Introduction
Lung cancer remains one of the malignancies with the
highest incidence worldwide and a major cause of cancer-related
deaths (1). Lung adenocarcinoma
(LUAD) is the most prevalent histological subtype of non-small cell
lung cancer (NSCLC), accounting for 40–55% of the cases (2,3).
Although great progress has been made in the diagnosis and
treatment of lung cancer, ~90% of lung cancer-related deaths are
caused by metastasis (4).
Currently, platinum compounds including cisplatin are first-line
chemotherapy drugs for the majority of metastatic LUAD cases
(5). However, LUAD can become
resistant to treatment with additional cisplatin treatment cycles.
Therefore, it is imperative to identify targets that improve LUAD
sensitivity to chemotherapy.
One mechanism by which tumors metastasize is
epithelial-mesenchymal transition (EMT). During this process,
epithelial cells gain characteristics of mesenchymal cells; this
process is associated with primary tumor formation, metastasis, and
drug resistance (6). When cells
undergo EMT, they lose cell polarity and the need for cell-cell
adhesion for continued survival. In addition, mesenchymal markers,
including N-cadherin, vimentin and α-smooth muscle actin, are
upregulated, whereas epithelial markers, including E-cadherin, are
downregulated; these post-EMT cells acquire migratory and invasive
capabilities typical of mesenchymal cells (7,8).
Because these changes enable tumor metastasis, EMT regulators
should be monitored in LUAD. For example, CD73, ELK1, and VPS33BD
have been shown to promote LUAD progression by regulating EMT
(9–11). Cancer cells that undergo EMT invade,
metastasize, and exhibit cancer stem cell-like properties,
conferring resistance to conventional and targeted therapies
(12). In addition,
mesenchymal-epithelial transition, the reverse process of EMT,
plays a vital role in stem cell differentiation and
dedifferentiation (13). Although
numerous EMT regulators have been identified, other proteins or
enzymes may contribute to this transformation.
Dipeptidase-2 (DPEP2), a member of the
membrane-binding dipeptidase family (DPEP), is an extracellular
enzyme fixed on the plasma membrane by glycosyl
phosphatidylinositol and highly expressed in the lung, heart, and
testis (14). The DPEP family is
responsible for hydrolyzing dipeptides. For example, DPEP1 and
DPEP2 can convert leukotriene D4 (LTD4) as a substrate to
leukotriene E4 (LTE4), reducing or eliminating leukotriene activity
(14–16). A previous study demonstrated that
the expression of E-cadherin is increased by DPEP1 by inhibiting
the LTD4 signaling pathway through the conversion of LTD4 to LTE4
(17). In addition, DPEP1 and DPEP3
have the ability to cleave cystinyl-bis-glycine to cysteine
and glycine (14,15,18).
Another related study suggested that DPEP1 is upregulated by
dexamethasone in a glucocorticoid receptor-dependent manner to
hydrolyze glutathione, thereby increasing dexamethasone sensitivity
to ferroptosis (19). Notably,
DPEP1 is the only enzyme known, to date, that is capable of
hydrolyzing β-lactam substrates (14,18).
However, the effects of members of the DPEP family, which hydrolyze
substrates, remain to be fully elucidated and require further
investigation.
A previous study indicated that macrophage DPEP2 can
alleviate coxsackievirus B3-induced myocarditis by acting as a
regulator of the NF-κB inflammatory signaling pathway
(20). Recent studies have revealed
that DPEP2 can be used as one of the risk-scoring factors of fatty
acid metabolism genes in LUAD (21)
and DPEP2 may constitute an immune indicator of LUAD (22). In addition, other DPEP family
members have also been identified to be highly expressed in
cancers. For instance, DPEP3 is associated with tumor-initiating
cells in epithelial ovarian carcinoma (23). In addition, studies have shown that
DPEP3 co-locates and forms a physical complex with TEX101 on the
surface of murine testicular germ cells, which may be related to
male infertility (24,25). Notably, DPEP1 exhibited the greatest
degree of overexpression in colorectal cancer and knockdown of
DPEP1 significantly increased cell apoptosis and attenuated cell
proliferation as well as invasion (26,27).
Similar studies have shown that DPEP1 is a biologically-related
gene in pancreatic ductal adenocarcinoma with prognostic and
therapeutic significance and overexpression of DPEP1 suppressed
tumor cell invasiveness and increased sensitivity to
chemotherapeutic agent gemcitabine (28). Furthermore, DPEP1 has been revealed
to be highly expressed in hepatoblastoma and promoted the
progression of hepatoblastoma via activating the
phosphatidylinositol-3-kinase/Akt/mammalian target of rapamycin
signaling (29). However, the
expression, functions and mechanisms of DPEP2 in cancer,
particularly in LUAD, remain poorly understood.
Therefore, in the present study the role of DPEP2 in
LUAD was explored. DPEP2 expression in LUAD samples was analyzed
with the use of public databases. The analysis included the gene,
mRNA, and protein levels, and assessed the prognostic capabilities
of DPEP2. Furthermore, the effects of DPEP2 in LUAD were
investigated, both in vitro and in vivo, for the
identification of targets which are able to disrupt EMT and
metastasis. The present study highlighted the key role of DPEP2 in
LUAD metastasis, and supports the clinical monitoring of this
marker for assessment of prognosis.
Materials and methods
Patient datasets
As previously described (30), the gene expression profile GSE31210
(including 226 LUAD samples and 20 adjacent non-tumor samples) was
downloaded from the Gene Expression Omnibus (GEO) database
(https://www.ncbi.nlm.nih.gov/) for
analysis (31,32). The inclusion criteria were as
follows: i) The dataset only included LUAD tissue; ii) only
included patients with LUAD; iii) was derived from human samples;
iv) included a large number of patients between the ages of 30 and
70, as required by the study design; v) inclusion of stage I and II
patients with LUAD, with ≥50 patient samples per stage; and vi)
used high-throughput gene chips or sequencing technology. The
exclusion criteria were as follows: i) The dataset contained a
large number of missing or abnormal values; ii) the number of
samples in each pathological stage of LUAD was limited in the
dataset.
Furthermore, mRNA expression data, matched genome
and clinical information (including 535 LUAD samples and 59
adjacent non-tumor samples) were downloaded from The Cancer Genome
Atlas (TCGA) database (http://cancergenome.nih.gov/) for further analytical
verification. The inclusion criteria were as follows: i) The data
of the samples were sourced from TCGA database; ii) consisted of
LUAD samples; iii) the inclusion of mRNA expression data, matched
genome data, and clinical information; iv) complete clinical
information including pathological stage, differentiation degree,
survival time, sex, age, etc.; v) the mRNA expression data was
assessed using RNA-seq technology and included high-quality
expression data (such as RPKM, FPKM, etc.); and vi) the genomic
data were matched and the gene annotation information was the
latest. The exclusion criteria were as follows: i) The data of
samples were limited; ii) the quality of the expression data was
poor, including lowly-expressed genes, missing values, etc.; and
iii) the genomic data did not match, or gene annotation information
was incomplete.
The gene amplification and mutation status of DPEP2
was obtained using cBioPortal for Cancer Genomics (http://www.cbioportal.org/). Briefly, the module of
‘Query-Lung-Lung Adenocarcinoma (TCGA, Nature 2014)’ was selected
on the homepage, ‘Query By Gene’ was clicked, ‘DPEP2’ was entered,
and ‘Submit Query’ was then clicked to submit the analysis. The
Human Protein Atlas (HPA) database (http://www.proteinatlas.org/) was used to verify the
expression of DPEP2 in LUAD. Briefly, ‘DPEP2’ was entered in the
homepage for query, ‘TISSUE’ and then ‘LUNG’ was selected to obtain
the normal group images; in addition, ‘PATHOLOGY’ and then
‘CANCER-LUNG CANCER’ were selected to obtain images.
Establishment and evaluation of LUAD
survival prediction
Cox regression analysis was used to screen
independent clinicopathological prognostic factors and construct a
contingency table to assess the 1-, 3-, and 5-year overall survival
(OS) probability in patients with LUAD. Its accuracy was verified
by comparing its predicted probability with the actual probability
observed in the correction curve. Receiver operating characteristic
(ROC) curve analysis was used to compare the predicted accuracy of
the combined model line chart and the clinicopathological
prognostic factor line chart.
TIMER2.0 database analysis
TIMER2.0 database (http://timer.cistrome.org/) was used to analyze the
expression levels of genes in normal and multiple tumor tissues
extracted from TCGA. Briefly, the module of ‘Cancer Exploration’ on
the homepage was selected, and then ‘DPEP2’ and ‘Submit’ were
selected in order to perform the analysis.
Cell culture
A549 (cat. no. CRL-7909; NSCLC cell line; was
initiated by explant culture of lung carcinomatous tissue from a
58-year-old Caucasian male; a hypotriploid human cell line with the
modal chromosome number of 66, occurring in 24% of cells;
mutations: KRAS, STK11, TP53) and H1650 (cat. no. CRL-5883; human
NSCLC cell line; this was a cell line exhibiting epithelial
morphology that was isolated in 1987 from the lung tissue of a
27-year-old, male smoker with stage 3B, bronchoalveolar carcinoma;
mutations: EGFR, TP53) cell lines were obtained from the American
Type Culture Collection. BEAS-2B (cat. no. F26092; Chengdu Feiouer
Biotechnology Co., Ltd.; a human lung bronchial epithelial cell
line; this was isolated from pathological sections of normal
bronchial epithelium from a non-cancerous individual) and H1299
(cat. no. F26035; Chengdu Feiouer Biotechnology Co., Ltd.; human
NSCLC cell line; this was isolated from the lung of a Caucasian,
43-year-old, male patient with carcinoma; gene deletion: TP53;
mutation: NRAS) cell lines were obtained from Feiouer Biological
Technology Co. The tissue origins of A549, H1650 and H1299 were
from patients with lung cancer, and the tissue origin of BEAS-2B
was from a non-lung cancer patient. All cells were authenticated
using short tandem repeat (STR) DNA profiling analysis. The STR
appraisal reports were issued by Procell Life Science &
Technology Co., Ltd. and Chengdu Feiouer Biotechnology Co., Ltd.
The cells were cultured in RPMI-1640 medium (Invitrogen; Thermo
Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum
(FBS; Procell Life Science & Technology Co., Ltd.) and 5 mg/ml
penicillin/streptomycin (Invitrogen; Thermo Fisher Scientific,
Inc.) and were maintained in a humidified incubator with 5%
CO2 at 37°C.
Cell transfection
DPEP2-overexpressing lentivirus
(PGMLV-CMV-H_DPEP2-3×Flag-PGK-Puro; transcript no. NM_022355.4;
http://www.ncbi.nlm.nih.gov/nuccore/NM_022355.4/)
and empty lentivirus (GM-18844: PGMLV-CMV-MCS-3×Flag-PGK-Puro) were
constructed and packaged by Genomeditech (Shanghai) Co., Ltd.
(https://www.genomeditech.com/). For this
experiment, the 2nd generation system, and the interim cell line,
293T cells (Genomeditech (Shanghai) Co., Ltd.), were used. A549 and
H1650 cell lines, were divided into an empty vector group (Vector)
and a pGMLV-CMV-DPEP2-overexpression group (DPEP2). Firstly,
2×105 cells/well were seeded in six-well plates, and
when the cell density reached ~70%, fresh medium was replaced.
Subsequently, 10 µl of lentivirus liquid (virus titer,
1×108 TU/ml; multiplicity of infection: 10) was added,
and polybrene [Genomeditech (Shanghai) Co., Ltd.] was added at the
same time to increase infection efficiency so that the final
concentration reached 8 µg/ml. After 24 h of transfection at 37°C,
the medium was changed, and the cells successfully transfected with
the virus were screened and maintained with a final concentration
of 1 µg/ml of puromycin dihydrochloride (cat. no. ST551-10 mg;
Beyotime Institute of Biotechnology). The survival state of the
cells was observed every 24 h. After two days, the cells of the
Vector and DPEP2 groups began to proliferate. After one week,
transfection efficiencies were evaluated using reverse
transcription-quantitative PCR (RT-qPCR) and western blotting.
Total RNA extraction and RT-qPCR
Total RNA was extracted from the cells on ice
according to the instructions of the Total RNA Extraction Kit
(Beijing Solarbio Science & Technology Co., Ltd.). RNA samples
were reverse transcribed using the iScript cDNA Synthesis Kit
(Bio-Rad Laboratories, Inc.) according to the manufacturer's
instructions. The cDNAs were then added to the reaction with the
indicated primers using SYBR Green Supermix (Bio-Rad Laboratories,
Inc.) on ice according to the manufacturer's instructions and
amplified using RT-qPCR, performed at 95°C for 2 min, followed by
40 cycles of amplification at 95°C for 10 sec, 62°C for 30 sec and
72°C for 30 sec using a CFX96 Real-Time PCR Detection System
(Bio-Rad Laboratories, Inc.). β-actin was used as an endogenous
control. Relative gene expression levels were calculated by
normalizing the transcript levels to the housekeeping gene using
the comparative threshold cycle 2−ΔΔCq method (33). The primers used were as follows:
DPEP2 forward, 5′-ATCATGCCCAGGCGGTTCATTTC-3′ and reverse,
5′-GGCGTCCACTCCTTCTACAACAAC-3′; β-actin forward,
5′-GGGCCGGACTCGTCATAC-3′ and reverse, 5′-CCTGGCACCCAGCACAAT-3′.
Western blotting
Cells were lysed with radioimmunoprecipitation assay
buffer containing the protease inhibitor phenylmethylsulfonyl
fluoride (Beyotime Institute of Biotechnology). The protein
concentration was determined using a BCA Protein Determination Kit
(Beyotime Institute of Biotechnology) according to the
manufacturer's instructions. Protein samples (30 µg) were separated
on 10% gels using sodium dodecyl sulfate-polyacrylamide gel
electrophoresis and transferred to 0.2-µm polyvinylidene fluoride
membranes (Immobilon-P PVDF; EMD Millipore). The membranes were
blocked using Tris-buffered saline containing 0.1% Tween-20 and 5%
skim milk for 1 h at 25°C. The membranes were incubated with the
primary antibodies, including anti-DPEP2 (1:1,000; cat. no.
16466-1-AP), anti-E-cadherin (1:2,000; cat. no. 60335-1-Ig),
anti-N-cadherin (1:2,000; cat. no. 66219-1-Ig), anti-vimentin
(1:20,000; cat. no. 60330-1-Ig), anti-α-SMA (1:1,000; cat. no.
14395-1-AP), anti-CD44 (1:2,000; cat. no. 15675-1-AP), anti-CD133
(1:2,000; cat. no. 18470-1-AP), anti-GADPH (1:50,000; cat. no.
60004-1-Ig; all from Proteintech Group, Inc.), overnight at 4°C.
After washing three times with Tris-buffered saline with 0.1%
Tween-20, the membranes were incubated with corresponding
horseradish peroxidase-conjugated secondary antibodies [cat. no.
SA00001-1 (mouse); SA00001-2 (rabbit); 1:4,000; both from
Proteintech Group, Inc.] for 1.5 h at 25°C. The bands were
visualized in conjunction with ECL Detection Reagents (EMD
Millipore) using the Quantity One 5.2 Software System (Bio-Rad
Laboratories, Inc.).
Wound-healing assays
A549 and H1650 cells (5×105; treatment
with or without DPEP2 overexpression) cultured in serum-free medium
for 24 h were seeded on a six-well plate and allowed to reach
nearly 100% confluence. The cell monolayer was scratched with a
200-µl sterile pipette tip to create an artificial wound, and the
cell debris was removed by repeated washes with PBS. Subsequently,
the cells were cultured in fresh serum-free medium and kept in a
humidified incubator with 5% CO2 at 37°C. At 0 and 12 h,
the wound-healing process was captured at a magnification of ×10
under a fluorescence microscope (Eclipse Ti Series; Nikon
Corporation).
Cell migration and invasion assay
As previously described (34), 1×105 A549 and H1650 cells
(treatment with or without DPEP2 overexpression) cultured in
serum-free medium for 24 h were seeded into the upper lumen of an
8-µm pore size Transwell insert (Corning, Inc.) for the migration
assay, and another set of 1×105 cells were seeded into
the upper lumen of pre-coated Matrigel (at 37°C for 1 h) (BD
Biosciences) for the invasion assay. Medium containing 10% FBS was
added to the lower lumen as an attractant. For both experiments,
cells were incubated for 24 h at 37°C and 5% CO2. For
the migration experiment, the Transwell container was removed and
washed with sterile PBS. For both experiments the cells were then
fixed with methanol for 20 min and stained with 0.1% crystal violet
for 20 min at 25°C. The cells that had not migrated were removed
with cotton swabs and images were captured at a magnification of
×20 under a fluorescence microscope, and were subsequently counted
(XI71; Olympus Corporation).
Sphere formation assay
A549 and H1650 cells (treatment with or without
DPEP2 overexpression) were digested with 0.25% Trypsin-EDTA
(Invitrogen; Thermo Fisher Scientific, Inc.), centrifuged with a
low centrifugation speed (141 × g for 3 min at 25°C; model no.
JW-1016; Anhui Jiawen Instrument Equipment Co., Ltd.), and washed
thrice with PBS. The cells were resuspended in Dulbecco's modified
Eagle's medium/F12 medium (Invitrogen; Thermo Fisher Scientific,
Inc.) supplemented with 20 ng/ml Epidermal Growth Factor
(MedChemExpress), 20 ng/ml Basic Fibroblast Growth Factor
(MedChemExpress), and 1X B-27 supplement (Invitrogen; Thermo Fisher
Scientific, Inc.). Cells were incubated at a density of
5×103 cells/well in a six-well ultra-low adhesion plate
for 7–10 days at 37°C with 5% CO2. Representative tumor
spheres were captured at a magnification of ×20 and quantified
under a fluorescence microscope (Eclipse Ti Series; Nikon
Corporation).
Immunofluorescence (IF)
The prepared cell slides were carefully washed with
PBS, fixed with 4% paraformaldehyde for 15 min at 25°C, washed
repeatedly with PBS, and blocked with PBS containing 10% normal
goat serum (cat. no. C0265; Beyotime Institute of Biotechnology)
for 1 h at 25°C. The slides were incubated with primary antibodies,
including anti-E-cadherin (1:200; cat. no. 60335-1-Ig),
anti-N-cadherin (1:50; cat. no. 66219-1-Ig), anti-vimentin (1:50;
cat. no. 60330-1-Ig), anti-α-SMA (1:800; cat. no. 14395-1-AP),
anti-CD44 (1:50; cat. no. 15675-1-AP), and anti-CD133 (1:50; cat.
no. 18470-1-AP; all from Proteintech Group, Inc.), overnight at
4°C. After multiple rinses with PBS, the slides were incubated with
Cy3 (red)-conjugated secondary antibodies [1:300; cat. no. A0516
(goat anti-rabbit); A0521 (goat anti-mouse); Beyotime Institute of
Biotechnology] at room temperature for 2 h in the dark. The samples
were washed three times with PBS and stained with DAPI (1:4,000;
cat. no. C1002; Beyotime Biotechnology) for 5 min at 25°C before
imaging. Random images were captured at a magnification of ×40 with
an inverted fluorescence microscope (IX71; Olympus
Corporation).
Cell viability assays
Cell viability was determined using a Cell Counting
Kit-8 (CCK-8) and colony formation assays. Briefly, for the colony
formation assay, the cells were cultured in a six-well plate at a
ratio of 600 cells/well with varying concentrations of cisplatin
(0, 2, 4, or 8 µg/ml; Jiangsu Hansoh Pharmaceutical Group Co.,
Ltd.). The medium containing varying concentrations of cisplatin
was changed every 3 days, and cells were cultured for 14 days at
37°C. After the colonies were washed with PBS, they were fixed with
methanol for 15 min at 25°C and stained with 0.1% crystal violet
for 20 min at 25°C. Colonies were defined as >50 ells and were
counted manually. The CCK-8 assay was performed according to the
instructions of the Enhanced Cell Counting Kit-8 (Beyotime
Institute of Biotechnology). A549 and H1650 cells
(5×103; treatment with or without DPEP2 overexpression)
were seeded into 96-well cell culture plates followed by treatment
with cisplatin (0, 2, 4 or 8 µg/ml) at 37°C for 24 h. The medium in
each well was then replaced with 100 µl fresh medium with 10% CCK-8
and the cells were incubated at 37°C for an additional 1 h. The
absorbance was measured at 450 nm.
Flow cytometry
A549 and H1650 cells (treatment with or without
DPEP2 overexpression) with a density of 70%, treated with or
without 2 µg/ml cisplatin for 24 h at 37°C, were digested with
0.25% Trypsin (Invitrogen; Thermo Fisher Scientific, Inc.), washed
twice with PBS and collected. Cells were stained for 15 min in the
dark at 25°C using the Annexin V-PE/7-AAD Apoptosis Detection Kit
(Nanjing KeyGen Biotech Co., Ltd.), according to the manufacturer's
instructions. Cells were analyzed using flow cytometry
(FACSCalibur; Becton, Dickinson and Company).
Immunohistochemistry (IHC)
IHC was performed using a Universal two-step
detection kit (mouse/rabbit enhanced polymer detection system; cat.
no. PV9000; ZSGB-BIO; OriGene Technologies, Inc.). The prepared
5-µm paraffin sections were dewaxed, dehydrated in a liquid
gradient, and washed several times with PBS. Sections were then
boiled in a pH 6.0 citric acid buffer for target antigen retrieval.
Subsequently, endogenous peroxidase was blocked according to the
manufacturer's instructions. The sections were incubated with the
primary antibodies, including anti-DPEP2 (1:200; cat. no.
16466-1-AP), anti-E-cadherin (1:1,000; cat. no. 60335-1-Ig),
anti-N-cadherin (1:8,000; cat. no. 66219-1-Ig), anti-vimentin
(1:4,000; cat. no. 60330-1-Ig), anti-α-SMA (1:2,000; cat. no.
14395-1-AP), anti-CD44 (1:500; cat. no. 15675-1-AP), anti-CD133
(1:4,000; cat. no. 18470-1-AP), anti-Ki67 (1:2,000; cat. no.
27309-1-AP; all from Proteintech Group, Inc.), at 4°C overnight.
After multiple rinses using PBS, the sections and reaction
enhancement solution were incubated at 25°C for 20 min. After
multiple rinses using PBS, the immune complexes were incubated with
the corresponding enzyme-conjugated secondary antibodies (cat. no.
PV9000; ZSGB-BIO; OriGene Technologies, Inc.) for 30 min at
37°C.
Finally, the sections were dyed with a
3,3′-diaminobenzene (DAB) Chromogenic kit (cat. no. ZLI-9019;
ZSGB-BIO; OriGene Technologies, Inc.). Briefly, DAB was used as the
chromogen to stain the reaction products for 3 min at room
temperature for visualization. The sections were then
counterstained with hematoxylin (Beyotime Institute of
Biotechnology) at room temperature for 1 min. Random images were
obatined at a magnification of ×20 using a fluorescence microscope
(IX71; Olympus Corporation).
Nude mouse subcutaneous tumor
model
A total of 16, five-week-old nude female BALB/c mice
with a weight of 18–20 g (GemPharmatech Co. Ltd.) were used for
in vivo experiments. All nude mice were kept at an
auto-controlled room (24±2°C; 50±10%, relative humidity) with 12-h
light/dark cycle. Nude mice were acclimatized for least 7 days
before experiments, and allowed free access to food and water. All
nude mice were randomly divided into 4 groups with 4 mice per group
(vector; DPEP2; vector + cisplatin; DPEP2 + cisplatin). Vector or
DPEP2-overexpressing A549 cells (2×106) were resuspended
in 100 µl of serum-free RPMI-1640 medium, and subcutaneously
injected into the left armpit of each nude mouse in each group.
Subsequently, seven days after A549 cell implantation, each group
was intraperitoneally injected with either 100 µl PBS or cisplatin
(20 mg/m2/mouse/week). The tumor growth in each group
(n=4/group) was measured with Vernier calipers every seven days.
The formula, V=(a × b2)/2 was used to calculate the
tumor volume, where a and b are the maximum and minimum diameters,
respectively, in millimeters. On day 28 after the cell injection,
all tumors images were captured, and the nude mice were then
euthanized by intraperitoneal injection of 100 mg/kg sodium
pentobarbital until cardiac arrest and spontaneous respiratory
arrest. The tumors were weighed and their images were captured
immediately after dissection.
Statistical analysis
The R software package ‘limma’ (version 3.40.6; R
Foundation for Statistical Computing) was used to perform gene
expression differential analysis. DPEP2 expression data were
divided using the median method, and duplicate samples were
excluded. The Wilcoxon signed-rank test and logistic regression was
used to assess the correlation between the clinicopathological
characteristics of LUAD and DPEP2 expression. Univariate and
multivariate Cox analyses were used to identify potential
prognostic factors. Kaplan-Meier plotter and log-rank testing were
used to evaluate the OS. Each in vitro experiment was
independently performed at least three times. Data were analyzed
using GraphPad Prism 8.0 statistical program (GraphPad Software,
Inc.). Results are presented as the mean ± standard deviation
unless otherwise stated. The Shapiro-Wilk normality test was used
to determine if the data followed normal distribution. If the data
followed normal distribution, the results were presented as the
mean ± standard, and the two-tailed Student's independent samples
t-test or one-way ANOVA was used to analyze the significance of the
differences between two groups or multiple groups. The post hoc
tests used following ANOVA were Dunnett's t-test to compare one
group with the other groups, or Newman-Keuls to determined which
specific pairs of means were different. When the normality
assumption was violated, Kruskal-Wallis test was used to determine
statistically significant differences between three or more
independent groups, and Bonferroni correction was applied for the
post hoc multiple comparisons. For all hypotheses tests, a
P<0.05 was considered to indicate a statistically significant
difference.
Results
Expression of DPEP2 is low in
LUAD
Differential expression analysis of RNA sequencing
data of LUAD samples was performed in GSE31210 to identify genes
that exhibited consistent expression changes with LUAD progression.
DPEP2 was among the genes that were notably negatively
associated with LUAD progression (Fig.
1A). DPEP2 mRNA levels in different tumors and
corresponding normal tissues were confirmed using the TIMER2.0
database. DPEP2 expression was significantly lower in LUAD
than in normal tissues (Fig. 1B).
To assess the mutation level of DPEP2, a cBioPortal map was
used to analyze the OncoPrint map of DPEP2 in patients with
LUAD in TCGA dataset (Fig. 1C),
which revealed that DPEP2 had less than 2.2% missense
mutations and gene amplifications.
DPEP2 mRNA expression levels were then
assessed in patients with LUAD and adjacent tissues. As a result,
DPEP2 expression was identified to be significantly lower in
patients with LUAD than in precancerous tissue (P<0.01; Fig. 1D). Similar results were obtained by
comparing DPEP2 expression levels in LUAD and paired
precancerous tissues (Fig. 1E). In
addition, DPEP2 transcription and protein expression levels were
analyzed in vitro, and it was demonstrated that the levels
in LUAD cells were significantly lower than that in a normal lung
cell line (P<0.001; Fig. 1F and
H). Based on the HPA database results, it was revealed that
DPEP2 levels were lower in the tumors than in precancerous tissues
(Fig. 1G). Therefore, these data
indicated that DPEP2 was considerably downregulated in LUAD.
DPEP2 is an independent prognostic
factor for the OS of patients with LUAD
The role of DPEP2 was investigated in patients with
LUAD. The results revealed that low DPEP2 expression was associated
with sex (P<0.05), age (P<0.05), smoking status (P<0.01),
tumor (T) stage (P<0.05), node (N) stage (P<0.05), and
clinical stage (P<0.01; Fig.
2A-F and Table I). These
results indicated that patients with LUAD with low DPEP2
expression progress to advanced disease faster than those with high
DPEP2 expression.
 | Figure 2.Association between DPEP2 expression
and clinicopathological features and its prognostic and diagnostic
value in LUAD. (A-H) The tumor tissues from patients with different
clinical characteristics in TCGA. (A) Sex, (B) age, (C) smoking
status, (D) T stage, (E) N stage, (F) pathologic stage, (G) primary
treatment outcome and (H) OS; A-C and H: *P<0.05, **P<0.01
and ***P<0.001, two-tailed Student's t-test; and D-G:
*P<0.05, **P<0.01 and ***P<0.001, one-way ANOVA with
Bonferroni correction. (I) Kaplan-Meier analysis of the OS
probability of patients with LUAD in TCGA. (J) Receiver operating
characteristic curve analysis for DPEP2 expression in LUAD and
adjacent tissues. (K) Nomogram survival prediction chart for
predicting the 1-, 3-, and 5-year OS rates. DPEP2, dipeptidase-2;
LUAD, lung adenocarcinoma; TCGA, The Cancer Genome Atlas; OS,
overall survival. |
 | Table I.Association between DPEP2 expression
and the clinicopathological features of lung adenocarcinoma cases
in The Cancer Genome Atlas. |
Table I.
Association between DPEP2 expression
and the clinicopathological features of lung adenocarcinoma cases
in The Cancer Genome Atlas.
|
Characteristics | Low expression of
DPEP2 (n=256) | High expression of
DPEP2 (n=257) | P-value |
|---|
| T stage, n (%) |
|
| 0.015 |
| T1 | 70 (13.7%) | 98 (19.2%) |
|
| T2 | 144 (28.2%) | 132 (25.9%) |
|
| T3 | 27 (5.3%) | 20 (3.9%) |
|
| T4 | 14 (2.7%) | 5 (1%) |
|
| N stage, n (%) |
|
| 0.023 |
| N0 | 152 (30.3%) | 178 (35.5%) |
|
| N1 | 52 (10.4%) | 43 (8.6%) |
|
| N2 | 47 (9.4%) | 27 (5.4%) |
|
| N3 | 1 (0.2%) | 1 (0.2%) |
|
| M stage, n (%) |
|
| 0.500 |
| M0 | 175 (47.4%) | 169 (45.8%) |
|
| M1 | 15 (4.1%) | 10 (2.7%) |
|
| Pathologic stage, n
(%) |
|
| 0.009 |
| Stage
I | 119 (23.6%) | 155 (30.7%) |
|
| Stage
II | 64 (12.7%) | 57 (11.3%) |
|
| Stage
III | 53 (10.5%) | 31 (6.1%) |
|
| Stage
IV | 15 (3%) | 11 (2.2%) |
|
| Primary therapy
outcome, n (%) |
|
| 0.031 |
| PD | 43 (10.1%) | 25 (5.9%) |
|
| SD | 19 (4.5%) | 18 (4.2%) |
|
| PR | 4 (0.9%) | 2 (0.5%) |
|
| CR | 141 (33.1%) | 174 (40.8%) |
|
| Residual tumor, n
(%) |
|
| 0.609 |
| R0 | 178 (49.3%) | 166 (46%) |
|
| R1 | 7 (1.9%) | 6 (1.7%) |
|
| R2 | 1 (0.3%) | 3 (0.8%) |
|
| Age, n (%) |
|
| 0.004 |
|
≤65 | 135 (27.3%) | 103 (20.9%) |
|
|
>65 | 111 (22.5%) | 145 (29.4%) |
|
| Sex, n (%) |
|
| 0.030 |
|
Female | 125 (24.4%) | 151 (29.4%) |
|
|
Male | 131 (25.5%) | 106 (20.7%) |
|
| Number pack years
smoked, n (%) |
|
| 0.492 |
|
<40 | 87 (24.8%) | 87 (24.8%) |
|
|
≥40 | 96 (27.4%) | 81 (23.1%) |
|
| Smoker, n (%) |
|
| 0.014 |
| No | 27 (5.4%) | 47 (9.4%) |
|
|
Yes | 224 (44.9%) | 201 (40.3%) |
|
| OS event, n
(%) |
|
| 0.016 |
|
Alive | 149 (29%) | 177 (34.5%) |
|
|
Dead | 107 (20.9%) | 80 (15.6%) |
|
| Age, median
(IQR) | 64 (58, 71) | 68 (59, 73.25) | 0.007 |
In addition, treatment outcomes (P<0.01) and OS
(P<0.001) were associated to low DPEP2 expression
(Fig. 2G and H). To further explore
the association between the survival rate and clinicopathological
features, univariate Cox regression analysis was conducted, and it
was revealed that DPEP2 was significantly associated with OS
(hazard ratio, 0.643; 95% confidence interval, 0.479–0.862;
P=0.003) (Table II). Multivariate
Cox regression analysis demonstrated that the primary therapeutic
outcome and residual tumors were independent prognostic factors
(Table II). Additionally, low
DPEP2 expression was associated with poor patient prognosis
(hazard ratio, 0.63; 95% confidence interval, 0.47–0.84; P=0.002)
(Fig. 2I). These results indicated
that DPEP2 was a prognostic factor for LUAD. To analyze the
diagnostic value of DPEP2 expression, a ROC curve was
constructed and nomogram analysis of DPEP2 expression data
was performed. The area under the ROC curve was 0.976 (Fig. 2J). DPEP2 expression levels
were combined with clinical variables to construct a nomogram to
predict patient survival at 1, 3, and 5 years. The predictive power
of DPEP2 expression was comparable to that of the T stage,
which is the most common tumor staging system worldwide (Fig. 2K). These findings indicated that
DPEP2 may serve as a prognostic biomarker in LUAD.
 | Table II.Univariate and multivariate Cox
regression analysis of clinical features associated with overall
survival of lung adenocarcinoma in The Cancer Genome Atlas. |
Table II.
Univariate and multivariate Cox
regression analysis of clinical features associated with overall
survival of lung adenocarcinoma in The Cancer Genome Atlas.
|
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|
|---|
|
Characteristics | Total (N) | Hazard ratio (95%
CI) | P-value | Hazard ratio (95%
CI) | P-value |
|---|
| Age (≤65 vs.
>65) | 494 | 1.228
(0.915–1.649) | 0.171 |
|
|
| Sex (female vs.
male) | 504 | 1.060
(0.792–1.418) | 0.694 |
|
|
| T stage (T1/T2 vs.
T3/T4) | 501 | 2.364
(1.621–3.448) | <0.001 | 1.882
(0.900–3.939) | 0.093 |
| N stage (N0 vs. N1
and N2 and N3) | 492 | 2.606
(1.939–3.503) | <0.001 | 1.593
(0.966–2.627) | 0.068 |
| M stage (M0 vs.
M1) | 360 | 2.111
(1.232–3.616) | 0.007 | 1.502
(0.558–4.044) | 0.420 |
| Pathologic stage
(stage I/stage II vs. stage III/stage IV) | 496 | 2.624
(1.926–3.576) | <0.001 | 1.471
(0.725–2.983) | 0.285 |
| Primary therapy
outcome (PR and CR vs. PD and SD) | 419 | 2.786
(1.978–3.924) | <0.001 | 3.129
(1.883–5.200) | <0.001 |
| Residual tumor (R0
vs. R1 and R2) | 352 | 3.973
(2.217–7.120) | <0.001 | 3.373
(1.287–8.839) | 0.013 |
| Number pack years
smoked (<40 vs. ≥40) | 345 | 1.038
(0.723–1.490) | 0.840 |
|
|
| DPEP2 (low vs.
high) | 504 | 0.643
(0.479–0.862) | 0.003 | 0.955
(0.603–1.512) | 0.843 |
DPEP2 inhibits cell migration,
invasion and EMT in LUAD
To determine the role of DPEP2 in the malignant
progression of LUAD, stable A549 and H1650 cell lines
overexpressing DPEP2 were constructed. Both RT-qPCR and western
blotting revealed that DPEP2 mRNA and protein expression were
considerably upregulated (Fig. 3A).
The wound-healing assay revealed that DPEP2 overexpression
inhibited the migratory abilities of cells (Fig. 3B). In Transwell assays, DPEP2
overexpression inhibited cell migration and invasion (Fig. 3C and D). Furthermore, western
blotting and IF results revealed that DPEP2 overexpression
increased E-cadherin levels and decreased N-cadherin, vimentin, and
α-SMA levels (Fig. 3E and F). These
data indicated that DPEP2 inhibited LUAD cell migration, invasion,
and EMT.
 | Figure 3.DPEP2 affects migration, invasion,
and EMT of lung adenocarcinoma cells. (A) Expression of DPEP2 in
empty vector control (Vector) and DPEP2-overexpressing cells
(DPEP2) was detected using western blotting and reverse
transcription-quantitative PCR assays. GAPDH served as a loading
control. (B) Representative images and quantitative analysis of
wound-healing assay of A549 and H1650 cells (scale bar, 200 µm). (C
and D) Transwell representative images and analysis of (C) cell
migration and (D) invasion (scale bar, 100 µm). (E and F) The EMT
markers (E-cadherin, N-cadherin, vimentin, and α-SMA) were analyzed
using western blot analysis and IF staining (scale bar, 50 µm).
Histograms represent the mean ± standard deviation based on three
independent experiments. *P<0.05, **P<0.01 and ***P<0.001,
two-tailed Student's t-test. DPEP2, dipeptidase-2; EMT,
epithelial-mesenchymal transition. |
DPEP2 enhances cell sensitivity to
cisplatin by regulating stem cell transformation
Research has revealed that tumor cell EMT can lead
to the same chemotherapy resistance as cancer stem cells (35). Therefore, it was hypothesized that
DPEP2 may affect the chemotherapy response by regulating EMT.
Consequently, the levels of DPEP2 and the lung cancer stem cell
biomarkers, CD44 and CD133, were assessed using western blot and IF
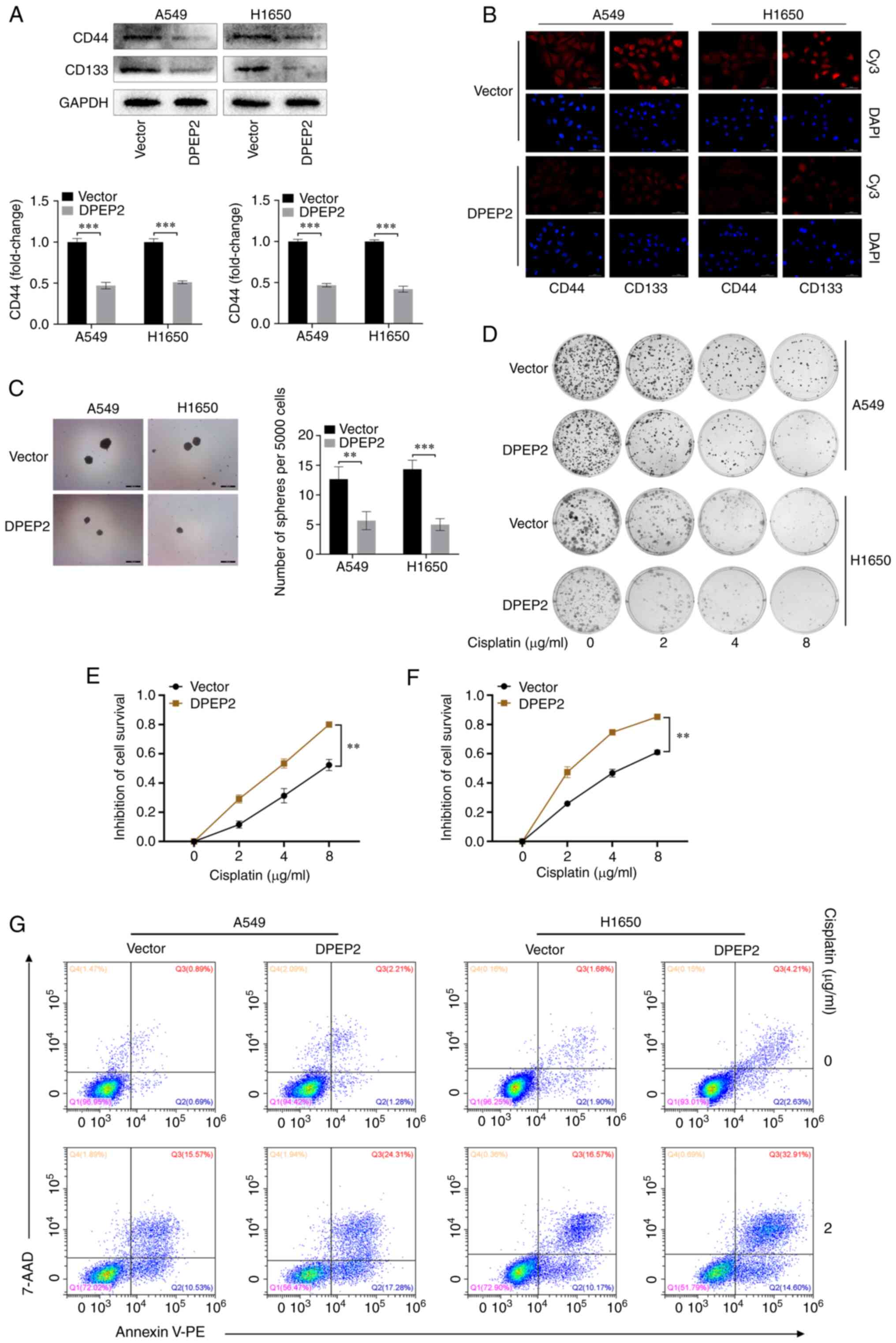
analyses. It was determined that DPEP2 overexpression significantly
decreased CD44 and CD133 in two LUAD cell lines (P<0.001;
Fig. 4A and B). In addition, DPEP2
overexpression significantly decreased sphere formation in A549
(P<0.01; Fig. 4C) and H1650
cells (P<0.001; Fig. 4C). These
results indicated that DPEP2 overexpression inhibited cancer stem
cell-like traits in LUAD.
The functional significance of DPEP2 expression in
cisplatin resistance was further explored. Notably, colony
formation and CCK-8 assays demonstrated that DPEP2 promoted
cisplatin-induced A549 and H1650 cell death (Fig. 4D-F). Apoptosis assays revealed
similar findings, wherein DPEP2-overexpressing A549 and H1650 cells
had higher apoptosis rates and higher sensitivity to cisplatin
(Fig. 4G).
DPEP2 enhances the sensitivity of LUAD
to cisplatin in vivo
To further investigate the effect of DPEP2 on
cisplatin treatment, a nude mouse subcutaneous tumor model was
established using A549 cells (Fig.
5A). It was demonstrated that DPEP2 overexpression
significantly increased cisplatin efficacy compared with the
control group, as revealed in the tumor chart (Fig. 5B and C). Similar results were
obtained by dissection and weight comparisons (Fig. 5D and E). In subsequent experiments,
IHC evaluation showed that DPEP2 overexpression markedly decreased
EMT marker genes (N-cadherin, vimentin, and α-SMA), stem cell
biomarkers (CD44 and CD133), and the proliferation marker Ki67, and
increased E-cadherin expression in tumor tissues (Fig. 5F and G).
 | Figure 5.DPEP2 enhances the sensitivity of
lung adenocarcinoma cells to cisplatin in vivo. (A) Flow
chart of the in vivo experiment with nude mice. (B and C)
Nude mice were subcutaneously injected with a vector or
DPEP2-overexpressing A549 stable strain. On days 7 and 14, mice
were injected with PBS or cisplatin (20 mg/m2). Every 7
days, the tumor volume was measured with calipers. Tumor volume on
day 28 was assessed using a two-tailed Student's t-test (vector vs.
DPEP2; and vector + cisplatin vs. DPEP2 + cisplatin). *P<0.05
and ***P<0.001. (D and E) On day 28, tumors were excised and
weighed. Representative tumors isolated from nude mice and average
tumor weights. **P<0.01 and ***P<0.001, one-way ANOVA with
post hoc Dunnett's t-test. (F and G) Immunohistochemical staining
of DPEP2, epithelial-mesenchymal transition-associated genes
(E-cadherin, N-cadherin, vimentin, and α-SMA), stem cell biomarkers
(CD44 and CD133), and the proliferation marker Ki67 in tumors of
mice injected with vector or DPEP2-overexpressing cells (scale bar,
100 µm). DPEP2, dipeptidase-2. |
Discussion
Lung cancer is the leading cause of cancer-related
deaths worldwide, and LUAD is the most prevalent subtype (1). Advances in LUAD treatment have
identified the histone chaperone ASF1A (36), methyltransferase-like 7B (37), and the long non-coding RNA HIF1A-AS2
(38) as targets to disrupt drug
resistance. Despite these advances, both metastasis and drug
resistance remain key characteristics of LUAD, resulting in high
rates of recurrence and mortality (39,40).
In the present study, differential expression
analysis of RNA sequencing data was performed and it was revealed
that DPEP2 was included in the genes that were substantially
downregulated during LUAD progression. Previous studies have
revealed that DPEP2 plays a role in immune cells, regulating
inflammation caused by macrophages (20) and can be used as an immune indicator
to identify lung adenocarcinoma (22). In addition, similar studies have
identified DPEP1 as a therapeutic target for neutrophil-driven
pulmonary inflammatory diseases (41) and DPEP3 may be associated with rat
rheumatoid arthritis (42).
Although DPEP2 has been studied in the metabolic and immune indexes
of LUAD, the specific mechanism of its effect on the progression of
LUAD remains unclear. Therefore, the expression and clinical
significance of DPEP2 in LUAD was investigated to determine its
functions and regulatory pathways.
In recent years, an increasing number of cancer
researchers have used data to reveal disease-related genes and
their potential functions via bioinformatics analysis (43,44).
Based on information from several major databases such as GEO, TCGA
and TIMER2.0 databases, both DPEP2 expression and the mutation rate
were low in LUAD, with similar results in vitro. In addition
to indicators such as age and sex, cancer grade and stage play a
significant role in influencing patient prognosis. The higher the
tumor grade and stage, the higher the likelihood of metastasis and
the worse the prognosis (45). In
the present study, it was demonstrated that low DPEP2 expression
was associated with poor clinicopathological features and poor OS
in patients with LUAD and that DPEP2 is an independent prognostic
factor, which was consistent with a previous study (21). In addition, the area under the ROC
curve of DPEP2 expression data was 0.976, which is close to the
maximum value of 1, suggesting good diagnostic value. These
findings indicated that DPEP2 may serve as a prognostic biomarker
in LUAD. However, the mechanism of action of DPEP2 on LUAD remains
unclear.
To better understand the role of DPEP2 in LUAD,
functional and mechanistic experiments were conducted. It was
revealed that DPEP2 overexpression inhibited cell migration and
invasion. Previous similar studies have also demonstrated that the
expression of DPEP1 in colon cancer and pancreatic ductal
adenocarcinoma cells influences the aggressiveness of cancer cells
(27,46,47).
Studies have shown that cell migration and invasion are a feature
of EMT. EMT is a reversible cellular program that transforms
epithelial cells into mesenchymal cell states (8) and is a key event in metastasis,
invasion, and diffusion to distant organs (48). In addition, EMT is highly associated
with tumor prognosis. For example, TFAP2A increased LUAD metastasis
by positively regulating the EMT process (49). FAM83A may play a key role in
promoting LUAD progression by influencing EMT (50). Similarly, the results of the present
study indicated that DPEP2 can affect the progression of LUAD by
inhibiting EMT, thus inhibiting the migration and invasion of LUAD
cells in vitro.
Previous studies have revealed a strong correlation
between EMT and cancer stem cell properties (51). EMT can enable cancer stem cells to
migrate to other organs and permit site colonization (52). In addition, cancer stem cells have
sufficient multidirectional differentiation ability to undergo EMT
(53). In lung cancer, a variety of
markers including CD44, CD133, CD24, ALCAM, OCT4, NANOG, and LGR5
are known to be related to cell stemness, with CD44 and CD133 being
more important (54–57). Previous studies have demonstrated
that EMT can affect the process of tumor formation through cell
stemness (8,35,58),
thus the effect of DPEP2 on the stem cell marker CD44/CD133 was
detected through western blotting and IF experiments, as a
superficial verification, without in-depth investigation of its
mechanism. Therefore, the expression of CD44/CD133 was not detected
by FACS analysis. The lack of FACS analysis of the stemness markers
is a limitation of this study. In the present study, it was
determined that DPEP2 overexpression decreased CD44 and CD133
protein levels. Moreover, sphere formation indicated that DPEP2
overexpression inhibited cell stemness. These results indicated
that DPEP2 may inhibit EMT in LUAD cells by regulating tumor
stemness.
Furthermore, the role of DPEP2 in chemotherapy
resistance was investigated in vitro and in vivo.
Cisplatin is a first-line chemotherapy drug for lung cancer
(59). Colony formation and flow
cytometry revealed that DPEP2 overexpression increased the
sensitivity of A549 and H1650 cells to cisplatin. Similar studies
on cisplatin resistance also revealed, for example, DPEP1 inhibited
invasiveness and enhanced the chemical sensitivity of pancreatic
ductal adenocarcinoma cells (28),
CCT3 promoted cisplatin resistance in LUAD cells (60), and TRIM44 overexpression conferred
cisplatin resistance in LUAD (61).
Therefore, a nude mouse subcutaneous tumor model was successfully
constructed and it was demonstrated that tumor nodules formed by
DPEP2-overexpressing cells decreased in size and weight, indicating
increased sensitivity to cisplatin. Moreover, IHC demonstrated that
DPEP2 overexpression could inhibit the expression of EMT- and stem
cell transformation-related markers. These results indicated that
DPEP2 enhanced the efficacy of cisplatin and reduced tumor
progression by inhibiting EMT.
However, despite the analytical exploration of DPEP2
and cross-validation using databases and in vitro and in
vivo experiments, there are some limitations to this study.
First, the number of cell lines and animal models used were
limited, which may in turn limit the clinical applicability of the
results. Thus, the results need to be validated in larger studies.
Second, the mechanism by which DPEP2 affects the stemness to
regulate the progression of LUAD has not been thoroughly discussed,
and will be further investigated through the search and
identification of functional targets in the future. These issues
are worthy of further investigation.
In conclusion, the present study revealed a novel
association between DPEP2 and LUAD progression, as low DPEP2
expression was positively associated with poor prognosis and poor
OS in patients with LUAD. DPEP2 inhibited migration, metastasis,
and EMT by regulating the transformation of cancer stem cells,
thereby increasing the sensitivity of LUAD to cisplatin. These
results indicated that DPEP2 may affect patient survival and serve
as a prognostic biomarker in LUAD. The findings of the present
study suggest that DPEP2 is a potential therapeutic target for
future therapeutic research in LUAD.
Acknowledgements
Not applicable.
Funding
The present study was funded by the National Natural Science
Foundation of China (grant nos. 81972977 and 81802955), the
Foundation of Health Commission of Chengdu (grant no. 2021001), the
Foundation of Chengdu Science and Technology Bureau (grant no.
2021-YF05-00291-SN), the Foundation of The First Affiliated
Hospital of Chengdu Medical College (grant no. CYFY2021LNZD01), the
Foundation of Chengdu Medical College and Chengdu Xindu District
People's Hospital Joint Research (grant no. 2021LHZD-07), and the
Foundation of Graduate Innovation and Chengdu Medical College
(grant no. YCX2022-03-03).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YW, TZ, HD and MY collected data. YW and TZ wrote,
reviewed, and edited the manuscript. GX, TL and SD revised the
study critically for important intellectual content. WY and SH
confirm the authenticity of all the raw data. DW and YX directed
the project and wrote, reviewed, and edited the manuscript. All
authors contributed to manuscript composition, and all authors read
and approved the final manuscript.
Ethics approval and consent to
participate
All xenograft experiments were performed in
accordance with the guidelines of the Laboratory Animal Ethics
Committee of Chengdu Medical College (Chengdu, China). All
experimental protocols were approved by the Experimental Animal
Ethics Committee of Chengdu Medical College (approval no.
2021-096).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
CCK-8
|
Cell Counting Kit-8
|
|
DPEP2
|
dipeptidase-2
|
|
DPEP
|
dipeptidase family
|
|
EMT
|
epithelial-mesenchymal transition
|
|
FBS
|
fetal bovine serum
|
|
GEO
|
Gene Expression Omnibus
|
|
LTD4
|
leukotriene D4
|
|
LTE4
|
leukotriene E4
|
|
HPA
|
Human Protein Atlas
|
|
IF
|
immunofluorescence
|
|
IHC
|
immunohistochemistry
|
|
LUAD
|
lung adenocarcinoma
|
|
NSCLC
|
non-small cell lung cancer
|
|
OS
|
overall survival
|
|
ROC
|
receiver operating characteristic
|
|
RT-qPCR
|
reverse transcription-quantitative
polymerase chain reaction
|
|
TCGA
|
The Cancer Genome Atlas
|
|
PD
|
progressive disease
|
|
SD
|
stable disease
|
|
PR
|
partial response
|
|
CR
|
complete response
|
|
IQR
|
interquartile range
|
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global Cancer Statistics 2020:
GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36
Cancers in 185 Countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gong L, Hu Y, He D, Zhu Y, Xiang L, Xiao
M, Bao Y, Liu X, Zeng Q, Liu J, et al: Ubiquitin ligase CHAF1B
induces cisplatin resistance in lung adenocarcinoma by promoting
NCOR2 degradation. Cancer Cell Int. 20:1942020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Esai Selvan M, Zauderer MG, Rudin CM,
Jones S, Mukherjee S, Offit K, Onel K, Rennert G, Velculescu VE,
Lipkin SM, et al: Inherited rare, deleterious variants in ATM
increase lung adenocarcinoma risk. J Thorac Oncol. 15:1871–1879.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Li Y, Zhang H, Fan L, Mou J, Yin Y, Peng
C, Chen Y, Lu H, Zhao L, Tao Z, et al: MiR-629-5p promotes the
invasion of lung adenocarcinoma via increasing both tumor cell
invasion and endothelial cell permeability. Oncogene. 39:3473–3488.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Rose MC, Kostyanovskaya E and Huang RS:
Pharmacogenomics of cisplatin sensitivity in non-small cell lung
cancer. Genomics Proteomics Bioinformatics. 12:198–209. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bakir B, Chiarella AM, Pitarresi JR and
Rustgi AK: EMT, MET, plasticity, and tumor metastasis. Trends Cell
Biol. 30:764–776. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Piera-Velazquez S and Jimenez SA:
Endothelial to mesenchymal transition: Role in physiology and in
the pathogenesis of human diseases. Physiol Rev. 99:1281–1324.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Dongre A and Weinberg RA: New insights
into the mechanisms of epithelial-mesenchymal transition and
implications for cancer. Nat Rev Mol Cell Biol. 20:69–84. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gao ZW, Liu C, Yang L, Chen HC, Yang LF,
Zhang HZ and Dong K: CD73 severed as a potential prognostic marker
and promote lung cancer cells migration via enhancing EMT
progression. Front Genet. 12:7282002021. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yu TT, Zhang T, Su F, Li YL, Shan L, Hou
XM and Wang RZ: ELK1 Promotes epithelial-mesenchymal transition and
the progression of lung adenocarcinoma by upregulating B7-H3. Oxid
Med Cell Longev. 2021:28055762021. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu Z, Liu J, Li Y, Wang H, Liang Z, Deng
X, Fu Q, Fang W and Xu P: VPS33B suppresses lung adenocarcinoma
metastasis and chemoresistance to cisplatin. Genes Dis. 8:307–319.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chua KN, Poon KL, Lim J, Sim WJ, Huang RY
and Thiery JP: Target cell movement in tumor and cardiovascular
diseases based on the epithelial-mesenchymal transition concept.
Adv Drug Deliv Rev. 63:558–567. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chen T, You Y, Jiang H and Wang ZZ:
Epithelial-mesenchymal transition (EMT): A biological process in
the development, stem cell differentiation, and tumorigenesis. J
Cell Physiol. 232:3261–3272. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Habib GM, Shi ZZ, Cuevas AA and Lieberman
MW: Identification of two additional members of the membrane-bound
dipeptidase family. FASEB J. 17:1313–1315. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kozak EM and Tate SS:
Glutathione-degrading enzymes of microvillus membranes. J Biol
Chem. 257:6322–6327. 1982. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Habib GM, Shi ZZ, Cuevas AA, Guo Q, Matzuk
MM and Lieberman MW: Leukotriene D4 and cystinyl-bis-glycine
metabolism in membrane-bound dipeptidase-deficient mice. Proc Natl
Acad Sci USA. 95:4859–63. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Park SY, Lee SJ, Cho HJ, Kim TW, Kim JT,
Kim JW, Lee CH, Kim BY, Yeom YI, Lim JS, et al: Dehydropeptidase 1
promotes metastasis through regulation of E-cadherin expression in
colon cancer. Oncotarget. 7:9501–9512. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hayashi K, Longenecker KL, Koenig P,
Prashar A, Hampl J, Stoll V and Vivona S: Structure of human DPEP3
in complex with the SC-003 antibody Fab fragment reveals basis for
lack of dipeptidase activity. J Struct Biol. 211:1075122020.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
von Mässenhausen A, Zamora Gonzalez N,
Maremonti F, Belavgeni A, Tonnus W, Meyer C, Beer K, Hannani MT,
Lau A, Peitzsch M, et al: Dexamethasone sensitizes to ferroptosis
by glucocorticoid receptor-induced dipeptidase-1 expression and
glutathione depletion. Sci Adv. 8:eabl89202022. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yang X, Yue Y and Xiong S: Dpep2 emerging
as a modulator of macrophage inflammation confers protection
against CVB3-induced viral myocarditis. Front Cell Infect
Microbiol. 9:572019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Huang G, Zhang J, Gong L, Wang X, Zhang B
and Liu D: Characterization of the fatty acid metabolism-related
genes in lung adenocarcinoma to guide clinical therapy. BMC Pulm
Med. 22:4862022. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Han T, Liu Y, Wu J, Bai Y, Zhou J, Hu C,
Zhang W, Guo J, Wang Q and Hu D: An immune indicator based on BTK
and DPEP2 identifies hot and cold tumors and clinical treatment
outcomes in lung adenocarcinoma. Sci Rep. 13:51532023. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hamilton E, O'Malley DM, O'Cearbhaill R,
Cristea M, Fleming GF, Tariq B, Fong A, French D, Rossi M, Brickman
D and Moore K: Tamrintamab pamozirine (SC-003) in patients with
platinum-resistant/refractory ovarian cancer: Findings of a phase 1
study. Gynecol Oncol. 158:640–645. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yoshitake H, Yanagida M, Maruyama M,
Takamori K, Hasegawa A and Araki Y: Molecular characterization and
expression of dipeptidase 3, a testis-specific membrane-bound
dipeptidase: Complex formation with TEX101, a germ-cell-specific
antigen in the mouse testis. J Reprod Immunol. 90:202–213. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Schiza C, Korbakis D, Panteleli E, Jarvi
K, Drabovich AP and Diamandis EP: Discovery of a human
testis-specific protein complex TEX101-DPEP3 and selection of its
disrupting antibodies. Mol Cell Proteomics. 17:2480–2495. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hao JJ, Zhi X, Wang Y, Zhang Z, Hao Z, Ye
R, Tang Z, Qian F, Wang Q and Zhu J: Comprehensive proteomic
characterization of the human colorectal carcinoma reveals
signature proteins and perturbed pathways. Sci Rep. 7:424362017.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wu YF, Wang CY, Tang WC, Lee YC, Ta HDK,
Lin LC, Pan SR, Ni YC, Anuraga G and Lee KH: Expression profile and
prognostic value of Wnt signaling pathway molecules in colorectal
cancer. Biomedicines. 9:13312021. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhang G, Schetter A, He P, Funamizu N,
Gaedcke J, Ghadimi BM, Ried T, Hassan R, Yfantis HG, Lee DH, et al:
DPEP1 inhibits tumor cell invasiveness, enhances chemosensitivity
and predicts clinical outcome in pancreatic ductal adenocarcinoma.
PLoS One. 7:e315072012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Cui X, Liu X, Han Q, Zhu J, Li J, Ren Z,
Liu L, Luo Y, Wang Z, Zhang D, et al: DPEP1 is a direct target of
miR-193a-5p and promotes hepatoblastoma progression by
PI3K/Akt/mTOR pathway. Cell Death Dis. 10:7012019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang W, Chen T, Liu J, Yu S, Liu L, Zheng
M, Liu Y, Zhang H, Bian T and Zhao X: RAB11FIP1: An indicator for
tumor immune microenvironment and prognosis of lung adenocarcinoma
from a comprehensive analysis of bioinformatics. Front Genet.
12:7571692021. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Okayama H, Kohno T, Ishii Y, Shimada Y,
Shiraishi K, Iwakawa R, Furuta K, Tsuta K, Shibata T, Yamamoto S,
et al: Identification of genes upregulated in ALK-positive and
EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer Res.
72:100–111. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yamauchi M, Yamaguchi R, Nakata A, Kohno
T, Nagasaki M, Shimamura T, Imoto S, Saito A, Ueno K, Hatanaka Y,
et al: Epidermal growth factor receptor tyrosine kinase defines
critical prognostic genes of stage I lung adenocarcinoma. PLoS One.
7:e439232012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhu G, Cheng Z, Huang Y, Zheng W, Yang S,
Lin C and Ye J: MyD88 mediates colorectal cancer cell
proliferation, migration and invasion via NF-κB/AP-1
signaling pathway. Int J Mol Med. 45:131–140. 2020.PubMed/NCBI
|
|
35
|
Wu DM, Zhang T, Liu YB, Deng SH, Han R,
Liu T, Li J and Xu Y: The PAX6-ZEB2 axis promotes metastasis and
cisplatin resistance in non-small cell lung cancer through PI3K/AKT
signaling. Cell Death Dis. 10:3492019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li F, Huang Q, Luster TA, Hu H, Zhang H,
Ng WL, Khodadadi-Jamayran A, Wang W, Chen T, Deng J, et al: In Vivo
Epigenetic CRISPR screen identifies Asf1a as an immunotherapeutic
target in Kras-Mutant lung adenocarcinoma. Cancer Discov.
10:270–287. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Song H, Liu D, Wang L, Liu K, Chen C, Wang
L, Ren Y, Ju B, Zhong F, Jiang X, et al: Methyltransferase like 7B
is a potential therapeutic target for reversing EGFR-TKIs
resistance in lung adenocarcinoma. Mol Cancer. 21:432022.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Si J, Ma Y, Lv C, Hong Y, Tan H and Yang
Y: HIF1A-AS2 induces osimertinib resistance in lung adenocarcinoma
patients by regulating the miR-146b-5p/IL-6/STAT3 axis. Mol Ther
Nucleic Acids. 26:613–624. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Chaffer CL and Weinberg RA: A perspective
on cancer cell metastasis. Science. 331:1559–1564. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Chen J, Liu X, Xu Y, Zhang K, Huang J, Pan
B, Chen D, Cui S, Song H, Wang R, et al: TFAP2C-Activated MALAT1
modulates the chemoresistance of docetaxel-resistant lung
adenocarcinoma cells. Mol Ther Nucleic Acids. 14:567–582. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Choudhury SR, Babes L, Rahn JJ, Ahn BY,
Goring KR, King JC, Lau A, Petri B, Hao X, Chojnacki AK, et al:
Dipeptidase-1 is an adhesion receptor for neutrophil recruitment in
lungs and liver. Cell. 178:1205–1221.e17. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Lai W, Wang C, Lai R, Peng X and Luo J:
Lycium barbarum polysaccharide modulates gut microbiota to
alleviate rheumatoid arthritis in a rat model. NPJ Sci Food.
6:342022. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Song X, Du R, Gui H, Zhou M, Zhong W, Mao
C and Ma J: Identification of potential hub genes related to the
progression and prognosis of hepatocellular carcinoma through
integrated bioinformatics analysis. Oncol Rep. 43:133–146.
2020.PubMed/NCBI
|
|
44
|
Chen J, Xu D, Wang T, Yang Z, Yang Y, He K
and Zhao L: HMGB1 promotes the development of castration resistant
prostate cancer by regulating androgen receptor activation. Oncol
Rep. 48:1972022. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Yamanashi K, Menju T, Hamaji M, Tanaka S,
Yutaka Y, Yamada Y, Nakajima D, Ohsumi A, Aoyama A, Sato T, et al:
Prognostic factors related to postoperative survival in the newly
classified clinical T4 lung cancer. Eur J Cardiothorac Surg.
57:754–761. 2020.PubMed/NCBI
|
|
46
|
Toiyama Y, Inoue Y, Yasuda H, Saigusa S,
Yokoe T, Okugawa Y, Tanaka K, Miki C and Kusunoki M: DPEP1,
expressed in the early stages of colon carcinogenesis, affects
cancer cell invasiveness. J Gastroenterol. 46:153–163. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zeng C, Qi G, Shen Y, Li W, Zhu Q, Yang C,
Deng J, Lu W, Liu Q and Jin J: DPEP1 promotes drug resistance in
colon cancer cells by forming a positive feedback loop with ASCL2.
Cancer Med. 12:412–424. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Mittal V: Epithelial mesenchymal
transition in tumor metastasis. Annu Rev Pathol. 13:395–412. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Xiong Y, Feng Y, Zhao J, Lei J, Qiao T,
Zhou Y, Lu Q, Jiang T, Jia L and Han Y: TFAP2A potentiates lung
adenocarcinoma metastasis by a novel miR-16
family/TFAP2A/PSG9/TGF-β signaling pathway. Cell Death Dis.
12:3522021. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Liu X, Fu M, Xia D, Ji Z, Hu N, Leng Z,
Xie W, Fang Y and Zhang J: Overexpression of FAM83A is associated
with poor prognosis of lung adenocarcinoma. J Oncol.
2022:87673332022. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Thiery JP, Acloque H, Huang RY and Nieto
MA: Epithelial-mesenchymal transitions in development and disease.
Cell. 139:871–890. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Kavanagh DP, Robinson J and Kalia N:
Mesenchymal stem cell priming: Fine-tuning adhesion and function.
Stem Cell Rev Rep. 10:587–599. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
McCabe EM and Rasmussen TP: lncRNA
involvement in cancer stem cell function and epithelial-mesenchymal
transitions. Semin Cancer Biol. 75:38–48. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Leung EL, Fiscus RR, Tung JW, Tin VP,
Cheng LC, Sihoe AD, Fink LM, Ma Y and Wong MP: Non-small cell lung
cancer cells expressing CD44 are enriched for stem cell-like
properties. PLoS One. 5:e140622010. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Hess DA, Wirthlin L, Craft TP, Herrbrich
PE, Hohm SA, Lahey R, Eades WC, Creer MH and Nolta JA: Selection
based on CD133 and high aldehyde dehydrogenase activity isolates
long-term reconstituting human hematopoietic stem cells. Blood.
107:2162–2169. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Prasetyanti PR and Medema JP: Intra-tumor
heterogeneity from a cancer stem cell perspective. Mol Cancer.
16:412017. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Jiang P, Li F, Liu Z, Hao S, Gao J and Li
S: BTB and CNC homology 1 (Bach1) induces lung cancer stem cell
phenotypes by stimulating CD44 expression. Respir Res. 22:3202021.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Pan G, Liu Y, Shang L, Zhou F and Yang S:
EMT-associated microRNAs and their roles in cancer stemness and
drug resistance. Cancer Commun (Lond). 41:199–217. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Konoshenko M, Lansukhay Y, Krasilnikov S
and Laktionov P: MicroRNAs as predictors of lung-cancer resistance
and sensitivity to cisplatin. Int J Mol Sci. 23:75942022.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Danni X, Jiangzheng Z, Huamao S, Yinglian
P, Changcheng Y and Yanda L: Chaperonin containing TCP1 subunit 3
(CCT3) promotes cisplatin resistance of lung adenocarcinoma cells
through targeting the Janus kinase 2/signal transducers and
activators of transcription 3 (JAK2/STAT3) pathway. Bioengineered.
12:7335–7347. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Zhang S, Cao M, Yan S, Liu Y, Fan W, Cui
Y, Tian F, Gu R, Cui Y, Zhan Y, et al: TRIM44 promotes BRCA1
functions in HR repair to induce Cisplatin Chemoresistance in Lung
Adenocarcinoma by Deubiquitinating FLNA. Int J Biol Sci.
18:2962–2979. 2022. View Article : Google Scholar : PubMed/NCBI
|