (Beijing Solarbio
Science & Technology Co., Ltd.). Protein concentrations were
measured using a BCA protein assay kit (Wuhan Servicebio Technology
Co., Ltd.). Total proteins (20 µg) were separated by 10% sodium
dodecyl-sulfate polyacrylamide gel electrophoresis. Then, the
proteins were transferred to nitrocellulose membranes and blocked
with 5% skim milk (Yili; http://oceaniadairy.co.nz/yili-group/) or 5% BSA
(Wuhan Servicebio Technology Co., Ltd.) at room temperature for 60
min. After that, the membrane was incubated with primary antibodies
against STAT3 (1:1,000), phosphorylated STAT3 (1:1,000), FUS
(1:1,000), Histone H3 (1:2,000), or GAPDH (1:2,000, cat. no. AC001;
Abclonal Biotech Co., Ltd.) at 4°C overnight. Next, membranes were
incubated with HRP-conjugated goat anti-rabbit (1:10,000; cat. no.
ab288151; Abcam) or goat anti-mouse secondary antibodies (1:10,000;
cat. no. ab97040; Abcam) at room temperature for 60 min. Finally,
the immunoreactive blot was visualized with enhanced
chemiluminescence reagent (Wuhan Servicebio Technology Co.,
Ltd.).
Co-immunoprecipitation (co-IP) was conducted with a
magnetic IP kit (cat. no. 88804; Thermo Fisher Scientific, Inc.).
Briefly, cell lysates were gently rotated overnight with anti-FUS
antibody, anti-STAT3 antibody or normal rabbit IgG (Beyotime
Institute of Biotechnology). Afterwards, pre-washed protein A/G
magnetic beads were incubated with rotating for 1 more h. The beads
were collected and washed three times. Western blotting was
performed after eluting bound proteins with elution buffer. All
antibodies used in the present study are listed in Table SIII.
Haematoxylin and eosin (H&E) and
immunohistochemical (IHC) staining
Tissues embedded in paraffin were sectioned to a
thickness of 5 µm, deparaffinized using xylene and rehydrated using
a graded series of ethanol. H&E was applied to one section
according to standard procedures. Other sections were stained for
IHC. IHC was performed by incubating sections with primary
antibodies at room temperature for 2 h. Anti-STAT3 rabbit
polyclonal antibody and anti-Ki67 rabbit polyclonal antibody were
used as primary antibodies at concentrations of 10 and 5 µg/ml,
respectively. The next step involved adding HRP-conjugated goat
anti-rabbit (1:500; cat. no. ab288151; Abcam) for 30 min at 37°C,
followed by streptavidin labelled with peroxidase. Antibody
staining was revealed using chromogen 3,3-diaminobenzidine.
Moreover, non-specific immunoglobulin was used as the negative
control. Finally, the slides were observed under a light microscope
(Olympus Corporation).
In vivo study
This animal experiment was approved (approval no.
TJH-202110004) by the Animal Research Ethics Committee of Tongji
Hospital (Wuhan, China). The experimental procedure and animal care
were all in line with the National Institutes of Health Guide for
the Care and Use of Laboratory Animals. Female nude mice (n=10, 4
weeks old, weight 19-25 g) were purchased from Beijing Vital River
Laboratory Animal Technology Co., Ltd. and housed in a
pathogen-free facility (26°C; 50% humidity; 12-h light/dark cycle
with food and water provided ad libitum). The experimental
animals were randomly divided into sh-circrRPPH1 group and sh-NC
group (5 mice per group). Cells (1×107) were suspended
in 200 µl PBS and injected into the right back region of nude mice.
Mice were under daily monitoring. The tumor size was measured with
calipers at 7, 14, 21 and 28 days after inoculation. The largest
tumor diameter allowed in this experiment was <20 mm. After 30
days, the nude mice were euthanized by pentobarbital sodium (100
mg/kg) via intravenous injection and verified the sacrifice by
cessation of breathing and heartbeat. Then, the weight of the
excised tumor was recorded. Moreover, HE and IHC staining were
applied to study tumor tissues.
To establish the lung metastasis model, 4 weeks old
BABL/C female nude mice were randomly divided into sh-circRPPH1
group and sh-NC group (5 mice per group). Single cells
(2×106) were suspended in 100 µl PBS and injected into
the tail vein of nude mice. After 6 weeks, mice were euthanized
with sodium pentobarbital (100 mg/kg), and lung tissue was
obtained. The resected lungs were subjected to H&E and IHC
staining. Then, the metastatic lesions in lung were carefully
examined by 3 pathologists.
Statistical analysis
Statistical analysis in the present study was
performed using the SPSS 26.0 package program (IBM Corp.). The
statistical significance between the two groups was calculated
using the unpaired Student's t-test, while the statistical
significance between the three or more groups was calculated using
one-way ANOVA followed by Tukey's multiple comparison test. Data
were expressed as the mean ± standard deviation (SD) of three
independent experiments. Moreover, differences were considered
statistically significant when P<0.05.
Results
The expression profiles and
characteristics of circRPPH1
An increasing number of public databases are
constructed to facilitate cancer research. To identify critical
circRNAs involved in BCa tumorigenesis, the expression patterns of
circRNAs were analyzed with two GEO datasets (GSE97239 and
GSE92675) and a variety of circRNAs aberrantly expressed in BCa
were found (Fig. 1A and B). A total
of 4 overlapping differentially expressed circRNAs exist between
these two databases, including hsa_circ_0000824, hsa_circ_0004368,
hsa_circ_0006117, and hsa_circ_0000512 (Fig. 1C). Among them, circRPPH1
(hsa_circ_0000512) was the only one upregulated circRNA and was
selected for further exploration (Fig.
1D).
Next, the expression levels of circRPPH1 were
compared between BCa cells and normal human bladder epithelial
cells. Compared with normal human bladder epithelial cells,
circRPPH1 was significantly higher in BCa cells (Fig. 2A). In addition, circRPPH1 is
generated from exon of the RPPH1 gene located on chr14:
20,811,283-20,811,436. The special sequences in the back-spliced
junction point of circRPPH1 were verified by Sanger sequencing
(Fig. 2B). Next, the circular
structure of circRPPH1 was confirmed by RT-qPCR and agarose gel
electrophoresis using convergent and divergent primers. As
expected, divergent primers could amplify circRPPH1 in cDNA rather
than genomic DNA (Fig. 2C).
Compared with parental linear genes, circRNAs are more resistant to
the degradation of RNase R because of their unique circular
structure (4). In fact, circRPPH1
also exhibited resistance to RNase R in the present study, while
the linear RPPH1 RNA or GAPDH mRNA were significantly degraded
after RNase R treatment (Fig. 2D).
Similarly, the results of RT-qPCR after actinomycin D treatment
showed that the degradation rate of circRPPH1 was lower than that
of the linear transcript (Fig. 2E).
Moreover, the nuclear-cytoplasmic separation assay and FISH
experiments indicated that most of the circRPPH1 was located in the
cytoplasm (Fig. 2F and G). In
brief, these results indicated that circRPPH1 is upregulated in BCa
cells and is primarily located in the cytoplasm.
CircRPPH1 promotes the proliferation
and invasion of BCa cells in vitro
To evaluate the biological functions of circRPPH1 in
BCa cells, three interference plasmids (sh-circRPPH1#1,
sh-circRPPH1#2 and sh-circRPPH1#3) and one overexpression plasmid
were constructed. It was found that transient transfection of
sh-circRPPH1 #2 or OE-circRPPH1 could significantly change the
expression level of circRPPH1, while sh-circRPPH1#1 and
sh-circRPPH1#3 had relatively weak effects. Then, the stable
interference or overexpression systems of circRPPH1 was
successfully established in T24 and 5637 cells using lentiviral
packaging plasmid. Notably, the RNA level of linear RPPH1 was
stable while circRPPH1 expression was altered. The aforementioned
results are shown in Fig.
S1A-E.
The cell viability of BCa cells was detected by
CCK-8 assay. The results indicated that circRPPH1 knockdown
significantly inhibited the cell viability of T24 and 5637 cells.
The clone formation experiments showed that interference of
circRPPH1 could significantly inhibit BCa cell proliferation.
Conversely, overexpression of circRPPH1 increased the proliferation
ability of BCa cells (Fig. 3A and
B). Transwell and Matrigel experiments indicated that the
migration and invasion abilities of BCa cells were significantly
inhibited following circRPPH1 knockdown, while the migration and
invasion abilities of BCa cells were enhanced by circRPPH1
overexpression (Fig. 3C and D).
CircRPPH1 sponges miR-296-5p in BCa
cells
Considering most circRNAs usually act as miRNA
sponges to exert their functions (23), CircInteractome (https://circinteractome.nia.nih.gov/)
and CIRCBANK (http://www.circbank.cn/) were searched to predict the
miRNAs that can interact with circRPPH1. MiR-615-5p and miR-296-5p
were predicted to be the potential target miRNAs (Fig. 4A), and were selected for subsequent
experiments. Notably, miR-296-5p but not miR-615-5p levels were
affected when circRPPH1 was overexpressed or interfered (Fig. 4B and C). Additionally, the targeted
relationship between circRPPH1 and miR-296-5p was further verified
by luciferase reporter assays. Compared with the negative control,
miR-296-5p mimic reduced the luciferase activity of WT-circRPPH1
plasmid in 293T cells, while the luciferase activity of
MUT-circRPPH1 plasmid had little change (Fig. 4D). With RIP assays, circRPPH1 and
miR-296-5p were significantly enriched by AGO2 antibodies (Fig. 4E) Collectively, circRPPH1 probably
functioned as a sponge of miR-296-5p.
 | Figure 4.CircRPPH1 directly targets to
miR-296-5p and suppresses miR-296-5p activity. (A) miR-296-5p and
miR-615-5p were predicted to be the potential targets of circRPPH1
in the Circinteractome and Circbank databases. (B and C) Bladder
cancer cells were transfected with OE-NC, OE-circRPPH1,
sh-circRPPH1, and sh-NC. After transfection, the expression levels
of miR-296-5p and miR-615-5p were analyzed by reverse
transcription-quantitative PCR. (D) After co-transfection of
luciferase reporter vectors and miR-296-5p mimics or miR-NC into
293T cells, the luciferase activities were detected and analyzed.
MiR-296-5p overexpression could significantly inhibit the
luciferase activities of WT vector but not MUT vector. (E)
CircRPPH1 and miR-296-5p were significantly enriched by AGO2
antibodies in RNA immunoprecipitation assays. Data are presented as
the mean ± SD. *P<0.05, **P<0.01 and ***P<0.001. circ-,
circular; miR, microRNA; OE, overexpression; sh-, short hairpin;
NC, negative control; WT, wild-type; MUT, mutant. |
MiR-296-5p targets STAT3 and
suppresses the proliferation and invasion of BCa cells
Based on the TargetScan (http://www.targetscan.org), miR-296-5p was predicted
to bind to the 3′ untranslated region of STAT3 mRNA with a
high-score. The luciferase reporter assays were applied to testify
this interaction. Compared with miR-NC, miR-296-5p mimic could
significantly reduce the luciferase activity of WT-STAT3, whereas
the luciferase activity of MUT-STAT3 was not affected by miR-296-5p
mimic (Fig. 5A). Furthermore, it
was found that miR-296-5p overexpression could significantly reduce
the expression of STAT3 by RT-qPCR and western blot analysis
(Fig. 5B and C).
 | Figure 5.miR-296-5p suppresses BCa cell
activities through STAT3. (A) miR-296-5p significantly inhibited
luciferase activity in the WT-STAT3 vector, but not in the
MUT-STAT3 vector. (B and C) The STAT3 expression was significantly
inhibited by miR-296-5p in BCa cells. (D and E) The proliferation
abilities of BCa cells transfected with miR-296-5p mimics or miR-NC
were assessed by Cell Counting Kit-8 assay and colony formation
assay. (F and G) Invasion or migration abilities of BCa cells
transfected with miR-296-5p mimics or miR-NC were examined through
Transwell and Matrigel assays (scale bar, 100 µm). Data are
presented as the mean ± SD. *P<0.05, **P<0.01 and
***P<0.001. miR, microRNA; BCa, bladder cancer; WT, wild-type;
MUT, mutant; OE, overexpression; sh-, short hairpin; NC, negative
control. |
The effect of miR-296-5p on the malignant biological
behaviors was explored. The results showed that overexpression of
miR-296-5p inhibited the proliferation (Fig. 5D and E), invasion and migration
(Fig. 5F and G) of BCa cells.
Additionally, miR-296-5p knockdown exerted an opposite effect
(Fig. S2A and B). The transfection
efficiencies of miR-296-5p mimic and inh-296-5p were verified with
RT-qPCR (Fig. S1H and I). Notably,
the transfection efficacy of inh-296-5p was verified by a known
target gene (NRG1) from a previous study (24). In conclusion, miR-296-5p could
suppress the development of BCa by targeting STAT3.
CircRPPH1 promotes the proliferation
and invasion of BCa by sponging miR-296-5p to regulate STAT3
The interaction among circRPPH1, miR-296-5p and
STAT3 was further explored. In the luciferase reporter assays,
circRPPH1 overexpression significantly increased the luciferase
activity of WT-STAT3 plasmid, whereas the miR-296-5p could
eliminate this effect. Furthermore, this phenomenon disappeared
with MUT-STAT3 plasmid (Fig. 6A).
Using western blot analysis and RT-qPCR, the STAT3 mRNA and protein
levels were significantly increased by circRPPH1 overexpression,
while miR-296-5p mimic could cancel out this effect (Fig. 6B and C). The aforementioned results
demonstrated that STAT3 expression was regulated by circRPPH1,
which is a competing endogenous RNA and sponges miR-296-5p. Several
rescue experiments were implemented to further verify this complex
relationship. The results indicated that circRPPH1 knockdown
significantly inhibited the proliferation, invasion and migration
abilities of BCa cells. However, co-transfection of sh-circRPPH1
and inh-296-5p may cancel out this effect (Fig. 6D-G). In summary, circRPPH1
accelerates the biological behaviors of BCa cells by sponging
miR-296-5p to regulate STAT3 expression.
 | Figure 6.CircRPPH1 promotes the proliferation
and invasion of BCa by sponging miR-296-5p to regulate STAT3. (A)
The luciferase assays were conducted to explore the interaction
among circRPPH1, miR-296-5p and STAT3. CirRPPH1 overexpression
significantly promoted the luciferase activities in the WT-STAT3
vectors, while the miR-296-5p mimic could counteract this effect.
(B and C) Single transfection of OE-circRPPH1 vectors could
significantly upregulate the STAT3 expression levels in BCa cells,
while the co-transfection of OE-circRPPH1 and miR-296-5p mimic
would cancel out this effect. (D and E) With Cell Counting Kit-8
and colony formation assays, the proliferation abilities of cells
were evaluated after co-transfection with circRPPH1 and miR-296-5p
mimics. (F and G) Cell invasion and migration abilities was
examined in cells co-transfected with circRPPH1 and miR-296-5p
mimics (scale bar, 100 µm). Data are presented as the mean ± SD.
**P<0.01 and ***P<0.001. circ-, circular; BCa, bladder
cancer; miR, microRNA; OE, overexpression; sh-, short hairpin; NC,
negative control; inh-, inhibitor. |
The interaction of circRPPH1 and FUS
facilitates the nuclear translocation of phosphorylated STAT3 in
BCa cells
Recently, two studies reported that the binding of
FUS to STAT3 promotes the translocation of p-STAT3 into the nuclei
(25,26). Moreover, three online databases
suggested a potential interaction between circRPPH1 and FUS
(Fig. 7A). Then, the interaction
between circRPPH1 and FUS was further analyzed on HDOCK (http://hdock.phys.hust.edu.cn/). Firstly, the
RNAfold Web Server and RNACOmposer were used to predict the
tertiary structure of circRPPH1. Next, the tertiary structure of
FUS was acquired from Protein Data Bank (27). Finally, this structure information
was imported into the HDOCK. As demonstrated in Fig. 7B, the predicted result further
indicated the interaction between circRPPH1 and FUS.
To further explore the interaction among circRPPH1,
FUS, STAT3 and p-STAT3, several experiments were conducted. RIP
assays were used to verify the interaction of circRPPH1 with FUS
protein (Fig. 7C). A co-IP assay
verified the interaction between FUS and STAT3 (Fig. 7D). Western blotting indicated that
FUS knockdown has no effect on the expression of total STAT3
(Fig. 7E). Similarly, FUS knockdown
has no effect on the circRPPH1 expression (Fig. S1F and G). It was observed that
circRPPH1 overexpression upregulated the STAT3 level and promoted
the nuclear translocation of p-STAT3, while FUS knockdown could
counteract this effect (Fig. 7F).
Next, CCK-8 and Transwell assays further confirmed that the
interaction between the FUS and circRPPH1 could promote the
proliferation and invasion abilities of BCa cells (Fig. S2C and D). In conclusion, it was
demonstrated that the interaction between circRPPH1 and FUS could
promote the nuclear translocation of p-STAT3, which further
explained the carcinogenic role of circRPPH1.
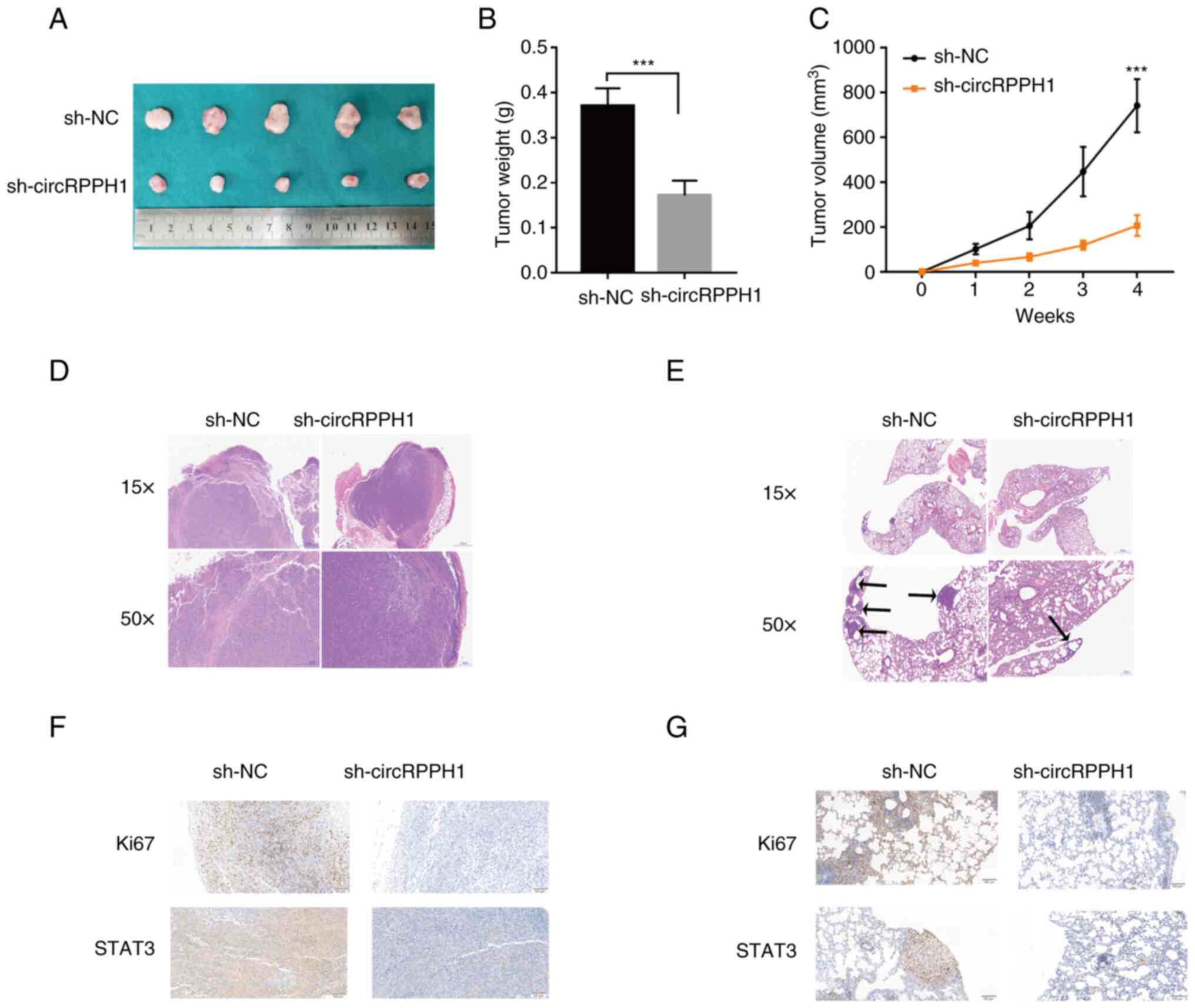
CircRPPH1 accelerates tumor growth and
metastasis in vivo
To further evaluate the effects of circRPPH1 on
growth or metastasis in vivo, stably transfected BCa cells
were injected into the dorsal and tail vein of nude mice,
respectively. Tumor volume and weight in the sh-circRPPH1 group
were significantly smaller than those in the sh-NC group (Fig. 8A-C). H&E staining was used to
further examine these subcutaneous tumors and lung metastases
(Fig. 8D and E). Compared with the
sh-NC group, the sh-circRPPH1 group had fewer lung metastases. In
addition, the results of IHC indicated that STAT3 expressions were
significantly downregulated in the sh-circrPPH1 group compared with
the sh-NC group (Fig. 8F and G).
Collectively, these results indicated that circRPPH1 still acted as
an oncogene in xenograft models.
Discussion
BCa is the most common malignant disease in the
urinary system with high incidence and recurrence (28). There is growing evidence that
circRNAs play important regulatory roles in the carcinogenesis of
BCa and are potential treatment targets for BCa (29,30).
With bioinformatics and cell biological analyses, it was revealed
that circRPPH1, a 154-bp exonic circRNA, is highly expressed in BCa
cells. However, its role and mechanism in BCa remain unclear.
Proliferation, migration and invasion are important
characteristics of tumor cells and have important effects on tumor
progression (31). The results of
the present study suggested that the high expression of circRPPH1
in BCa could promote the proliferation, migration and invasion of
cancer cells, while the downregulation of circRPPH1 could inhibit
the proliferation, migration and invasion of BCa cells. It is worth
noting that circRPPH1 has been reported to play an oncogenic role
in breast cancer and glioblastoma, which indicates that the role of
circRPPH1 in different types of tumors has certain commonalities
(32–35).
miR-296-5p is considered to be an important tumor
suppressor in colorectal cancer, non-small cell lung cancer, liver
cancer and nasopharyngeal carcinoma, inhibiting cell proliferation,
migration, invasion and epithelial-mesenchymal transition
((24,36–38).
Particularly, circRPPH1 promotes cancer progression through the
miR-296-5p/FOXP4 axis in breast cancer and through the
miR-296-5p/RUNX1 axis in colorectal cancer, indicating that
circRPPH1/miR-296-5p axis plays an important role in cancer
malignancies (32,39). However, the role of
circRPPH1/miR-296-5p axis in the malignant phenotype of BCa cells
remains unclear. The present study confirmed that miR-296-5p could
inhibit the proliferation, migration and invasion of BCa cells.
Furthermore, circRPPH1 could exert oncogenic functions through
miR-296-5p/STAT3 axis.
The JAK2/STAT3 signaling pathway, as a classical
oncogenic signaling pathway, is widely involved in the migration,
growth and differentiation of cancer cells (40). After JAK2 activation, downstream
STAT3 can be activated to form p-STAT3 and promote the expression
of downstream oncogenes. Numerous studies have reported that STAT3
can promote cell invasion by regulating EMT-related genes and
promote cell proliferation by inducing c-Myc transcription
(41,42). In addition, the results of
bioinformatics analysis in the present study showed that the
expression of STAT3 was closely related to the pathological
classification, pathological stage, histological grade and overall
survival of BCa, which confirmed the cancer-promoting role of STAT3
in BCa.
Currently, there are certain studies suggesting that
the interaction between RNA and protein may pose different effects
on the nuclear translocation of related proteins, including the
ability to aid or inhibit the nuclear translocation of proteins.
For example, lncRNA OLA1P2 and circ-Amotl1 could inhibit or
accelerate the nuclear translocation of the related binding
proteins, respectively (9,43). FUS is a multi-function RNA-binding
protein. In a recent study, the interaction between circSPARC and
FUS was revealed to contribute to the nuclear translocation of
p-STAT3 (26). In the present
study, FUS is the only one RNA-binding protein (RBP) that binds
with circRPPH1, and the interaction was confirmed by RIP assays.
Furthermore, the interaction was also confirmed to promote the
nuclear translocation of p-STAT3. Considering the complex and
colorful functions of circRNAs, circRPPH1 may also exert its
oncogene role in BCa by other pathways, which needs to be further
confirmed.
There exist certain limitations to the present
study. Firstly, only circRPPH1, a circRNA, was detected. However,
there are a large number of differentially expressed circRNAs in
BCa, and the role of these circRNAs in the occurrence and
development of BCa requires further study. Secondly, whether
circRPPH1 is involved in the occurrence and development of BCa
through other mechanisms, such as regulating RNA selective
splicing, still requires further investigation. It is expected that
subsequent studies will further elucidate the role and mechanism of
circRNA in BCa. In conclusion, circRPPH1 was firstly identified to
play an oncogenic role in BCa progression. It was identified that
circRPPH1 could upregulate STAT3 expression through sponging
miR-296-5p, and interact with FUS to promote p-STAT3 nuclear
translocation, thereby accelerating the proliferation, migration
and invasion of BCa cells. CircRPPH1 could be a potential and
promising treatment target for BCa.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Natural Science
Foundation of China (grant nos. 81900645, 82170779 and 82270804),
the Natural Science Foundation of Hubei Province (gran no.
2021CFB366), the 2019 Wuhan Yellow Crane Talent Program
(Outstanding Young Talents) and the Tongji Hospital (HUST)
Foundation for Excellent Young Scientist (grant no. 2020YQ15).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
KT, ZC and XL designed the study. XL, YT and QH
implemented the experiments. YH, HS and XL analyzed the data. TY
and XL created the figures and tables. KT and XL drafted the
article. KT and ZC confirm the authenticity of all the raw data.
All authors revised the paper and read and approved the final
version of the manuscript, and agreed to be accountable for all
aspects of the work.
Ethics approval and consent to
participate
This animal experiment was approved by the Animal
Research Ethics Committee of Tongji Hospital (Wuhan, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
BCa
|
bladder cancer
|
|
circRNAs
|
circular RNAs
|
|
JAK
|
Janus kinase
|
|
STAT3
|
signal transducer and activator of
transcription 3
|
|
p-STAT3
|
phosphorylated STAT3
|
|
RT-qPCR
|
reverse transcription-quantitative
polymerase chain reaction
|
|
cDNA
|
complementary DNA
|
|
WT
|
wild-type
|
|
MUT
|
mutant-type
|
|
FISH
|
fluorescence in situ
hybridization
|
|
PBS
|
phosphate-buffered saline
|
|
CCK-8 assay
|
Cell Counting Kit-8 assay
|
|
RIP assay
|
RNA immunoprecipitation assay
|
|
co-IP assay
|
co-immunoprecipitation assay
|
|
H&E
|
Haematoxylin and eosin
|
|
IHC
|
immunohistochemistry
|
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2020. CA Cancer J Clin. 70:7–30. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Luo C, Lei T, Zhao M, Meng Q and Zhang M:
CD40 is positively correlated with the expression of nucleophosmin
in cisplatin-resistant bladder cancer. J Oncol. 2020:36767512020.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Knowles MA and Hurst CD: Molecular biology
of bladder cancer: New insights into pathogenesis and clinical
diversity. Nat Rev Cancer. 15:25–41. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kristensen LS, Hansen TB, Venø MT and
Kjems J: Circular RNAs in cancer: Opportunities and challenges in
the field. Oncogene. 37:555–565. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tong Y, Liu X, Xia D, Peng E, Yang X, Liu
H, Ye T, Wang X, He Y, Xu H, et al: Biological roles and clinical
significance of exosome-derived noncoding RNAs in bladder cancer.
Front Oncol. 11:7047032021. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang H, Xiao X, Wei W, Huang C, Wang M,
Wang L, He Y, Sun J, Jiang Y, Jiang G and Zhang X: CircLIFR
synergizes with MSH2 to attenuate chemoresistance via MutSα/ATM-p73
axis in bladder cancer. Mol Cancer. 20:702021. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Xie F, Li Y, Wang M, Huang C, Tao D, Zheng
F, Zhang H, Zeng F, Xiao X and Jiang G: Circular RNA BCRC-3
suppresses bladder cancer proliferation through miR-182-5p/p27
axis. Mol Cancer. 17:1442018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zeng Y, Du WW, Wu Y, Yang Z, Awan FM, Li
X, Yang W, Zhang C, Yang Q, Yee A, et al: A circular RNA binds to
and activates AKT phosphorylation and nuclear localization reducing
apoptosis and enhancing cardiac repair. Theranostics. 7:3842–3855.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ashwal-Fluss R, Meyer M, Pamudurti NR,
Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N and
Kadener S: circRNA biogenesis competes with pre-mRNA splicing. Mol
Cell. 56:55–66. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Legnini I, Di Timoteo G, Rossi F, Morlando
M, Briganti F, Sthandier O, Fatica A, Santini T, Andronache A, Wade
M, et al: Circ-ZNF609 is a circular RNA that can be translated and
functions in myogenesis. Mol Cell. 66:22–37.e9. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bromberg JF, Wrzeszczynska MH, Devgan G,
Zhao Y, Pestell RG, Albanese C and Darnell JE Jr: Stat3 as an
oncogene. Cell. 98:295–303. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Huynh J, Etemadi N, Hollande F, Ernst M
and Buchert M: The JAK/STAT3 axis: A comprehensive drug target for
solid malignancies. Semin Cancer Biol. 45:13–22. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Mirzaei S, Gholami MH, Mahabady MK, Nabavi
N, Zabolian A, Banihashemi SM, Haddadi A, Entezari M, Hushmandi K,
Makvandi P, et al: Pre-clinical investigation of STAT3 pathway in
bladder cancer: Paving the way for clinical translation. Biomed
Pharmacother. 133:1110772021. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hindupur SV, Schmid SC, Koch JA, Youssef
A, Baur EM, Wang D, Horn T, Slotta-Huspenina J, Gschwend JE, Holm
PS and Nawroth R: STAT3/5 inhibitors suppress proliferation in
bladder cancer and enhance oncolytic adenovirus therapy. Int J Mol
Sci. 21:11062020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhong Z, Lv M and Chen J: Screening
differential circular RNA expression profiles reveals the
regulatory role of circTCF25-miR-103a-3p/miR-107-CDK6 pathway in
bladder carcinoma. Sci Rep. 6:309192016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li Y, Zheng F, Xiao X, Xie F, Tao D, Huang
C, Liu D, Wang M, Wang L, Zeng F and Jiang G: CircHIPK3 sponges
miR-558 to suppress heparanase expression in bladder cancer cells.
EMBO Rep. 18:1646–1659. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liu M, Wang Q, Shen J, Yang BB and Ding X:
Circbank: A comprehensive database for circRNA with standard
nomenclature. RNA Biol. 16:899–905. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Dudekula DB, Panda AC, Grammatikakis I, De
S, Abdelmohsen K and Gorospe M: CircInteractome: A web tool for
exploring circular RNAs and their interacting proteins and
microRNAs. RNA Biol. 13:34–42. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
McGeary SE, Lin KS, Shi CY, Pham TM,
Bisaria N, Kelley GM and Bartel DP: The biochemical basis of
microRNA targeting efficacy. Science. 366:eaav17412019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zeng J, Xiang W, Zhang Y, Huang C, Chen K
and Chen Z: Ubiquitous expressed transcript promotes tumorigenesis
by acting as a positive modulator of the polycomb repressive
complex 2 in clear cell renal cell carcinoma. BMC Cancer.
19:8742019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yu CY and Kuo HC: The emerging roles and
functions of circular RNAs and their generation. J Biomed Sci.
26:292019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Shi DM, Li LX, Bian XY, Shi XJ, Lu LL,
Zhou HX, Pan TJ, Zhou J, Fan J and Wu WZ: miR-296-5p suppresses EMT
of hepatocellular carcinoma via attenuating NRG1/ERBB2/ERBB3
signaling. J Exp Clin Cancer Res. 37:2942018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liang C, Zhao T, Li H, He F, Zhao X, Zhang
Y, Chu X, Hua C, Qu Y, Duan Y, et al: Long non-coding RNA ITIH4-AS1
accelerates the proliferation and metastasis of colorectal cancer
by activating JAK/STAT3 signaling. Mol Ther Nucleic Acids.
18:183–193. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang J, Zhang Y, Song H, Yin H, Jiang T,
Xu Y, Liu L, Wang H, Gao H, Wang R and Song J: The circular RNA
circSPARC enhances the migration and proliferation of colorectal
cancer by regulating the JAK/STAT pathway. Mol Cancer. 20:812021.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Berman HM, Westbrook J, Feng Z, Gilliland
G, Bhat TN, Weissig H, Shindyalov IN and Bourne PE: The protein
data bank. Nucleic Acids Res. 28:235–242. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Antoni S, Ferlay J, Soerjomataram I, Znaor
A, Jemal A and Bray F: Bladder cancer incidence and mortality: A
global overview and recent trends. Eur Urol. 71:96–108. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sun K, Wang D, Yang BB and Ma J: The
emerging functions of circular RNAs in bladder cancer. Cancers
(Basel). 13:46182021. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Su Y, Feng W, Shi J, Chen L, Huang J and
Lin T: circRIP2 accelerates bladder cancer progression via
miR-1305/Tgf-β2/smad3 pathway. Mol Cancer. 19:232020. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang L, Wu H, Chu F, Zhang L and Xiao X:
Knockdown of circ_0000512 inhibits cell proliferation and promotes
apoptosis in colorectal cancer by regulating miR-296-5p/RUNX1 axis.
Onco Targets Ther. 13:7357–7368. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhou Y, Liu X, Lan J, Wan Y and Zhu X:
Circular RNA circRPPH1 promotes triple-negative breast cancer
progression via the miR-556-5p/YAP1 axis. Am J Transl Res.
12:6220–6234. 2020.PubMed/NCBI
|
|
34
|
Zhao C, Li L, Li Z, Xu J, Yang Q, Shi P,
Zhang K and Jiang R: A novel circular RNA hsa_circRPPH1_015 exerts
an oncogenic role in breast cancer by impairing miRNA-326-Mediated
ELK1 inhibition. Front Oncol. 10:9062020. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Xiong JW, Song SB, Xiong LM, Duan CH, Song
Q, Yu DL and Zhang XQ: CircRPPH1 promotes cell proliferation,
migration and invasion of non-small cell lung cancer via the
PI3K/AKT and JAK2/STAT3 signaling axes. J Biochem. 171:245–252.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Rong D, Lu C, Zhang B, Fu K, Zhao S, Tang
W and Cao H: CircPSMC3 suppresses the proliferation and metastasis
of gastric cancer by acting as a competitive endogenous RNA through
sponging miR-296-5p. Mol Cancer. 18:252019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Savi F, Forno I, Faversani A, Luciani A,
Caldiera S, Gatti S, Foa P, Ricca D, Bulfamante G, Vaira V and
Bosari S: miR-296/Scribble axis is deregulated in human breast
cancer and miR-296 restoration reduces tumour growth in vivo. Clin
Sci (Lond). 127:233–242. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Han W, Kong D, Lu Q, Zhang W and Fan Z:
Aloperine inhibits proliferation and promotes apoptosis in
colorectal cancer cells by regulating the
circNSUN2/miR-296-5p/STAT3 pathway. Drug Des Devel Ther.
15:857–870. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yang L, Liu Z, Ma J, Wang H, Gao D, Zhang
C and Ma Q: CircRPPH1 serves as a sponge for miR-296-5p to enhance
progression of breast cancer by regulating FOXP4 expression. Am J
Transl Res. 13:7556–7573. 2021.PubMed/NCBI
|
|
40
|
Yu H, Lee H, Herrmann A, Buettner R and
Jove R: Revisiting STAT3 signalling in cancer: New and unexpected
biological functions. Nat Rev Cancer. 14:736–746. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Shen M, Xu Z, Xu W, Jiang K, Zhang F, Ding
Q, Xu Z and Chen Y: Inhibition of ATM reverses EMT and decreases
metastatic potential of cisplatin-resistant lung cancer cells
through JAK/STAT3/PD-L1 pathway. J Exp Clin Cancer Res. 38:1492019.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Jin W: Role of JAK/STAT3 signaling in the
regulation of metastasis, the transition of cancer stem cells, and
chemoresistance of cancer by epithelial-mesenchymal transition.
Cells. 9:2172020. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Guo H, Liu J, Ben Q, Qu Y, Li M, Wang Y,
Chen W and Zhang J: The aspirin-induced long non-coding RNA OLA1P2
blocks phosphorylated STAT3 homodimer formation. Genome Biol.
17:242016. View Article : Google Scholar : PubMed/NCBI
|